Abstract
The light-emitting chemical reaction catalyzed by the enzyme firefly luciferase is widely used for noninvasive imaging in live mice. However, photon emission from the luciferase is critically dependent on the chemical properties of its substrate, D-luciferin. In this review, we describe recent work to replace the natural luciferase substrate with synthetic analogs that extend the scope of bioluminescence imaging.
Introduction
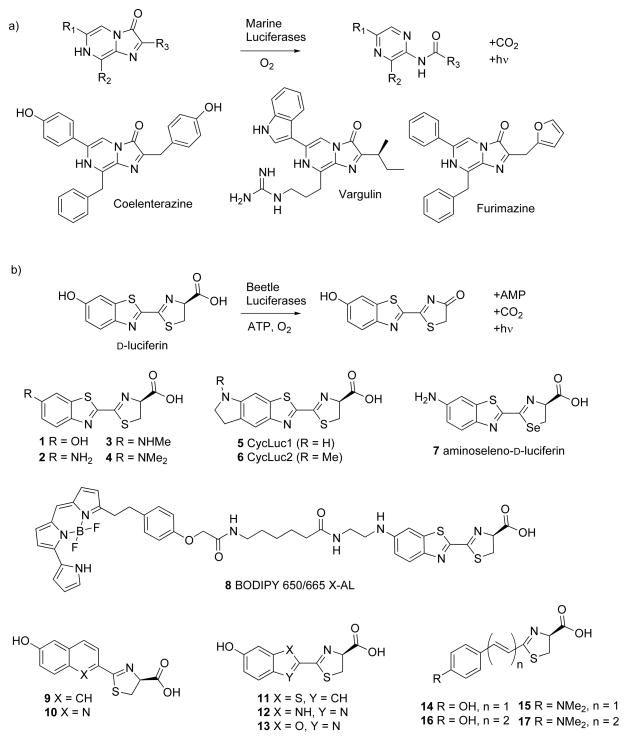
Bioluminescence is the chemical production of light by a living organism. Central to the bioluminescent reaction are an enzyme (luciferase) and a substrate (luciferin) which can be oxidized by the luciferase to generate an excited state molecule that emits light. Although there are many luminescent organisms, particularly among marine life [1], there are only a handful of luciferins known. The most widely studied luciferins are the imidazopyrazinone coelenterazine and the benzothiazole D-luciferin (Figure 1). These luciferins and their respective luciferases have found ubiquitous use as biological reporters.
Figure 1.
Luciferases oxidize their luciferin substrates to access an excited-state molecule that emits light. a) Many luciferases utilize imidazopyrazinone luciferins, which are directly oxidized by the luciferase. Coelenterazine is the substrate for Renilla, Gaussia, and many other marine luciferases [1]. Vargulin is used by Cypridina and some fish [1]. Furimazine is the synthetic imidazopyrazinone substrate for NanoLuc [10*]. b) Beetle luciferases all use the same substrate, D-luciferin, which must first be activated to an AMP ester before oxidation to the excited-state oxyluciferin. Consequently, D-luciferin is much more stable toward oxidation than imidazopyrazinones. Many synthetic modifications are tolerated by firefly luciferase.
Bioluminescence imaging (BLI) in living animals
Pioneering work by Contag and coworkers first established that bioluminescent bacteria could be imaged in live mice using a sensitive CCD camera, and then extended these results to other luciferases [2]. Many different luciferase-expressing cell lines, transgenic luciferase-expressing animals, and other bioluminescent reporters are now available for noninvasive imaging in live mice as has been extensively reviewed elsewhere [2,3].
The most common choices of luciferase and luciferin for in vivo BLI are firefly luciferase and its substrate D-luciferin. Photon emission from this pair extends into tissue-penetrating red and near-infrared wavelengths [4], the substrate is nontoxic and stable in cells and live animals, and bioluminescence can be readily imaged several minutes after routine intraperitoneal (IP) injection of the substrate. Coelenterazine-utilizing enzymes such as Renilla and Gaussia are less commonly used due to the poor tissue penetration of the blue-green light emitted by these marine luciferases [1] and the high cost and inherent instability of their imidazopyrazinone-based luciferin, which is prone to auto-oxidation [5]. Typically, these luciferins must be prepared immediately prior to use, injected intravenously (IV), and rapidly imaged [6–8]. Although significant effort has been aimed at mitigating the deficiencies of imidazopyrazinones and the luciferases that use them [9, 10*, 11], the remainder of this review will primarily be focused on D-luciferin and its analogs.
Seeing Red
Because light beyond the visible range is more tissue penetrant [12], much emphasis has been placed on finding ways to modulate luciferases to increase light emission in the red and near-IR. While the broad emission of firefly luciferase already contains a significant near-IR component over 650 nm, shifting the emission to even longer wavelengths would in theory enhance our ability to image deeply within living organisms. However, luciferase mutants and homologs with red-shifted spectra do not significantly increase the total emission of red light from D-luciferin compared to firefly luciferase [13–15]. As the peak emission is red-shifted, there is a concomitant reduction in light intensity, due in part to a lower quantum yield at the longer wavelength. Unfortunately, in vivo studies that control for luciferase expression levels have not shown improvements in sensitivity for red-shifted luciferases over the standard codon-optimized firefly luciferase luc2 [8,15].
