Abstract
While all viruses must transit the plasma membrane of mammalian cells to initiate infection, we know little about the complex processes involved in viral attachment, which commonly involve recognition of glycans by viral proteins. Glycan microarrays derived from both synthetic glycans and natural glycans isolated through shotgun glycomics approaches provide novel platforms for interrogating diverse glycans as potential viral receptors. Recent studies with influenza and rotaviruses using such glycan microarrays provide examples of their utility in exploring the challenging questions raised in efforts to define the complex mechanistic protein-glycan interactions that regulate virus attachment to host cells.
Introduction
The initial step in viral infections of living cells typically involves the interaction of a virus with a cell surface receptor in order to be eventually transported through the plasma membrane into the cell interior. The plasma membrane andexterior of all living cells is comprised of a thick wrapping of complex cell surface glycans on glycoproteins and glycolipids, sometimes referred to as the glycocalyx (1). The surface of cells has evolved in an environment of constant exposure to pathogens that bind to specific glycans on cells, thus evolutionarily helping to drive the creation of a hugely diverse set of glycan structures, as cells balance their glycan functions and structures with pressures to evade pathogen recognition. At the same time pathogens evolve by changing their glycan coat to appear more host-like, and in addition pathogens exploit host glycans for initial interaction by constantly modifying their glycan recognition molecules in response to glycan structural changes at the host cell surface. This complex interplay is a driving force for molecular evolution at the glycan and protein levels (2–4), reflecting the literal war between viruses and animal cells that is fought on a battlefield of cell surface glycans.
Observations in the 1930’s and 1940’s on many viruses including influenza viruses indicated that viruses could agglutinate vertebrate erythrocytes, and it was subsequently shown for influenza virus that the receptors and hemagglutinating potential of the erythrocytes were eventually lost upon long exposure to virus (5, 6). The erythrocyte receptors recognized by influenza virus were eventually identified as N-acetylneuraminic acid (sialic acid), and the “receptor-destroying enzyme” was discovered to be a neuraminidase (NA or sialidase), which was independent of the hemagglutinin (HA)(7, 8). Remarkably and despite these early observations and many years of research on virus-host interactions, it is still not clear exactly how these processes operate in the mechanisms of viral infection, but an optimal balance between the HA and the NA is probably key (9).
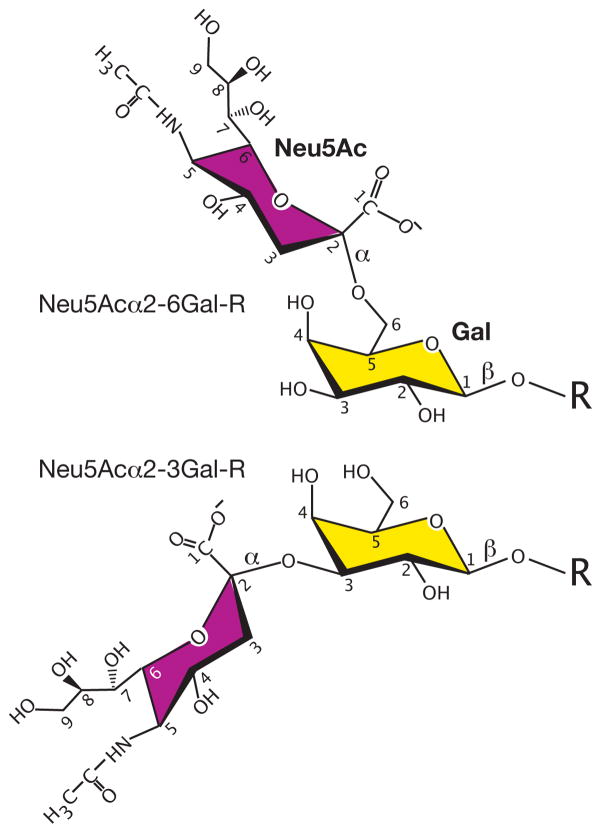
Cell surface carbohydrate, however, appears to be a major pathway of entry for influenza as well as other viruses, and the initial virus-host interactions involve recognition among a diverse set of glycan structures; thus, the specificities of viral surface adhesion molecules are thought to play important roles in viral tropism. This concept was demonstrated by the observation in the early 1980’s showing that the hemagglutinins (HA) of human influenza viruses prefer α2–6 linked sialic acid while avian viruses prefer the α2–3 linked form (Figure 1), and that a single amino acid substitution was responsible for the specificity switch (10–12). The proposed location of these linkages in the intestinal tract of birds and upper respiratory tract of humans is thought to correspond with the specificity of the appropriate HA (13). Although many viruses including norovirus, polyomavirus, rotavirus, and paramyxovirus exploit surface carbohydrate to facilitate entry into cells (14, 15), we will specifically focus on influenza viruses and rotaviruses, which have received significant attention in recent years.
Figure 1.
Depiction of sialic acid linked to galactose in either α2–6 or α2–3 linkages.
Analysis of Influenza A binding specificity on defined glycan microarrays
The Consortium for Functional Glycomics (CFG), a National Institute of General Medical Sciences-funded program, developed a glycan microarray of >600 defined glycans including >160 sialylated structures that could be interrogated with fluorescence labeled glycan binding proteins or intact viruses simultaneously generated significant interest in defining the fine specificity of influenza HAs to determine the relationship of the glycan structures underlying the terminal sialic acid to infection and transmission of influenza (16–18) (www.functionalglycomics.org/). This resource was made available to investigators worldwide and was a revolutionary development in the area of protein-glycan interactions, since both the glycan microarray resources as well as database of results were freely available.
