Abstract
Epstein-Barr virus is a gammaherpes virus that is causally associated with several malignancies and expresses multiple miRNAs in both normal and tumor cells. Since the identification of virally-encoded miRNAs, various mRNAs have been identified as targets for regulation by EBV’s miRNAs in host cells. We shall summarize these targets, the robustness of their identification, and examine how the regulation of these targets by EBV contributes to the successful infection of its host.
Introduction
Epstein-Barr Virus (EBV) is a successful human pathogen, and is now known to cause at least 200,000 new cancers per year [1]. While initially identified in the cells of a Burkitt’s lymphoma, much evidence has implicated EBV as causing several additional lymphomas and carcinomas (reviewed in [2]). A new layer of virus-host interaction has emerged with the discovery of virally-encoded miRNAs. MicroRNAs and their biogenesis have been extensively reviewed previously [3]–[5]. Briefly, they are short (19–24 nt) single-stranded RNA molecules that post-transcriptionally regulate gene expression by recruiting the RNA-induced silencing complex (RISC) to target mRNAs [6]–[8]. Multiple studies using computational and molecular biology techniques as well as deep sequencing have led to the identification of at least 40 viral miRNAs encoded within 25 precursor transcripts [3], [9], [10]. They are encoded within two regions of EBV’s genome: BART (Bam HI-A region rightward transcript) and BHRF1 (Bam HI fragment H rightward open reading frame 1) (Figure 1). The BHRF1 transcript also encodes the BHRF1 ORF, while the BART transcript has not been confirmed to express other functional products besides its miRNAs. Since their identification, the expression of these miRNAs has been extensively profiled in various EBV-infected cells lines including Burkitt’s lymphomas, lymphoblastoid cell lines, carcinomas as well as in tumor biopsies [11]–[18]. The abundance of individual miRNAs within cell lines varies widely and is cell type specific. BART miRNAs were found to be expressed in all types of EBV-associated latency, whereas expression of the BHRF1 miRNAs appears to be more restricted (ibid). The levels of the miRNAs measured in carcinoma biopsies can exceed those in cell lines by more than 100-fold [18] making it plausible that in vivo EBV’s miRNAs have more functions than found in cell culture.
Figure 1. Non-coding EBV RNAs.
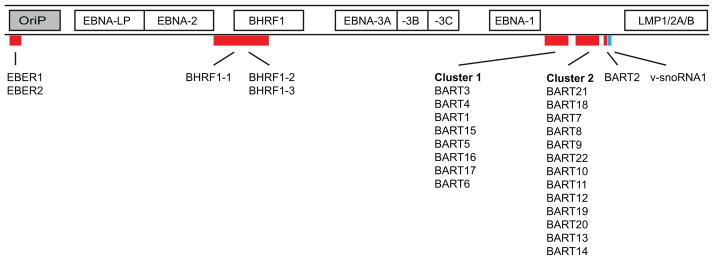
This schematic illustration shows the locations of non-coding RNAs within EBV’s genome. Along with the non-coding nuclear EBER RNAs, and a snoRNA [36], EBV encodes at least 25 pre-miRNAs located within two regions of the genome. The smaller subset of BHRF1 miRNAs is derived from transcripts mapping approximately between 53,000 and 56,000 bps. The majority of EBV miRNAs arise from highly spliced transcripts mapping approximately between 139,000 and 153,000 bps. Also shown are the latent origin of replication (OriP) and genes expressed in latently infected cells. Note: the figure is not drawn to scale and is a linear representation of EBV’s circular genome which encompasses approximately 165,000 bps.
MiRNAs provide a potent mechanism for EBV to modulate the cellular environment: they are thought not to elicit an immunogenic response; they take up little genomic space; and they also have the potential to regulate hundreds of targeted genes. Here we focus on specific cellular processes that appear to be modulated by the currently identified targets of viral miRNAs and explore their possible contributions to EBV’s lifecycle. We have also used insights from HITS- and PAR-CLIP experiments to gauge the robustness of these identifications.