Synthetic luciferins
In another approach, synthetic modification of the substrate changes the inherent chemical properties of the light emitter. In the last several years, it has become clear that firefly luciferase will tolerate many modifications to its luciferin (Figure 1). Analogs with mono- and di-alkylation of the amino group (e.g., 3, 4) as well as cyclic alkylamino modifications (5, 6) retain luminogenic activity [16–19]. Even large fluorescent dyes can be appended to aminoluciferins (8) to red-shift emission over 650 nm by bioluminescence resonance energy transfer (BRET), yet not destroy the ability of luciferase to catalyze light emission [20]. Moreover, the core benzothiazole can be replaced with other heterocycles (9–13) [21–24], or be removed altogether and replaced by extended π-conjugation (14–17) [25*,26]. Of particular interest, Iwano et al. have reported a luciferin analog with maximal bioluminescence emission over 650 nm (17, Figure 1) [25*].
Although substrates 2–8, 10, 16, and 17 can red-shift the peak emission of luciferase [17,20,21,25–27], none have yet shown improved light emission over D-luciferin under conditions of saturating luciferin and ATP. The reasons for this behavior are manifold. In the case of aminoluciferins, one limiting factor is product inhibition [16,17,27]. For substrates that replace the benzothiazole ring (9–17), a reduced rate of AMP ester formation and/or oxidation is a likely contributor [21,22,25*]. In both cases, a lowered quantum yield from the excited state may also play a role. D-luciferin remains the optimal substrate in vitro, particularly when the concentration of luciferin is not a limiting factor (e.g., gene reporter assays in lysed cells). Yet the superior performance of D-luciferin in vitro may be a moot point for in vivo imaging, because the modest cell permeability and mid-micromolar Km of D-luciferin limits access to the intracellular luciferase [27,28].
Bioluminescence in live cells: it’s all about access
Just as the emission wavelength of firefly luciferase is fundamentally dictated by the chemical properties of the luciferin substrate, so too is the affinity of the substrate for the luciferase, the cell-permeability, and pharmacokinetic properties of the luciferin. For alkylated aminoluciferins, superior photon flux relative to D-luciferin is observed in live luciferase-expressing cells at low substrate concentrations, likely because of a lower Km and improved cell permeability [27,29**]. On the other hand, BRET-based luciferins such as 8 yield much lower photon flux in live cells (0.1–0.4% of that of aminoluciferin) [20], undoubtedly affected by the permeability properties of the attached acceptor dye, presence of amide bonds in the linker, and larger overall size of the molecules (Figure 1). Although comparisons of many luciferin analogs to D-luciferin have not yet been performed in live cells [21,22,25], substrates with few hydrogen bond donors and acceptors (e.g., 17) are anticipated to be more cell-permeable than polar luciferin analogs (e.g., 12). The Km values of 14–17 have not been reported [25*], but 9–13 have Km values that are comparable or higher than D-luciferin [21,22], which is likely to limit light emission.
In vivo imaging with synthetic luciferins
The most common method for BLI with D-luciferin is to inject 150 mg/kg intraperitoneally (IP), and to image the mice roughly 10 minutes later, when emission typically is at its peak. For the average mouse, this is 0.1 mL of a 100 mM D-luciferin solution. It is not clear how much luciferin actually reaches luciferase-expressing cells and tissues. The biodistribution of D-luciferin in the mouse is not homogenous, and access to some tissues (e.g., the brain) is relatively low [30].
Synthetic luciferins, due to their chemical modification, are acknowledged to possibly influence tissue distribution in ways that differ from D-luciferin. Aminoluciferin (2) has been shown to emit 25% greater photon flux than D-luciferin from the ubiquitously-expressing transgenic luciferase mouse L2G85 when compared at a low IP injection dose of 0.1 mL of 1 mM substrate [31]. Aminoseleno-D-luciferin (7) yields lower peak photon flux than aminoluciferin after IV injection of a 2.5 mM solution (0.1 mL) [24]. BODIPY 650/665 X-AL (8) was compared to aminoluciferin by injecting a mouse with a 0.1 mM solution (0.1 mL) into subcutaneous luciferase-expressing tumor cells [20]. While the light emission was red-shifted, the overall photon flux was lower than aminoluciferin and no comparison was made to D-luciferin or using standard IP or IV injection methods.
Most recently, the synthetic luciferin CycLuc1 (5) was found to show improvements over the standard D-luciferin imaging conditions [29**]. This substrate allowed imaging of luciferase-expressing tumor cells with photon flux equivalent to the standard D-luciferin IP imaging conditions of 150 mg/kg, while using 20–200 fold lower doses of CycLuc1. Even doses 2000-fold lower could be imaged, a concentration that yielded no signal with D-luciferin [29**]. When compared in L2G85 transgenic luciferase mice, the substrate was readily bioavailable by both IP and IV injection methods, yielding brighter and more persistent photon flux than D-luciferin.