Investigating the binding specificity of different virus strains on a glycan microarray is typically accomplished by analysis of recombinant HA or intact viruses. Since the interaction of the recombinant HA with receptor is relatively weak with dissociation constant in the mM range (19), which is below the detection limit of a GBP on the CFG array, the avidity of this system is increased by pre-complexing the His-tagged trimer with fluorescence labeled mouse anti-HisTag-IgG and fluorescence labeled anti-mouse-IgGin approximate molar ratio of 4:2:1 (16, 20). This protocol has been generally used for analyzing the specificity of recombinant, His-tagged HA in large-scale screening. An alternate approach has been the analysis of fluorescent labeled whole virus (21, 22) that is directly labeled with a fluorescent dye and assayed post labeling with hemagglutination assays to ensure no loss of binding activity during the labeling procedure and to have an estimate of the amount of virus added to the array so that analyses can be carried out at multiple concentrations of virus for detection of strongest binding glycans (19). Both of these approaches have been used extensively by the CFG Protein-Glycan Interaction Resource at Emory University and by the Centers for Disease Control and Prevention in Atlanta, GA for determining specificities of virus binding to arrays of glycans with comparable results (19).
After nearly a decade of studies on influenza virus recognition of glycans using defined glycan microarrays, the general conclusion is that most, but not all, avian and human virus HA retain their canonical specificity for Siaα2-3Gal- and Siaα2-6Gal-, respectively. However, the underlying glycan structure is increasingly recognized as also being critical to recognition. For example, human virus HAs may prefer the presentation or topology of Siaα2-6 expression on extended poly-N-acetyllactosamine (PL) [-3Galβ1-4GlcNAcβ1-]n glycans (23, 24). Other studies evaluating the binding of influenza virus to synthetic sialylated PLs noted that while most influenza virus strains demonstrated differences in their preference for the structures presented on glycan arrays, there appears to be no consistent recognition pattern associated with the underlying glycan structures. Such results suggest that the fine specificity of the receptors may drift due to antigenic selective pressure, while binding glycans with a Siaα2-6 determinant might be sufficient for infection and transmission (25). This observation is in agreement with the results of a systematic investigation of human H3N2 influenza viruses (26). In that study the binding properties of all major variants of human H3N2 viruses from their appearance in 1968 to 2012 were characterized on the large CFG defined glycan microarray. The binding specificity of viruses isolated in the same season was similar, but the specificities showed significant variation from year-to-year (26). Since all viruses were representative of epidemic strains, they all could infect and transmit efficiently among humans. The results indicate that the year-to-year variation in binding specificity is due to amino acid sequence changes driven by antigenic drift, and that in spite of having different binding specificity and avidity the viruses were still able to infect and transmit in the population.
Thus, the idea that influenza viruses simply recognize the terminal sialic acid in a specific linkage to galactose without regard for the underlying glycan structure is no longer tenable. Many additional studies using infection-based approaches have also identified interesting exceptions to the current paradigm. For example, influenza virus can infect cells from which surface Sia has enzymatically removed (27, 28), and avian viruses with Siaα2-3 specificity have recently been shown to replicate and transmit in mice (29). Human influenza virus with specificity for Siaα2-6 effectively replicate in mice lacking Siaα2-6 linkages on N-glycans in the respiratory tract (30). Furthermore, a laboratory mutant (Y98F at a conserved residue in the receptor binding pocket of HA) of influenza replicates efficiently in MDCK cells, but fails to agglutinate erythrocytes and is highly attenuated for replication in mice following intranasal infection (31, 32). The novel H7N9 viruses, through substitutions at residues 186 and 226 in their HA have acquired the ability to bind to human-like Siaα2-6 receptors, and such mutations occur in most novel human H7N9 viruses as well as in the majority of novel avian H7N9 IAVs (33). Finally, several mutant viruses derived from the Y98F mutant restored the capacity for hemagglutination and replication in the mouse respiratory tract, yet none of these viruses bind to any glycans present on the CFG microarrays (31).
The distribution of the glycan receptors in animal tissues has been based largely on histochemical studies using lectins, such as the plant lectins Sambucus nigra agglutinin (SNA) (for Siaα2-6)(34, 35) and Maackia amurensis leukoagglutinin (MAL, also termed MAA) (for Siaα2-3)(36). But while these lectins can recognize many glycans containing that simple terminal substituent, their recognition is also dependent on the glycan substructure (37). Interestingly, a recent glycomic analysis of human respiratory tissue indicated the presence of both Siaα2-3- and Siaα2-6-terminated glycans in both lung and bronchus (38). These and other observations raise questions regarding the current paradigms in this field, and support the notion that binding of influenza virus to cells in host tissues is a complex process involving not only sialic acid recognition and removal together with recognition of the underlying glycan structures, but also the presentation of glycans, their specific location in the glycocalyx, and the glycans associated with the mucins and other glycoconjugates in the immediate environment of the infection/uptake process.