Identifying and Validating Targets of EBV’s miRNAs
Leads to identifying the targets of cellular miRNAs can be found using bioinformatics programs such as TargetScan [19]. Because these programs use the evolutionary conservation of the seed sequences recognized in the target mRNAs in their identification, they are not readily applicable to studies of EBV’s miRNA targets given that EBV has evolved to infect a single host species. Therefore, validating the targets of EBV’s miRNAs requires a set of successively more stringent tests. These include assays in which sequences encoding the 3′ UTR of the presumptive mRNA target are ligated to luciferase as a reporter and its regulation recorded in the presence of physiological levels of the miRNAs to be tested. A necessary control experiment is to change the seed sequences in the 3′UTR and demonstrate that the changes abrogate any inhibition of the luciferase activity by the test miRNA. Finally experiments with PAR-CLIP allow identification of sites in mRNAs that are modified by the juxtaposition of Argonaute in the RISC complex. If these sites are found and correspond to the seed sequence recognized by an EBV miRNA, then we can conclude that the mRNA is targeted by that EBV miRNA. Table 1 documents currently identified target mRNAs of EBV miRNAs and the robustness of the experiments validating these identifications. One caveat to consider is that the levels of EBV’s miRNAs in cell in culture are far lower than in biopsies isolated from carcinomas so that studies of cells in culture may miss many mRNAs that are targeted by EBV’s miRNAs in vivo.
Table 1.
Possible Validations for Candidate Targets of EBV’s miRNAs
| MRNA | miRNA | mRNA Functions | PAR-CLIP | Ref. | |
|---|---|---|---|---|---|
| Gottwein et al. | Skalsky et al. | ||||
| Bim (BCL2L11) | Various (BART9, 11+12) | Pro-apoptotic; inhibits Bcl-2 | − | ? | [26] |
| BRUCE (birc6) | BART 15-3p | Inhibitor of apoptosis (IAP) | − | − | [29] |
| C1orf109 | BART 2-5p# | Cancer cell proliferation (proposed) | − | + | [38] |
| CAPRIN2 | BART 13-3p | Wnt signaling | − | ? | [27] |
| CASP3 | BART 1-3p | Pro-apoptotic; death protease | + | − | [24]† |
| CLEC2D(LLT1) | BART 1-3p, 3-3p | Ligand for NK receptor | − | + | [28] |
| CXCL11 | BHRF1-3 | IFN-inducible chemokine | ? | − | [32] |
| DAZAP2 | BART 3 | Wnt signaling | + | + | [28] |
| DICE1 | BART 3* | DEAD box protein; tumor suppressor | − | − | [39] |
| DICER1 | BART 6-5p | RNAi processing (miRNA maturation) | − | ? | [40] |
| IPO7 | BART 3 | Nuclear import | + | + | [24], [38]† |
| LY75 | BART 1-5p | Mediates antigen presentation | − | + | [28] |
| MICB | BART 2-5p | Activates NK cell response | − | − | [34] |
| NLRP3 | BART 11-5p, 15 | Pro-apoptotic; release of IL-1B | − | ? | [35] |
| PDCD1LG2 | BHRF1-2-5p, BART 1-5p, 15-3p# | Ligand for PD-1; Immune regulation | ? | + | [38] |
| PDE7A | BART 3 | Cytokine production | − | + | [38] |
| PELI1 | BART 3 | E3 ubiquitin ligase; activation of NF-kB | − | + | [28] |
| PUMA (BBC3) | BART 5 | Pro-apoptotic; inhibits Bcl-2 | − | ? | [25] |
| SP100 | BART 1-5p | Anti-viral response; part of PML-NB | + | + | [28] |
| T-bet (TBX21) | BART 20-5p | Transcription factor, T cell development | − | ? | [41] |
| TOMM22 | BART 16 | Mitochondrial import | + | ? | [38] |
| ZNF451 | BHRF1-2 | Transcriptional regulation? | ? | + | [28] |
+ Clusters of sequence changes in RNAs in RISC consistent with binding of a given EBV miRNA with high confidence (for details see Gottwein et al. [37] and Skalsky et al. [28])
− No clusters of sequence changes in RNAs in RISC consistent with binding of a given EBV miRNA with high confidence (for details see Gottwein et al. [37] and Skalsky et al. [28])
Reference proposes additional EBV miRNAs target given mRNA that are not confirmed by PAR-CLIP.
? Some EBV miRNAs are not expressed in the cells analyzed by PAR-CLIP making the PAR-CLIP uninformative for this mRNA.