The blood-brain barrier poses an obstacle to many small molecules, including D-luciferin. When an IP injected dose of 0.1 mL of 5 mM CycLuc1 was compared to the equivalent volume of 100 mM D-luciferin for imaging luciferase-expressing cells in the brain striatum, eight-fold higher photon flux was observed for CycLuc1 (Figure 2) [29**]. Furthermore, the use of 5 mM CycLuc1 enabled imaging of low-level luciferase expression in dopaminergic neurons that could not be imaged with 100 mM D-luciferin, thereby expanding the scope of what is possible to image with in vivo BLI (Figure 2).
Figure 2.
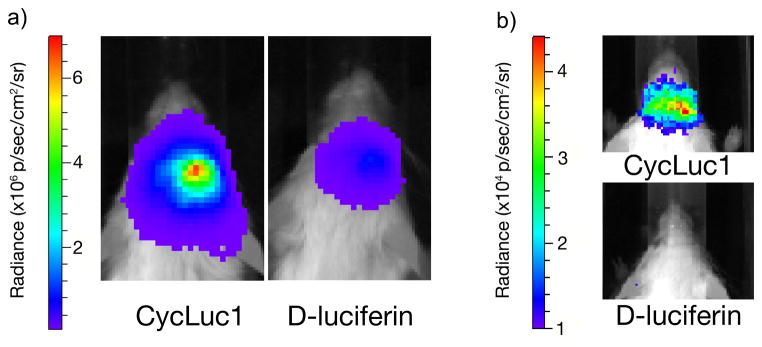
CycLuc1 compared to D-luciferin for BLI in the brain. a) Mice expressing luc2 in the brain striatum were injected IP with 100 μl of CycLuc1 (5 mM) or D-luciferin (100 mM) and imaged. Photon flux from CycLuc1-treated mice was eight-fold higher. b) Mice expressing luciferase at low levels in dopaminergic neurons were injected IP with CycLuc1 or D-luciferin as above and imaged. CycLuc1 enabled luciferase detection in live mice, while D-luciferin did not. Figure adapted from [29**].
The improved performance of CycLuc1 is likely a result of a lower Km and improved cell permeability, pharmacokinetics, and/or biodistribution. Another potential contributing factor is the action (or inaction) of efflux pumps and transporters [32,33]. D-luciferin is a substrate for ABCG2 [32], and there is evidence that the action of ABCG2 at the blood-brain barrier contributes to the lowered bioluminescent signal in this organ [34]. CycLuc1 and other synthetic luciferins could potentially be poorer substrates for ABCG2. At the same time, chemical modification of luciferin substrates could modulate affinity for organic anion transporters [33], or render them substrates for efflux pumps such as PgP and MRPs that are not known to recognize D-luciferin.
Because CycLuc1 red-shifts luciferase light emission relative to D-luciferin in vitro, it was surprising that the in vivo emission wavelength from CycLuc1 was not red-shifted compared to D-luciferin [29**]. This may be because the yellow-green emission wavelength of firefly luciferase with D-luciferin in vitro (~555 nm) is red-shifted to longer wavelengths at 37 °C in vivo (~612 nm) [4]. Therefore, substrates with similar cell-permeability, Km, and pharmacokinetic properties to CycLuc1 but emission wavelengths over 612 nm may offer further improvements.
Caged luciferin reporters
Geiger, Miska and coworkers pioneered the concept of bioluminogenic substrates that release D-luciferin or 6′-aminoluciferin upon the action of a hydrolytic enzyme (e.g., phosphatase, esterase, protease, β-galactosidase, or sulfatase) [35–38]. Prior to enzyme activation, these molecules are not light-emitting substrates for luciferase. Enzymatic release of the luciferin substrate therefore reports the presence of this enzyme activity. Other workers have extended this “caged” or “pro-luciferin” concept to allow in vivo imaging of the enzymatic activity of β-galactosidase [39], proteases [40*,41*], and cytochrome P450s [42] as well as the detection of reactive small molecules such as hydrogen peroxide [40*,43]. However, in employing this strategy, care must be taken to ensure that the reporter is actually specific for the desired analyte and is generally bioavailable [44]. Furthermore, caged luciferins that do not emit light with luciferase could still potentially be inhibitors [35] or even non-luminogenic substrates, a possibility that has not been universally explored.
Caged luciferins are generally less soluble than their parent luciferin and cannot be supplied at the same high dose typically used for imaging with D-luciferin. Furthermore, signal is not detected immediately upon release of D-luciferin or 6′-aminoluciferin, but rather once the released luciferin contacts the luciferase enzyme. The construction of caged luciferins that release luciferin analogs possessing higher cell permeability, lower Km values for luciferase and/or improved pharmacokinetic properties may therefore prove useful for increasing the sensitivity of these reporters.