Shotgun Glycan Microarrays
While defined glycan microarrays have generated a large body of data demonstrating that there are indeed differences between the specificity of if various isolates of Influenza virus, conclusions regarding the role of the underlying glycan structures are lacking, as well as the actual glycan structures in endogenous tissues that are recognized by viruses. This may be related to the fact that the specificities of HAs are deduced from glycan microarray data comprised of common, mammalian cell glycans that actually represent far less than 10% of the glycans proposed to comprise the human glycome (39, 40); thus, any conclusions relative to physiologically relevant glycans must be cautiously considered. This limitation of defined glycan microarrays is often overlooked and the assumption is made that the CFG glycan microarray and other such glycan microarrays (41, 42), while not complete, represent all possible glycan determinants present in the human glycome; however, this assumption suffers from our lack of knowledge of the human glycome or any glycome. Thus, current analyses using defined microarrays are limited to what structures are known and printed on the array and how these compare to the glycomic profiles by MS analyses of a cell or tissue of interest. Significant interest in the glycomics of the swine respiratory tract stems from the fact that pigs serve as host for both avian and human viruses, and the pig has been proposed as a “mixing vessel” for the generation of pandemic viruses (43, 44). Several recent studies have analyzed the glycans in pig lung and trachea and cell-derived materials. One of these studies involving glycomic analysis of pig trachea and lung indicated a predominance of Siaα2-6Gal in these tissues and suggested this could provide selective pressure for generation of Influenza variants that prefer the human Siaα2-6Gal receptor (45). That study identified the predominant structures as being disialylated biantennary complex type N-glycans similar to structures that bound human influenza viruses on glycan arrays. An earlier analysis showing the glycomic profile of primary swine respiratory epithelial cells also demonstrated that these cultured cells presented a larger proportion of Siaα2-6Gal compared to Siaα3-6Gal, but proposed that Siaα2-6-PL glycans play a major role in human virus infection based on their presence in the pig respiratory cells and the observation that the synthetic Siaα2-6-PL containing glycans on the defined CFG microarray also bound human influenza virus (46). A second study in 2013 also reported the analyses of sialylated N-glycans and O-glycans of isolated pig tracheobronchial epithelium and lung parenchymal mucosa and interestingly correlations were reported between the glycomic analyses and the ex-vivo avian influenza virus infection of pig cells (47). Infection with avian viruses was confined primarily to lung bronchioles rather than trachea and parenchyma. Major differences in N-glycan structures were noted, with the pig glycans having higher relative ratios of glycans terminating in Neu5Gc, Siaα2-6Gal-R and Galα1-3Gal-R than those reported for the human respiratory tract. Interestingly, similar to what was found in other studies, they also found PL sequences in sialylated multi-antennary glycans (47). These results provide indirect evidence that sialylated N-glycans and perhaps those containing sialylated PL sequences may be important in virus binding.
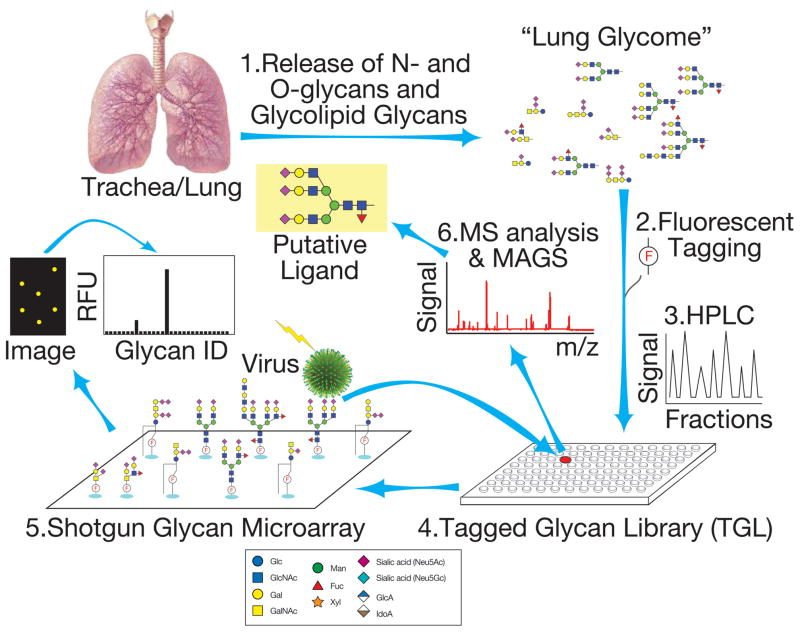
In order to directly address the question of endogenous glycan ligands for influenza viruses, we have taken a functional approach to glycomic analysis using a shotgun glycomics (48) (Figure 2). For the analysis of pig lung glycans that might be recognized by influenza viruses, the total released N- and O-glycans from pig lung glycoproteins and free glycans derived from glycolipids were fluorescently-tagged and separated by multidimensional HPLC to generated a tagged glycan library (TGL). The individual glycans were covalently printed to generate pig lung shotgun glycan microarrays, which were then interrogated with selected influenza virus strains as summarized in Figure 2 (49). In shotgun glycomics the glycans of interest can be retrieved from the TGL for the difficult task of defining structure. All viruses assayed bound one or more sialylated N-glycans, but not O-glycans or glycolipid-derived glycans, and each virus was different with respect to endogenous N-glycan recognition. The results illustrated the repertoire of N-glycans in pig lung glycoproteins that can bind influenza virus recognition, and this approach provides a new direction for identifying endogenous glycans that may functions in viral pathogenesis (49). In this study the structures, selected based on their binding by influenza virus and availability, were di- and tri-sialylated bi- and tri-antennary complex type N-glycans, which was consistent with the data obtained for fresh pig trachea and lung mentioned above, but different in conclusions with the earlier study that identified Siaα2-6-PL glycans as important virus-binding glycans. However, the latter glycomic analysis was of cultured explants and not fresh tissue. Nevertheless, taken together the results suggest that a variety of porcine-derived N-glycan structures, some with and without PL sequences may be selectively recognized by different strains of influenza virus. Clearly, more work remains to thoroughly identify the lung glycomes of humans and other influenza virus host animals, and to define the spatial and temporal nature of the glycan expression, as well as developing physiological approaches to assess the functional importance of glycans in actual cell infections.
Figure 2.