EBV’s miRNAs Contribute to its Transformation of B lymphocytes
While the majority of functions ascribed to EBV’s miRNAs are to sustain latently infected cells, several studies have expanded on their importance during the initial stages of infection of B-cells. Mature BART and BHRF1 miRNAs were detected in primary B- cells infected with either of two strains of EBV at two days post infection (dpi) and increased in expression through at least the first eight dpi [13]. Cells infected with derivatives of EBV null for the BHRF1 miRNA cluster grew more slowly when exposed to the same multiplicity of infection relative to wild type genomes [20]–[22]. The BHRF1 cluster of miRNAs appeared to promote both transformation of infected cells and cell-cycle progression [20]–[22]. In vivo studies using humanized mice to monitor EBV infection and tumorigenesis revealed significant delays in viremia in mice infected with a derivative of EBV lacking BHRF1 miRNAs [23]. Absence of the BHRF1 miRNAs however had no effect on tumor formation and survival of mice relative to those infected with wild type virus [23].
Regulation of Apoptosis by EBV miRNAs
A common fate for B-lymphocytes in vivo is death by apoptosis. EBV infects B-cells and evades apoptosis in its host cell by multiple means including its miRNAs (Table 1). The BHRF1 miRNAs inhibited apoptosis early during infection of primary B cells and promoted their proliferation as shown by infection with derivatives of EBV [22]. The BART miRNAs sustained Burkitt’s lymphomas in part by inhibiting Caspase 3 [24]. The BART miRNAs also have been reported to target the pro-apoptotic proteins PUMA (p53-upregulated modulator of apoptosis) and Bim (BCL2L11) [25], [26]. Recent comprehensive HITS- and PAR-CLIP analyses have identified CAPRIN2 and DAZAP2 which are involved in Wnt signaling as targets and which may also function in apoptosis[27], [28]. In contrast, Choi and colleagues reported that miR-BART15-3p promoted apoptosis in part by targeting BRUCE (BIRC6), a member of the inhibitor of apoptosis (IAP) family in gastric carcinoma cells [29]. The functional consequences of BRUCE inhibition are currently unclear but appear inconsistent with the association of EBV with its host cell’s survival and proliferation.
Role of EBV’s miRNAs in Immune Evasion
While EBV infects the majority of the adult population of the world, most of these infections are asymptomatic and persist for the lifetime of the host. EBV has evolved multiple strategies to avoid immune recognition in order to establish life-long, latent infections in B-cells (reviewed in [30], [31]). New findings indicate that viral miRNAs also attenuate the host’s antiviral immune response (Table 1). One of the earliest targets identified for miR-BHRF1-3 was CXCL-11, an IFN-inducible T-cell attracting chemokine [32]. CXCL-11 is one of the more abundantly expressed chemokines that interacts selectively with CXCR3, a chemokine receptor expressed on T cells [32], [33]. These findings show that viral miRNAs may contribute to immune evasion by modulating host cytokine networks.
Nachmani et al. reported that a stress-induced Natural Killer (NK) cell ligand, MICB, was targeted by miR-BART2-5p which could allow EBV-infected cells to escape recognition and subsequent elimination [34]. NK cells play a critical role in detection of virus-infected cells in part by using NKG2D receptors to detect release of molecules such as MICB, MICA and members of ULBP family in response to viral infections ([34] and references therein). A related virus KSHV (Kaposi’s Sarcoma-associated herpesvirus) was also found to regulate expression of MICB through its miRNA, miR-K12-7 emphasizing the importance of escaping NKG2D-mediated recognition and NK cell attack. Along with dampening NK cell response, viral miRNAs may also regulate activation of the NLRP3 inflammasome and subsequent production of pro-inflammatory cytokines such as IL-1β and IL-18 [35]. miR-BART15 was found to decrease expression of NLRP3 in reporter assays; its transient transfection reduced endogenous levels of NLRP3 as well as IL-1β production following inflammasome activation (ibid). Notably, miR-BART15 targets NLRP3 through the same site as a cellular miR-223. Several additional targets of EBV’s miRNAs that could contribute to viral immune evasion were identified through comprehensive PAR-CLIP analyses and confirmed using reporter assays. Among these are SP100, LY75, PDE7A, PELI1 though functional studies remain necessary to understand their biological significance [28].
Conclusions
EBV is exceptional in encoding so many miRNAs. It is a successful human pathogen that usually persists in people without causing disease. This success likely reflects its evolutionary fitness as a virus to infect human beings. By studying EBV’s miRNAs, and identifying their biological targets, we shall gain insights into how EBV succeeds as a pathogen both at the cellular and organismal levels.
Highlights.
EBV encodes at least 25 pre-miRNAs encoded within two gene clusters.
Steady-state levels of these miRNAs vary significantly among EBV-positive cell lines and tumor biopsies.