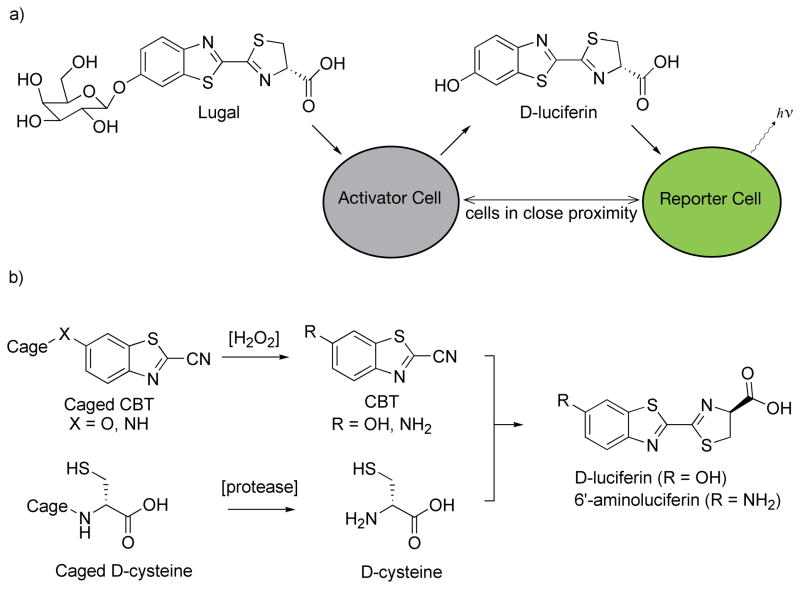
Typically, caged luciferins have been used in transgenic mice that ubiquitously express luciferase [45]. In one interesting variant of this approach, BLI was used as a proximity reporter for two different cell types – an “activator” cell and a “reporter” cell [46*]. An activator cell expressing β-galactosidase converts the pro-luciferin Lugal into D-luciferin (Figure 3). Bioluminescence is only observed if the released D-luciferin diffuses out of the cell and into a reporter cell expressing firefly luciferase. Thus bioluminescence can be used to detect the proximity of the activator cells to luciferase-expressing reporter cells.
Figure 3.
Caged luciferin reporters. a) Upon enzymatic removal of galactose from Lugal in an activator cell expressing β-galactosidase, D-luciferin is produced. Bioluminescent light emission is detected only if the liberated D-luciferin can access a luciferase-expressing reporter cell [46*]. b) Luciferin substrates can be formed in vivo from the reaction of a cyanobenzothiazole (CBT) with D-cysteine [40*,41*]. Either or both of these precursors can be caged, thereby linking light emission to enzymatic activity (e.g., a caspase) [40*,41*] and/or the presence of an analyte (e.g., hydrogen peroxide) [40*].
Caged luciferins could also potentially be used to improve delivery of the luciferin substrate. For example, esters of D-luciferin have been used in attempts to improve cellular delivery [28]. However, so far this approach has met with limited success, perhaps due to poor rates of esterase cleavage, low solubility, and/or the inhibition of luciferase by uncleaved esters. Luciferin esters are also inherently more reactive toward oxygen and prone to chemiluminescence than the parent luciferin.
In vivo synthesis of luciferins
In an amazing feat of in vivo chemistry that remains somewhat mysterious, the firefly synthesizes D-luciferin in a multi-step process from L-cysteine and benzoquinone [47]. In the laboratory, D-luciferin and aminoluciferin analogs are typically synthesized by the condensation of an electron-deficient nitrile with D-cysteine (Figure 3). Impressively, recent work has shown that luciferin substrates can be formed in live mice using this reaction, despite the presence of endogenous L-cysteine [40*,41*]. BLI thus does not require a pre-formed luciferin, but can be performed using component parts which react in vivo (Figure 3). Both D-luciferin and 6′-aminoluciferin can be formed, and by caging one or both components, protease activity and/or hydrogen peroxide can be detected [40*,41*].
Because firefly luciferase will only emit light with the D-enantiomer of luciferin or its analogs, the formation of L-luciferins and other products are largely invisible to bioluminescence imaging. However, the ultimate fate(s) of the reactive nitrile, which is biocompatible but not strictly bioorthogonal, has not been fully described. Besides L-cysteine, other potential endogenous reaction partners include homocysteine [48], proteins with N-terminal cysteine residues [48,49], and cysteine proteases [50]. Some of these products may have effects on the ability to faithfully and noninvasively image biological processes, and differences in the metabolism and cellular uptake of the nitrile and D-cysteine components could lead to cell or tissue-specific differences in the sensitivity of detection for a particular enzymatic activity or analyte. Competing reactions with endogenous molecules also poses challenges for the application of this chemistry for in vivo bioconjugation, although some success has been reported in the case of intramolecular reactions [51,52].
Substrate selectivity and the development of new luciferases
The noninvasive interrogation of multiple features in living animals can be achieved by combining luciferases that use D-luciferin with luciferases that use coelenterazine or other imidazopyrazinones (Figure 1) [7,8,53]. However, these approaches suffer the inherent shortcomings of imidazopyrazinones outlined in the introduction.