Shotgun Glycomics is a functional approach to glycomic analysis of cells and tissues. We used it to explore the repertoire of glycans in pig lung that may function as productive influenza virus receptors. The N- and O-linked glycans and glycolipid-derived glycans are released from tissue by chemical or enzymatic methods (1), labeled with a fluorescent tag (2) and separated into individual glycan components by multidimensional chromatography (3) to produce a TGL (4). The components of the TGL are then printed as a microarray and interrogated to identify relevant glycans (5). Glycans of interest are then retrieved from the TGL and subjected to structural analysis using Metadata Assisted Glycan Sequencing (MAGS) techniques (6). As the glycans in the TGL are structurally defined, the Shotgun Glycan Array becomes a Defined Glycan Array and a resource for determining specificities of other viruses and glycan binding proteins.
In spite of significant data accumulated from many investigators on analyses of influenza A virus binding to defined sialylated glycans on glycan microarrays, the exact structures of glycan receptors that function in productive infection of cells have not been defined. In a recent extensive, multidisciplinary study of influenza A virus-glycan interactions using a glycomics approach (38), the N-glycan and O-glycan composition of human lung and bronchus were analyzed by mass spectrometry (MS). The MS data permitted structural predictions that were used in combination with influenza A virus binding data from four published defined glycan arrays to determine if any array could predict replication of human, avian, and swine viruses in human ex vivo respiratory tract tissues. While the CFG array possessed the greatest diversity of sialylated glycans, the influenza A binding data from this array was not predictive of productive virus replication in human respiratory tract tissue. The authors concluded that more comprehensive and focused arrays need to be developed to evaluate emerging influenza A viruses. One solution to this dilemma is simply to synthesize the glycans corresponding to the sialylated glycans predicted by glycomics analyses; however, the glycan structures based on profiling are predicted structures and the synthesis of all possible structures is a formidable task that is currently not possible. Thus, the shotgun glycomics approach, which will obtain and identify the glycans from the natural source of tissue or glycoconjugate, has a high potential of presenting cellular glycans that function as viral receptors. The success of the Shotgun glycomics approach has been demonstrated using rotavirus virus spike protein for seeking specific virus-binding glycans.
Analysis of Rotavirus binding specificity on glycan microarrays
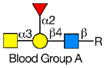
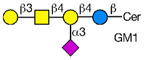
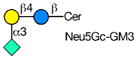
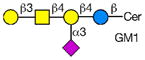
Rotaviruses are the major cause of severe diarrhea in infants and children worldwide causing more than 500,000 deaths in children under 5 years old. They are among the viruses that use attachment to cell surface glycans as their initial interaction with host cells, a process that is mediated by the VP8* domain of the rotavirus spike protein VP4 (50). In spite of significant investigation on the glycan specificity of rotavirus interactions, like Influenza A virus, the picture is murky (51, 52). The early observation that some rotavirus had hemagglutination activity led to studies indicating that binding and infectivity could be destroyed by treatment of the host cells with various neuraminidases or could be inhibited by incubating virus with sialic acid containing compounds. These types of viruses were originally called sialic acid-dependent, but that term was changed to neuraminidase-sensitive to accommodate the observation that some sialic acids (the internal Siaα2-3Gal in GM1) may be resistant to neuraminidase and thus continue to be available for binding after neuraminidase treatment (51, 53) (see Table 1). The uncertainty and controversy over the role of sialic acid in rotavirus binding and infection, similar to the current status of our understanding of Influenza virus binding and infection, continues due to the complexity of the glycan ligands and the difficulty of technically separating the various steps in the process. Clearly there are differences in animal rotaviruses versus human rotaviruses, and the available evidence suggests that while many animal rotaviruses require sialylated glycans for binding, most human rotaviruses do not (54, 55).
Table 1.
Examples of Glycan Ligands for Animal and Human Rotaviruses
The defined glycan microarray developed by the CFG has been shown to be a useful platform for investigating this question. The availability of the recombinant VP8* domain as a GST-fusion protein, provides a convenient GBP for interaction with solid phase glycans as well as a source of protein for structural detailed structural analyses (50, 52, 55). Sensitive detection of glycan binding on solid phases is straightforward using fluorescent-labeled anti-GST antibody. In a systematic analysis of a large number of rotavirus strains from humans and animals (56), it was shown that in most cases cell surface sialic acid was not required for infection, which led to investigations of what alternative cell surface glycan structures might be involved in this process. The human histo-blood group antigens (HBGAs) were likely candidates based on their presence in mucosal epithelia of the digestive tract and the ability of many rotavirus to agglutinate human erythrocytes (57). The first application of the defined glycan array to a rotavirus was the analysis of the VP8*-GST fusion protein of the human rotavirus strain HAL1166 (55). Comparison of the structure of the HAL1166 VP8* with other VP8* structures previously determined indicated that it was not compatible with Sia binding. Thus, with 511 glycans including >150 sialylated structures on that version of the CFG glycan microarray it was possible to determine if this neuraminidase-insensitive human rotavirus recognized ganglioside-derived glycans with an internal Sia or other glycans. The only structures bound by the HAL1166 VP8* were glycans with terminal GalNAcα1-3 (Fucα1-2) Galβ1-4GlcNAc, the A-type HBGA. The HAL1166 VP* was co-crystallized with the tri- and tetra-saccharides of the A-type HBGA to explain the detail interactions supporting the conclusion that this glycan structure is a cell attachment factor for HAL1166 representing the P[14] genotype. These conclusions were further supported by demonstrating that HT29 cells that express human blood group A were infected in a dose dependent manner by this strain of virus and that infectivity could be specifically inhibited by anti-A antibody, demonstrating a structural interaction between a human rotavirus VP8* and a cell surface glycan as well as demonstrating that sialylated glycans are not required for binding by neuraminidase-insensitive strains.