Targets of EBV’s miRNAs mediate various cellular processes to promote cell survival and proliferation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer J Int Cancer. 2006 Jun;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 2.Taylor GS, Blackbourn DJ. Infectious agents in human cancers: lessons in immunity and immunomodulation from gamma herpesviruses EBV and KSHV. Cancer Lett. 2011 Jun;305(2):263–278. doi: 10.1016/j.canlet.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 3.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005 Apr;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 4.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011 Mar;411(2):325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kincaid RP, Sullivan CS. Virus-encoded microRNAs: an overview and a look to the future. PLoS Pathog. 2012 Dec;8(12):e1003018. doi: 10.1371/journal.ppat.1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009 Jan;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010 Sep;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Cai X, Schäfer A, Lu S, Bilello JP, Desrosiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLoS Pathog. 2006 Mar;2(3):e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007 Jun;129(7):1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng Z, Huang H, Huang L, Sun M, Yan Q, Song Y, Wei F, Bo H, Gong Z, Zeng Y, Li Q, Zhang W, Li X, Xiang B, Li X, Li Y, Xiong W, Li G. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci China Life Sci. 2014 Feb; doi: 10.1007/s11427-013-4577-y. [DOI] [PubMed] [Google Scholar]
- 12.Qiu J, Cosmopoulos K, Pegtel M, Hopmans E, Murray P, Middeldorp J, Shapiro M, Thorley-Lawson DA. A novel persistence associated EBV miRNA expression profile is disrupted in neoplasia. PLoS Pathog. 2011 Aug;7(8):e1002193. doi: 10.1371/journal.ppat.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratt ZL, Kuzembayeva M, Sengupta S, Sugden B. The microRNAs of Epstein-Barr Virus are expressed at dramatically differing levels among cell lines. Virology. 2009 Apr;386(2):387–397. doi: 10.1016/j.virol.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marquitz AR, Mathur A, Chugh PE, Dittmer DP, Raab-Traub N. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J Virol. 2014 Jan;88(2):1389–1393. doi: 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imig J, Motsch N, Zhu JY, Barth S, Okoniewski M, Reineke T, Tinguely M, Faggioni A, Trivedi P, Meister G, Renner C, Grässer FA. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011 Mar;39(5):1880–1893. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosmopoulos K, Pegtel M, Hawkins J, Moffett H, Novina C, Middeldorp J, Thorley-Lawson DA. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J Virol. 2009 Mar;83(5):2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amoroso R, Fitzsimmons L, Thomas WA, Kelly GL, Rowe M, Bell AI. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J Virol. 2011 Jan;85(2):996–1010. doi: 10.1128/JVI.01528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S-J, Chen G-H, Chen Y-H, Liu C-Y, Chang K-P, Chang Y-S, Chen H-C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PloS One. 2010;5(9) doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witkos TM, Koscianska E, Krzyzosiak WJ. Practical Aspects of microRNA Target Prediction. Curr Mol Med. 2011 Mar;11(2):93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feederle R, Linnstaedt SD, Bannert H, Lips H, Bencun M, Cullen BR, Delecluse H-J. A Viral microRNA Cluster Strongly Potentiates the Transforming Properties of a Human Herpesvirus. PLoS Pathog. 2011 Feb;7(2) doi: 10.1371/journal.ppat.1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feederle R, Haar J, Bernhardt K, Linnstaedt SD, Bannert H, Lips H, Cullen BR, Delecluse HJ. The Members of an Epstein-Barr Virus MicroRNA Cluster Cooperate To Transform B Lymphocytes. J Virol. 2011 Oct;85(19):9801–9810. doi: 10.1128/JVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr Virus Promote Cell Cycle Progression and Prevent Apoptosis of Primary Human B Cells. PLoS Pathog. 2010 Aug;6(8) doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wahl A, Linnstaedt SD, Esoda C, Krisko JF, Martinez-Torres F, Delecluse HJ, Cullen BR, Garcia JV. A Cluster of Virus-Encoded MicroRNAs Accelerates Acute Systemic Epstein-Barr Virus Infection but Does Not Significantly Enhance Virus-Induced Oncogenesis In Vivo. J Virol. 2013 May;87(10):5437–5446. doi: 10.1128/JVI.00281-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *24.Vereide DT, Seto E, Chiu Y-F, Hayes M, Tagawa T, Grundhoff A, Hammerschmidt W, Sugden B. Epstein–Barr virus maintains lymphomas via its miRNAs. Oncogene. 