Alternatively, the complementary modification of luciferin substrates and mutation of luciferases could potentially allow the use of two (or more) selective beetle luciferin-luciferase pairs in vivo. Mutagenesis of firefly luciferase can improve the utilization and selectivity for synthetic aminoluciferins over D-luciferin in vitro and in live cells, suggesting that this approach is feasible [27].
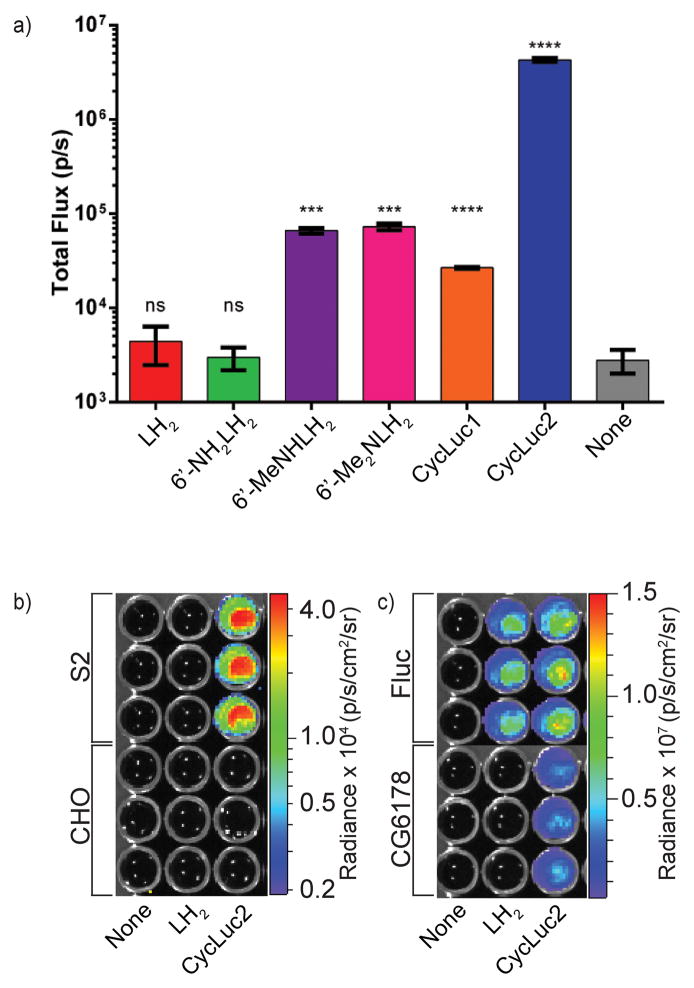
To engineer new firefly luciferase homologs that selectively utilize synthetic luciferins, it is worthwhile to ask: what are the fundamental requirements for luciferase activity? Beetle luciferases are all homologous members of the acyl-adenylate superfamily [54,55], share high homology to fatty acyl-CoA synthetases, and in fact retain fatty acyl-CoA synthetase activity [56]. Fatty acyl-CoA synthetases from nonluminescent organisms, on the other hand, do not possess luciferase activity with D-luciferin [57,58]. In an exciting recent development, a fatty acyl-CoA synthetase from the fruit fly Drosophila melanogaster was found to possess latent luciferase activity with the synthetic luciferin CycLuc2 (Figure 4) [59*]. Expression of this protein in mammalian cells allowed bioluminescence imaging of these cells only in the presence of CycLuc2 – no light emission was seen when D-luciferin was employed (Figure 4). This suggests that the fundamental chemistry of bioluminescence is broader than previously thought, and that the chemistry of existing adenylating enzymes could be exploited to create new substrate-selective luciferases. Furthermore, the ability of Drosophila S2 cells to emit light simply upon the addition of CycLuc2 (Figure 4) suggests that it may be possible to utilize endogenous enzymes to enable bioluminescence imaging. An area of particular interest would be the application of caged luciferins, where the detection of a particular enzymatic activity or analyte could potentially be performed in the absence of genetic manipulation, foregoing a canonical exogenous luciferase for an endogenous fatty acyl-CoA synthetase that moonlights as a luciferase.
Figure 4.
Latent luciferase activity in a fatty acyl-CoA synthetase from Drosophila. a) CycLuc2, but not D-luciferin, is a light-emitting substrate for the fatty acyl-CoA synthetase CG6178. b) Drosophila S2 cells, but not mammalian Chinese hamster ovary (CHO) cells, glow when treated with CycLuc2. c) Transfection of CG6178 into CHO cells confers bioluminescence in the presence of CycLuc2 but not D-luciferin. Figure adapted from [59*].