The defined glycan microarray has been used to screening a number of different rotavirus fusion proteins including the VP8* of a neonatal G10P[11] rotavirus, identified to be a bovine reassortant possessing a bovine rotavirus derived VP8* (58). The glycan array analyses indicated that this VP8*-domain showed a binding motif primarily toward glycans containing Galβ1-4GlcNAc presented as polylactosamine with 2 or more LacNAc units. An independent analysis of the VP8*-GST fusion protein from the identical strain generated identical results (59), and both groups interpreted the data to indicate that the binding was directed toward long, linear sequences of PL that may be biologically significant in the propensity of this rotavirus strain to infect neonates. They speculated that based on reports of developmental changes of linear to branched PL in blood and saliva of developing infants, infection by this strain of rotavirus might be restricted to neonates.
To date there are limited published studies of rotavirus attachment proteins on the defined CFG glycan array, but it is clear that this platform can provide useful screening data for on many defined glycan structures to generate useful hypotheses on the structure and function of these viruses with cellular glycans. However, the limitation of the defined array is primarily in the paucity of glycan structures represented on the array compared the thousands of glycan structures estimated to comprise the human glycome. As described above for Influenza virus, an alternative approach is a more functional shotgun glycomic analyses using arrays of glycans comprising complete metaglycomes look for biologically relevant structures, which can then be structurally characterized to define binding motifs and identify novel structures as opposed to arrays comprised of known structures.
For example, it has been speculated for a long time that the free glycans of human milk, which are thought to be similar in structure to many glycans on epithelial cell surfaces, might play a protective role in the infant intestine as receptor decoys for certain pathogens (60). The human milk free glycans, larger than lactose, are biosynthesized as linear and branched type 2 (Galβ1-4GlcNAc) or type 1 (Galβ1-3GlcNAc) LacNAc extensions of lactose and substituted with fucose and sialic acid (61). Since these structures are not metabolized, many of them are present in relatively high concentrations and may interfere with infections of pathogens that exploit cell surface glycans as receptors.
As a resource of the National Center for Functional Glycomics (www.biochem.emory.edu/emory-ncfg/) to evaluate potential functions of human milk glycans, we have developed a human milk shotgun glycan microarray comprised of over 240 human milk free glycan targets. The glycans were isolated from a pool of 10 milk donors representing all of the Lewis blood groups to encompass the entire human milk free glycan metaglycome. The neutral, monosialyl- and disialylglycans were then labeled with a fluorescent linker and each class of glycans was separated into individual components using multidimensional chromatography. MS analysis of the isolated glycans indicated that most of the glycan in the tagged glycan library (TGL) used for printing the array were comprised of single glycans or mixtures of seldom more than 3 structures. This array is now available for analysis with glycan binding proteins, viruses, or microorganisms, and we have initiated collaborative projects using recombinant VP8*-GST fusion proteins of the rotavirus outer capsid spike protein VP4 from human neonatal strains, N155 (G10P[11]) and RV3 (G3P[6]), and a bovine strain, B223 (G10P[11]) provided by B.V.V. Prasad and M.K. Estes (Baylor College of Medicine, Houston, TX). The results of this study, which also included collaborative studies on the milk glycan structures with Vernon Reinhold and David Ashline (University of New Hampshire), have been submitted for publication.
We were surprised to find that while all of these attachment proteins from neonatal strains bound polylactosamines on the defined CFG array, each could distinguish different synthetic polylactosamine structures. However, the significance of these observations are not known, since little is known about either the quantitative expression, occurrence, or distribution of such multi-antennary N-glycans with extended PL. On the human milk array, which does not present PL structures, the three proteins analyzed bound to three distinct groups of glycans. None of the proteins required sialic acid for binding; in fact, the presence of terminal sialic acid, in some cases, prevented binding. Since the structures of the bound glycans were not known, we selected 32 glycan targets and determined the structures bound by each individual VP8*. Remarkably, each VP8* recognized specific glycan determinants within a unique subset of related glycan structures and the specificity differences were a result of subtle differences in glycan structures. Among the 32 glycan targets analyzed, 40 structures were reported, 21 of which had not been previously described as free glycans in human milk, and may not have been identified if we had not observed them as isolated glycans binding to these VP8* domains.
In the near future we will make the human milk shotgun glycan microarray and other similar glycan arrays publicly available for the screening of viruses and viral attachment proteins to address questions related to the mechanisms of binding and infection of viruses that exploit cell surface glycans for host cell attachment.
Concluding Remarks
The use of glycan microarrays to explore glycan recognition by viruses is growing in importance as the technology is becoming more robust and publicly available. Both structurally defined glycan microarrays as well as shotgun glycomics microarrays of natural glycans in target tissues are now leading the way to a better understanding of potential glycan recognition important for virus attachment and entry. However, much remains to be done in further exploring the physiological roles of glycans identified through array screening and further defining the molecular interactions of viral adhesion molecules for specific glycans, as well as the native glycoconjugates, e.g. glycoproteins and glycolipids, which carry the glycans.
Highlights.
Glycan Microarrays have revolutionized studies of protein-glycan interactions and are used to define specificity of glycan binding proteins.
Glycan microarrays are used to explore glycan recognition by viruses, such as rotaviruses and influenza viruses.
Functional glycomics using shotgun glycan microarrays of endogenous glycans can identify cellular glycans that bind viruses.
Glycan microarrays can help to identify potential glycans that may be important for virus attachment and entry.
An array of >240 human milk glycans representing a portion of the human milk free glycan glycome is now available for interrogation.