2013 Mar; doi: 10.1038/onc.2013.71. This work uses several functional studies to identify mRNA targets of EBV’s miRNAs, one of which likely supports the maintenance of Burkitt’s Lymphomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choy EYW, Siu KL, Kok KH, Lung RWM, Tsang CM, To KF, Kwong DLW, Tsao SW, Jin DY. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J Exp Med. 2008 Oct;205(11):2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquitz AR, Mathur A, Nam CS, Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011 Apr;412(2):392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *27.Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012 May;31(9):2207–2221. doi: 10.1038/emboj.2012.63. This study identifies mRNAs potentially targeted by EBV’s miRNAs using HITS-CLIP in the Burkitt’s Lymphoma cell line, Jijoye, that encodes all EBV miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *28.Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse H-J, Luftig MA, Tuschl T, Ohler U, Cullen BR. The Viral and Cellular MicroRNA Targetome in Lymphoblastoid Cell Lines. PLoS Pathog. 2012 Jan;8(1) doi: 10.1371/journal.ppat.1002484. This survey is a broad comprehensive identification using PAR-CLIP of potential targets for EBV’s miRNAs expressed in B-cell lines transformed by the common lab strain of EBV that lacks some viral miRNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi H, Lee H, Kim SR, Gho YS, Lee SK. Epstein-Barr Virus-Encoded MicroRNA BART15-3p Promotes Cell Apoptosis Partially by Targeting BRUCE. J Virol. 2013 Jul;87(14):8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cullen BR. MicroRNAs as Mediators of Viral Immune Evasion. Nat Immunol. 2013 Mar;14(3):205–210. doi: 10.1038/ni.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ning S. Innate immune modulation in EBV infection. Herpesviridae. 2011 Jan;2(1):1. doi: 10.1186/2042-4280-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia T, O’Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ. EBV MicroRNAs in Primary Lymphomas and Targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008 Mar;68(5):1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, Gladue RP, Lin W, Boyd JG, Moser B, Wood DE, Sahagan BG, Neote K. Interferon-inducible T Cell Alpha Chemoattractant (I-TAC): A Novel Non-ELR CXC Chemokine with Potent Activity on Activated T Cells through Selective High Affinity Binding to CXCR3. J Exp Med. 1998 Jun;187(12):2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse Herpesvirus MicroRNAs Target the Stress-Induced Immune Ligand MICB to Escape Recognition by Natural Killer Cells. Cell Host Microbe. 2009 Apr;5(4):376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O’Neill LAJ, Masters SL. Cutting Edge: miR-223 and EBV miR-BART15 Regulate the NLRP3 Inflammasome and IL-1β Production. J Immunol. 2012 Oct;189(8):3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 36.Hutzinger R, Feederle R, Mrazek J, Schiefermeier N, Balwierz PJ, Zavolan M, Polacek N, Delecluse HJ, Hüttenhofer A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009 Aug;5(8):e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell Host Microbe. 2011 Nov;10(5):515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dölken L, Malterer G, Erhard F, Kothe S, Friedel CC, Suffert G, Marcinowski L, Motsch N, Barth S, Beitzinger M, Lieber D, Bailer SM, Hoffmann R, Ruzsics Z, Kremmer E, Pfeffer S, Zimmer R, Koszinowski UH, Grässer F, Meister G, Haas J. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010 Apr;7(4):324–334. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Lei T, Yuen KS, Xu R, Tsao SW, Chen H, Li M, Kok KH, Jin DY. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int J Cancer J Int Cancer. 2013 Jul;133(1):79–87. doi: 10.1002/ijc.28007. [DOI] [PubMed] [Google Scholar]
- 40.Iizasa H, Wulff BE, Alla NR, Maragkakis M, Megraw M, Hatzigeorgiou A, Iwakiri D, Takada K, Wiedmer A, Showe L, Lieberman P, Nishikura K. Editing of Epstein-Barr virus-encoded BART6 microRNAs controls their dicer targeting and consequently affects viral latency. J Biol Chem. 2010 Oct;285(43):33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin TC, Liu TY, Hsu SM, Lin CW. Epstein-Barr virus-encoded miR-BART20-5p inhibits T-bet translation with secondary suppression of p53 in invasive nasal NK/T-cell lymphoma. Am J Pathol. 2013 May;182(5):1865–1875. doi: 10.1016/j.ajpath.2013.01.025. [DOI] [PubMed] [Google Scholar]