Conclusions and outlook
Bioluminescence is a powerful and versatile technique for noninvasive imaging in live animals. Because it requires an enzyme (luciferase) and a substrate (luciferin), it combines genetically-encoded specificity with the flexibility of a small molecule. Conventional uses of bioluminescence include imaging of gene expression and tumor burden. Synthetic pro-luciferins can extend this repertoire to include imaging of enzymatic activity, small molecule analytes, and cellular proximity. It has also become clear that firefly luciferase will tolerate many chemical modifications to its luciferin that broaden the scope of BLI. While particular emphasis has been placed on finding ways to increase light emission at tissue-penetrating near-IR wavelengths, another important consideration is improving access of the substrate to the luciferase in vivo, which can enhance detection beyond what is possible with D-luciferin. Furthermore, luciferase mutants display selectivity for synthetic luciferins that could potentially allow the use of two (or more) beetle luciferin-luciferase pairs for multiplexed imaging, and luciferase homologs from insects that do not emit light with D-luciferin are potential substrate-selective latent luciferases. The breadth of BLI reporters is thus expanding beyond naturally-occurring luciferins and luciferases into a world of luminogenic small molecules and their activating enzymes. The combination of new luciferins, caged luciferins, and luciferin precursors with mutant and latent luciferases is expected to greatly enhance our ability to study basic biology and disease pathogenesis in living organisms, promising a bright future for bioluminescence.
Highlights.
CycLuc1 outperforms D-luciferin for bioluminescence imaging in live mice.
Luciferin substrates can be formed by reaction of component parts in vivo.
Caged luciferins can report on cellular proximity.
Caged luciferins can report on multiple enzymatic activities or analytes.
Enzymes from nonluminescent organisms can function as luciferases.
Acknowledgments
This work was supported by a grant from the National Institutes of Health (R01EB013270).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Haddock SHD, Moline MA, Case JF. Bioluminescence in the Sea. Annual Review of Marine Science. 2010;2:443–493. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- 2.Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr Op Chem Biol. 2010;14:80–89. doi: 10.1016/j.cbpa.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Dothager RS, Flentie K, Moss B, Pan M-H, Kesarwala A, Piwnica-Worms D. Advances in bioluminescence imaging of live animal models. Curr Opin Biotechnol. 2009;20:45–53. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao H, Doyle TC, Coquoz O, Kalish F, Rice BW, Contag CH. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 5.Otto-Duessel M, Khankaldyyan V, Gonzalez-Gomez I, Jensen MC, Laug WE, Rosol M. In vivo testing of Renilla luciferase substrate analogs in an orthotopic murine model of human glioblastoma. Mol Imaging. 2006;5:57–64. [PubMed] [Google Scholar]
- 6.Gil JS, Machado HB, Herschman HR. A Method to Rapidly and Accurately Compare the Relative Efficacies of Non-invasive Imaging Reporter Genes in a Mouse Model and its Application to Luciferase Reporters. Mol Imaging Biol. 2012;14:462–471. doi: 10.1007/s11307-011-0515-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stacer AC, Nyati S, Moudgil P, Iyengar R, Luker KE, Rehemtulla A, Luker GD. NanoLuc reporter for dual luciferase imaging in living animals. Mol Imaging. 2013;12:1–13. [PMC free article] [PubMed] [Google Scholar]
- 8.Mezzanotte L, Aswendt M, Tennstaedt A, Hoeben R, Hoehn M, Löwik C. Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast Media Mol Imaging. 2013;8:505–513. doi: 10.1002/cmmi.1549. [DOI] [PubMed] [Google Scholar]
- 9.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7:5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Hall MP, Unch J, Binkowski BF, Valley MP, Butler BL, Wood MG, Otto P, Zimmerman K, Vidugiris G, Machleidt T, et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. This paper describes the optimization of a small luciferase subunit derived from Oplophorus gracilirostris, and its engineering to selectively utilize a synthetic imidazopyrazinone luciferin, furimazine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindberg E, Mizukami S, Ibata K, Miyawaki A, Kikuchi K. Development of Luminescent Coelenterazine Derivatives Activatable by β-Galactosidase for Monitoring Dual Gene Expression. Chem Eur J. 2013;19:14970–14976. doi: 10.1002/chem.201302002. [DOI] [PubMed] [Google Scholar]
- 12.Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- 13.Viviani VR, Bechara EJ, Ohmiya Y. Cloning, sequence analysis, and expression of active Phrixothrix railroad-worms luciferases: relationship between bioluminescence spectra and primary structures. Biochemistry. 1999;38:8271–8279. doi: 10.1021/bi9900830. [DOI] [PubMed] [Google Scholar]