Acknowledgments
This work was support by NIH Grants R01GM085448 (DFS), P41GM103694 and R24GM098791, HHSN272201400006C (NIAID Centers of Excellence for Influenza Research and Surveillance)(RDC), and U54GM62116 to the Consortium for Functional Glycomics. The authors thank Dr. Jamie Heimburg-Molinaro for critical editing assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, published within the period of the review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Rambourg A, Leblond CP. Electron microscope observations on the carbohydrate-rich cell coat present at the surface of cells in the rat. J Cell Biol. 1967;32:27–53. doi: 10.1083/jcb.32.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Varki A. Nothing in glycobiology makes sense, except in the light of evolution. Cell. 2006;126:841–845. doi: 10.1016/j.cell.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 3.Varki A. Evolutionary Forces Shaping the Golgi Glycosylation Machinery: Why Cell Surface Glycans Are Universal to Living Cells. Csh Perspect Biol. 2011:3. doi: 10.1101/cshperspect.a005462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Springer SA, Gagneux P. Glycan evolution in response to collaboration, conflict, and constraint. J Biol Chem. 2013;288:6904–6911. doi: 10.1074/jbc.R112.424523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirst GK. The Agglutination of Red Cells by Allantoic Fluid of Chick Embryos Infected with Influenza Virus. Science (New York, N Y. 1941;94:22–23. doi: 10.1126/science.94.2427.22. [DOI] [PubMed] [Google Scholar]
- 6.Burnet FM, Stone JD. The receptor-destroying enzyme of V. cholerae. Aust J Exp Biol Med Sci. 1947;25:227–233. doi: 10.1038/icb.1947.33. [DOI] [PubMed] [Google Scholar]
- 7.Klenk E, Fallard H, Lempfrid H. Über die enzymatische Wirkung von Influenzaviren. Hoppe Seyler’s Z Physiol Chem. 1955;301:235–246. [PubMed] [Google Scholar]
- 8.Gottschalk A. Neuraminidase: the specific enzyme of influenza virus and Vibrio cholerae. Biochim Biophys Acta. 1957;23:645–646. doi: 10.1016/0006-3002(57)90389-x. [DOI] [PubMed] [Google Scholar]
- 9.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12:159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 10.Rogers GN, Paulson JC. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 11.Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 12.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 13.Couceiro JN, Paulson JC, Baum LG. Influenza virus strains selectively recognize sialyloligosaccharides on human respiratory epithelium; the role of the host cell in selection of hemagglutinin receptor specificity. Virus Res. 1993;29:155–165. doi: 10.1016/0168-1702(93)90056-s. [DOI] [PubMed] [Google Scholar]
- 14.Van Breedam W, Pohlmann S, Favoreel HW, de Groot RJ, Nauwynck HJ. Bitter-sweet symphony: glycan-lectin interactions in virus biology. FEMS Microbiol Rev. 2013 doi: 10.1111/1574-6976.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neu U, Bauer J, Stehle T. Viruses and sialic acids: rules of engagement. Curr Opin Struct Biol. 2011;21:610–618. doi: 10.1016/j.sbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, Razi N, Stevens DJ, Skehel JJ, van Die I, Burton DR, Wilson IA, Cummings R, Bovin N, Wong CH, Paulson JC. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevens J, Blixt O, Tumpey TM, Taubenberger JK, Paulson JC, Wilson IA. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science (New York, N Y. 2006;312:404–410. doi: 10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 18.Amonsen M, Smith DF, Cummings RD, Air GM. Human parainfluenza viruses hPIV1 and hPIV3 bind oligosaccharides with alpha2-3-linked sialic acids that are distinct from those bound by H5 avian influenza virus hemagglutinin. Journal of virology. 2007;81:8341–8345. doi: 10.1128/JVI.00718-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulati S, Lasanajak Y, Smith DF, Cummings RD, Air GM. Glycan array analysis of influenza H1N1 binding and release. Cancer Biomark. 2014;14:43–53. doi: 10.3233/CBM-130376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •20.Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–1155. doi: 10.1016/j.jmb.2005.11.002. This study provides the detailed protocol for preparing multivalent recombinant HA for detecting HA-binding glycans on microarrays. [DOI] [PubMed] [Google Scholar]
- •21.Gulati S, Smith DF, Air GM. Deletions of neuraminidase and resistance to oseltamivir may be a consequence of restricted receptor specificity in recent H3N2 influenza viruses. Virol J. 2009;6:22. doi: 10.1186/1743-422X-6-22. This study provides the detailed protocol for labeling intact viruses with fluorescent dyes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heimburg-Molinaro J, Tappert M, Song X, Lasanajak Y, Air G, Smith DF, Cummings RD. Probing virus-glycan interactions using glycan microarrays. Methods Mol Biol. 2012;808:251–267. doi: 10.1007/978-1-61779-373-8_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrasekaran A, Srinivasan A, Raman R, Viswanathan K, Raguram S, Tumpey TM, Sasisekharan V, Sasisekharan R. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nature biotechnology. 2008;26:107–113. doi: 10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 24.Viswanathan K, Chandrasekaran A, Srinivasan A, Raman R, Sasisekharan V, Sasisekharan R. Glycans as receptors for influenza pathogenesis. Glycoconj J. 2010;27:561–570. doi: 10.1007/s10719-010-9303-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nycholat CM, McBride R, Ekiert DC, Xu R, Rangarajan J, Peng W, Razi N, Gilbert M, Wakarchuk W, Wilson IA, Paulson JC. Recognition of sialylated poly-N-acetyllactosamine chains on N- and O-linked glycans by human and avian influenza A virus hemagglutinins. Angew Chem Int Ed Engl. 2012;51:4860–4863. doi: 10.1002/anie.201200596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••26.Gulati S, Smith DF, Cummings RD, Couch RB, Griesemer SB, St George K, Webster RG, Air GM. Human H3N2 Influenza Viruses Isolated from 1968 To 2012 Show Varying Preference for Receptor Substructures with No Apparent Consequences for Disease or Spread. PloS one. 2013;8:e66325. doi: 10.1371/journal.pone.0066325. In this comprehensive study the authors demonstrate that variation in receptor binding specificity is a consequence of antigenic drift, and that viruses with quite different binding specificity and avidity are equally fit to infect and transmit in the human population. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stray SJ, Cummings RD, Air GM. Influenza virus infection of desialylated cells. Glycobiology. 2000;10:649–658. doi: 10.1093/glycob/10.7.649. [DOI] [PubMed] [Google Scholar]
- 28.Oshansky CM, Pickens JA, Bradley KC, Jones LP, Saavedra-Ebner GM, Barber JP, Crabtree JM, Steinhauer DA, Tompkins SM, Tripp RA. Avian influenza viruses infect primary human bronchial epithelial cells unconstrained by sialic acid alpha2,3 residues. PloS one. 2011;6:e21183. doi: 10.1371/journal.pone.0021183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Driskell EA, Jones CA, Stallknecht DE, Howerth EW, Tompkins SM. Avian influenza virus isolates from wild birds replicate and cause disease in a mouse model of infection. Virology. 2010;399:280–289. doi: 10.1016/j.virol.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Glaser L, Conenello G, Paulson J, Palese P. Effective replication of human influenza viruses in mice lacking a major alpha2,6 sialyltransferase. Virus Res. 2007;126:9–18. doi: 10.1016/j.virusres.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 31.Bradley KC, Galloway SE, Lasanajak Y, Song X, Heimburg-Molinaro J, Yu H, Chen X, Talekar GR, Smith DF, Cummings RD, Steinhauer DA. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J Virol. 2011;85:12387–12398. doi: 10.1128/JVI.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meisner J, Szretter KJ, Bradley KC, Langley WA, Li ZN, Lee BJ, Thoennes S, Martin J, Skehel JJ, Russell RJ, Katz JM, Steinhauer DA. Infectivity studies of influenza virus hemagglutinin receptor binding site mutants in mice. J Virol. 2008;82:5079–5083. doi: 10.1128/JVI.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dortmans JC, Dekkers J, Wickramasinghe IN, Verheije MH, Rottier PJ, van Kuppeveld FJ, de Vries E, de Haan CA. Adaptation of novel H7N9 influenza A virus to human receptors. Sci Rep. 2013;3:3058. doi: 10.1038/srep03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. Fractionation of sialylated oligosaccharides, glycopeptides, and glycoproteins on immobilized elderberry (Sambucus nigra L.) bark lectin. Arch Biochem Biophys. 1987;254:1–8. doi: 10.1016/0003-9861(87)90074-9. [DOI] [PubMed] [Google Scholar]
- 35.Shibuya N, Goldstein IJ, Broekaert WF, Nsimba-Lubaki M, Peeters B, Peumans WJ. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac (alpha 2-6) Gal/GalNAc sequence. J Biol Chem. 1987;262:1596–1601. [PubMed] [Google Scholar]
- 36.Wang WC, Cummings RD. The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked alpha-2,3 to penultimate galactose residues. J Biol Chem. 1988;263:4576–4585. [PubMed] [Google Scholar]
- 37.Nicholls JM, Chan RW, Russell RJ, Air GM, Peiris JS. Evolving complexities of influenza virus and its receptors. Trends Microbiol. 2008;16:149–157. doi: 10.1016/j.tim.2008.01.008. [DOI] [PubMed] [Google Scholar]
- ••38.Walther T, Karamanska R, Chan RWY, Chan MCW, Jia N, Air G, Hopton C, Wong MP, Dell A, Peiris JSM, Haslam SM, Nicholls JM. Glycomic Analysis of Human Respiratory Tract Tissues and Correlation with Influenza Virus Infection. Plos Pathog. 2013:9. doi: 10.1371/journal.ppat.1003223. In this extensive, multidisciplinary study of influenza A virus-glycan interactions using a glycomics profiling approach the authors conclude that more comprehensive and focused defined glycan arrays need to be developed to investigate influenza virus binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5:1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 40.Cmmings RD, Pierce JM. The challenge and promise of glycomics. Chem Biol. 2014;21:1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crusat M, Liu J, Palma AS, Childs RA, Liu Y, Wharton SA, Lin YP, Coombs PJ, Martin SR, Matrosovich M, Chen Z, Stevens DJ, Hien VM, Thanh TT, Nhu le NT, Nguyet LA, Ha do Q, van Doorn HR, Hien TT, Conradt HS, Kiso M, Gamblin SJ, Chai W, Skehel JJ, Hay AJ, Farrar J, de Jong MD, Feizi T. Changes in the hemagglutinin of H5N1 viruses during human infection--influence on receptor binding. Virology. 2013;447:326–337. doi: 10.1016/j.virol.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Childs RA, Palma AS, Wharton S, Matrosovich T, Liu Y, Chai W, Campanero-Rhodes MA, Zhang Y, Eickmann M, Kiso M, Hay A, Matrosovich M, Feizi T. Receptor-binding specificity of pandemic influenza A (H1N1) 2009 virus determined by carbohydrate microarray. Nature biotechnology. 2009;27:797–799. doi: 10.1038/nbt0909-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. Journal of virology. 1998;72:7367–7373. doi: 10.1128/jvi.72.9.7367-7373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholtissek C, Burger H, Kistner O, Shortridge KF. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–294. doi: 10.1016/0042-6822(85)90131-x. [DOI] [PubMed] [Google Scholar]