- 14.Branchini BR, Ablamsky DM, Davis AL, Southworth TL, Butler B, Fan F, Jathoul AP, Pule MA. Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal Biochem. 2010;396:290–297. doi: 10.1016/j.ab.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Liang Y, Walczak P, Bulte JWM. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt. 2012;17:016004. doi: 10.1117/1.JBO.17.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodroofe CC, Shultz JW, Wood MG, Osterman J, Cali JJ, Daily WJ, Meisenheimer PL, Klaubert DH. N-Alkylated 6′-aminoluciferins are bioluminescent substrates for Ultra-Glo and QuantiLum luciferase: new potential scaffolds for bioluminescent assays. Biochemistry. 2008;47:10383–10393. doi: 10.1021/bi800505u. [DOI] [PubMed] [Google Scholar]
- 17.Reddy GR, Thompson WC, Miller SC. Robust light emission from cyclic alkylaminoluciferin substrates for firefly luciferase. J Am Chem Soc. 2010;132:13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takakura H, Sasakura K, Ueno T, Urano Y, Terai T, Hanaoka K, Tsuboi T, Nagano T. Development of luciferin analogues bearing an amino group and their application as BRET donors. Chem Asian J. 2010;5:2053–2061. doi: 10.1002/asia.201000219. [DOI] [PubMed] [Google Scholar]
- 19.Takakura H, Kojima R, Urano Y, Terai T, Hanaoka K, Nagano T. Aminoluciferins as functional bioluminogenic substrates of firefly luciferase. Chem Asian J. 2011;6:1800–1810. doi: 10.1002/asia.201000873. [DOI] [PubMed] [Google Scholar]
- 20.Kojima R, Takakura H, Ozawa T, Tada Y, Nagano T, Urano Y. Rational design and development of near-infrared-emitting firefly luciferins available in vivo. Angew Chem Int Ed Engl. 2013;52:1175–1179. doi: 10.1002/anie.201205151. [DOI] [PubMed] [Google Scholar]
- 21.Branchini BR, Hayward MM, Bamford S, Brennan PM, Lajiness EJ. Naphthyl- and quinolylluciferin: green and red light emitting firefly luciferin analogues. Photochem Photobiol. 1989;49:689–695. doi: 10.1111/j.1751-1097.1989.tb08442.x. [DOI] [PubMed] [Google Scholar]
- 22.Woodroofe CC, Meisenheimer PL, Klaubert DH, Kovic Y, Rosenberg JC, Behney CE, Southworth TL, Branchini BR. Novel heterocyclic analogues of firefly luciferin. Biochemistry. 2012;51:9807–9813. doi: 10.1021/bi301411d. [DOI] [PubMed] [Google Scholar]
- 23.McCutcheon DC, Paley MA, Steinhardt RC, Prescher JA. Expedient synthesis of electronically modified luciferins for bioluminescence imaging. J Am Chem Soc. 2012;134:7604–7607. doi: 10.1021/ja301493d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conley NR, Dragulescu-Andrasi A, Rao J, Moerner WE. A Selenium Analogue of Firefly D-Luciferin with Red-Shifted Bioluminescence Emission. Angew Chem Int Ed. 2012;51:3350–3353. doi: 10.1002/anie.201105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25*.Iwano S, Obata R, Miura C, Kiyama M, Hama K, Nakamura M, Amano Y, Kojima S, Hirano T, Maki S, et al. Development of simple firefly luciferin analogs emitting blue, green, red, and near-infrared biological window light. Tetrahedron. 2013;69:3847–3856. This work describes the most red-shifted luciferin substrate to date by replacing the benzothiazole and extending conjugation. [Google Scholar]
- 26.Miura C, Kiyama M, Iwano S, Ito K, Obata R, Hirano T, Maki S, Niwa H. Synthesis and luminescence properties of biphenyl-type firefly luciferin analogs with a new, near-infrared light-emitting bioluminophore. Tetrahedron. 2013;69:9726–9734. [Google Scholar]
- 27.Harwood KR, Mofford DM, Reddy GR, Miller SC. Identification of mutant firefly luciferases that efficiently utilize aminoluciferins. Chem Biol. 2011;18:1649–1657. doi: 10.1016/j.chembiol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Craig FF, Simmonds AC, Watmore D, McCapra F, White MR. Membrane-permeable luciferin esters for assay of firefly luciferase in live intact cells. Biochem J. 1991;276 (Pt 3):637–641. doi: 10.1042/bj2760637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29**.Evans MS, Chaurette JP, Adams ST, Jr, Reddy GR, Paley MA, Aronin N, Prescher JA, Miller SC. A synthetic luciferin improves bioluminescence imaging in live mice. Nat Methods. 2014;11:393–395. doi: 10.1038/nmeth.2839. This work describes the first synthetic luciferin to perform better than the standard imaging conditions with D-luciferin for BLI in live mice. Less substrate is required, the signal is more persistent, and imaging in the brain that could not be performed with D-luciferin is enabled. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berger F, Paulmurugan R, Bhaumik S, Gambhir SS. Uptake kinetics and biodistribution of 14C-D-luciferin--a radiolabeled substrate for the firefly luciferase catalyzed bioluminescence reaction: impact on bioluminescence based reporter gene imaging. Eur J Nucl Med Mol Imaging. 2008;35:2275–2285. doi: 10.1007/s00259-008-0870-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinde R, Perkins J, Contag CH. Luciferin derivatives for enhanced in vitro and in vivo bioluminescence assays. Biochemistry. 2006;45:11103–12. doi: 10.1021/bi060475o. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Bressler JP, Neal J, Lal B, Bhang H-EC, Laterra J, Pomper MG. ABCG2/BCRP expression modulates D-Luciferin based bioluminescence imaging. Cancer research. 2007;67:9389–9397. doi: 10.1158/0008-5472.CAN-07-0944. [DOI] [PubMed] [Google Scholar]