- 45.Sriwilaijaroen N, Kondo S, Yagi H, Takemae N, Saito T, Hiramatsu H, Kato K, Suzuki Y. N-glycans from porcine trachea and lung: predominant NeuAcalpha2-6Gal could be a selective pressure for influenza variants in favor of human-type receptor. PloS one. 2011;6:e16302. doi: 10.1371/journal.pone.0016302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bateman AC, Karamanska R, Busch MG, Dell A, Olsen CW, Haslam SM. Glycan analysis and influenza A virus infection of primary swine respiratory epithelial cells: the importance of NeuAc{alpha}2-6 glycans. J Biol Chem. 2010;285:34016–34026. doi: 10.1074/jbc.M110.115998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan RW, Karamanska R, Van Poucke S, Van Reeth K, Chan IW, Chan MC, Dell A, Peiris JS, Haslam SM, Guan Y, Nicholls JM. Infection of swine ex vivo tissues with avian viruses including H7N9 and correlation with glycomic analysis. Influenza Other Respir Viruses. 2013;7:1269–1282. doi: 10.1111/irv.12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •48.Song X, Lasanajak Y, Xia B, Heimburg-Molinaro J, Rhea JM, Ju H, Zhao C, Molinaro RJ, Cummings RD, Smith DF. Shotgun glycomics: a microarray strategy for functional glycomics. Nat Methods. 2011;8:85–90. doi: 10.1038/nmeth.1540. In this study, the authors describe how to harvest and separatea wide variety of glycans obtained from natural sources. This approach not only expands the repertoire of glycans on microarrays, but permits interrogating native glycans with biologically relevant glycan binding proteins and organisms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••49.Byrd-Leotis LLR, Bradley K, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, Steinhauer DA, Cummings RD. Shotgun Glycomics of Pig Lung Identifies Natural Endogenous Receptors for Influenza Viruses. Proceedings of the National Academy of Sciences of the United States of America; 2014. Studies using novel “shotgun glycan microarray” technology identify for the first time, the endogenous receptors for influenza viruses from a natural host, the pig. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Settembre EC, Chen JZ, Dormitzer PR, Grigorieff N, Harrison SC. Atomic model of an infectious rotavirus particle. The EMBO journal. 2011;30:408–416. doi: 10.1038/emboj.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isa P, Arias CF, Lopez S. Role of sialic acids in rotavirus infection. Glycoconj J. 2006;23:27–37. doi: 10.1007/s10719-006-5435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, von Itzstein M. Sialic acid dependence in rotavirus host cell invasion. Nature chemical biology. 2009;5:91–93. doi: 10.1038/nchembio.134. [DOI] [PubMed] [Google Scholar]
- 53.Fukudome K, Yoshie O, Konno T. Comparison of human, simian, and bovine rotaviruses for requirement of sialic acid in hemagglutination and cell adsorption. Virology. 1989;172:196–205. doi: 10.1016/0042-6822(89)90121-9. [DOI] [PubMed] [Google Scholar]
- 54.Fleming FE, Bohm R, Dang VT, Holloway G, Haselhorst T, Madge PD, Deveryshetty J, Yu X, Blanchard H, von Itzstein M, Coulson BS. Relative Roles of GM1 Ganglioside, N-Acylneuraminic Acids, and alpha2beta1 Integrin in Mediating Rotavirus Infection. Journal of virology. 2014;88:4558–4571. doi: 10.1128/JVI.03431-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••55.Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature. 2012;485:256–259. doi: 10.1038/nature10996. In this study the structural interactions between a human VP8* and a cellular glycan are described for the first time. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ciarlet M, Estes MK. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. The Journal of general virology. 1999;80 (Pt 4):943–948. doi: 10.1099/0022-1317-80-4-943. [DOI] [PubMed] [Google Scholar]
- 57.Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, Morrow A, Jiang X. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. Journal of virology. 2012;86:4833–4843. doi: 10.1128/JVI.05507-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ramani S, Cortes-Penfield NW, Hu L, Crawford SE, Czako R, Smith DF, Kang G, Ramig RF, Le Pendu J, Prasad BV, Estes MK. The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. Journal of virology. 2013;87:7255–7264. doi: 10.1128/JVI.03518-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, Huang P, Jiang B, Tan M, Morrow AL, Jiang X. Poly-LacNAc as an age-specific ligand for rotavirus P[11] in neonates and infants. PloS one. 2013;8:e78113. doi: 10.1371/journal.pone.0078113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bode L. Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology. 2012;22:1147–1162. doi: 10.1093/glycob/cws074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tadasu Urashima MK, Terabayashi Takashi, Fukuda Kenji, Ohnishi Masao, Kobata Akira. Milk Oligosaccharides. In: Gordon NS, editor. Oligosaccharides: sources, Properties, and applications. Nova Science Publishers, Inc; New York: 2011. pp. 1–58. [Google Scholar]
- 62.Delorme C, Brussow H, Sidoti J, Roche N, Karlsson KA, Neeser JR, Teneberg S. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. Journal of virology. 2001;75:2276–2287. doi: 10.1128/JVI.75.5.2276-2287.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Superti F, Donelli G. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. The Journal of general virology. 1991;72 (Pt 10):2467–2474. doi: 10.1099/0022-1317-72-10-2467. [DOI] [PubMed] [Google Scholar]
- 64.Rolsma MD, Kuhlenschmidt TB, Gelberg HB, Kuhlenschmidt MS. Structure and function of a ganglioside receptor for porcine rotavirus. Journal of virology. 1998;72:9079–9091. doi: 10.1128/jvi.72.11.9079-9091.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo CT, Nakagomi O, Mochizuki M, Ishida H, Kiso M, Ohta Y, Suzuki T, Miyamoto D, Hidari KI, Suzuki Y. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J Biochem. 1999;126:683–688. doi: 10.1093/oxfordjournals.jbchem.a022503. [DOI] [PubMed] [Google Scholar]