- 33.Patrick PS, Lyons SK, Rodrigues TB, Brindle KM. Oatp1 Enhances Bioluminescence by Acting as a Plasma Membrane Transporter for d-luciferin. Mol Imaging Biol. 2014 doi: 10.1007/s11307-014-0741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakhsheshian J, Wei B-R, Chang K-E, Shukla S, Ambudkar SV, Simpson RM, Gottesman MM, Hall MD. Bioluminescent imaging of drug efflux at the blood-brain barrier mediated by the transporter ABCG2. Proc Natl Acad Sci US A. 2013;110:20801–20806. doi: 10.1073/pnas.1312159110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miska W, Geiger R. Synthesis and characterization of luciferin derivatives for use in bioluminescence enhanced enzyme immunoassays. New ultrasensitive detection systems for enzyme immunoassays, I. J Clin Chem Clin Biochem. 1987;25:23–30. doi: 10.1515/cclm.1987.25.1.23. [DOI] [PubMed] [Google Scholar]
- 36.Miska W, Geiger R. A new type of ultrasensitive bioluminogenic enzyme substrates. I. Enzyme substrates with D-luciferin as leaving group. Biol Chem Hoppe-Seyler. 1988;369:407–411. doi: 10.1515/bchm3.1988.369.1.407. [DOI] [PubMed] [Google Scholar]
- 37.Geiger R, Schneider E, Wallenfels K, Miska W. A new ultrasensitive bioluminogenic enzyme substrate for beta-galactosidase. Biol Chem Hoppe-Seyler. 1992;373:1187–1191. doi: 10.1515/bchm3.1992.373.2.1187. [DOI] [PubMed] [Google Scholar]
- 38.Monsees T, Miska W, Geiger R. Synthesis and characterization of a bioluminogenic substrate for alpha-chymotrypsin. Anal Biochem. 1994;221:329–34. doi: 10.1006/abio.1994.1421. [DOI] [PubMed] [Google Scholar]
- 39.Wehrman TS, von Degenfeld G, Krutzik PO, Nolan GP, Blau HM. Luminescent imaging of beta-galactosidase activity in living subjects using sequential reporter-enzyme luminescence. Nat Methods. 2006;3:295–301. doi: 10.1038/nmeth868. [DOI] [PubMed] [Google Scholar]
- 40*.Van de Bittner GC, Bertozzi CR, Chang CJ. Strategy for dual-analyte luciferin imaging: in vivo bioluminescence detection of hydrogen peroxide and caspase activity in a murine model of acute inflammation. J Am Chem Soc. 2013;135:1783–1795. doi: 10.1021/ja309078t. This paper established that D-luciferin could be formed in vivo from a 2-cyanobenzothiazole and D-cysteine. Furthermore, both components could be caged to report on both protease activity and the presence of hydrogen peroxide. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41*.Godinat A, Park HM, Miller SC, Cheng K, Hanahan D, Sanman LE, Bogyo M, Yu A, Nikitin GF, Stahl A, et al. A biocompatible in vivo ligation reaction and its application for noninvasive bioluminescent imaging of protease activity in living mice. ACS Chem Biol. 2013;8:987–999. doi: 10.1021/cb3007314. This paper established that D-luciferin and aminoluciferin could be formed in vivo from D-cysteine and a 2-cyanobenzothiazole. Furthermore, D-cysteine could be caged to report on protease activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roncoroni C, Rizzi N, Brunialti E, Cali JJ, Klaubert DH, Maggi A, Ciana P. Molecular imaging of cytochrome P450 activity in mice. Pharm Res. 2012;65:531–536. doi: 10.1016/j.phrs.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 43.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. In vivo imaging of hydrogen peroxide production in a murine tumor model with a chemoselective bioluminescent reporter. Proc Natl Acad Sci US A. 2010;107:21316–21321. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, Chen L, Wu W, Zhang W, Ma Z, Cheng Y, Du L, Li M. Discovery of bioluminogenic probes for aminopeptidase N imaging. Anal Chem. 2014;86:2747–2751. doi: 10.1021/ac404176x. [DOI] [PubMed] [Google Scholar]
- 45.Cao Y-A, Wagers AJ, Beilhack A, Dusich J, Bachmann MH, Negrin RS, Weissman IL, Contag CH. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci US A. 2004;101:221–226. doi: 10.1073/pnas.2637010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Sellmyer MA, Bronsart L, Imoto H, Contag CH, Wandless TJ, Prescher JA. Visualizing cellular interactions with a generalized proximity reporter. Proc Natl Acad Sci US A. 2013;110:8567–8572. doi: 10.1073/pnas.1218336110. This work showed that bioluminescence imaging with a caged luciferin could be used to report on cellular proximity in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oba Y, Yoshida N, Kanie S, Ojika M, Inouye S. Biosynthesis of Firefly Luciferin in Adult Lantern: Decarboxylation of L-Cysteine is a Key Step for Benzothiazole Ring Formation in Firefly Luciferin Synthesis. PLoS ONE. 2013;8:e84023. doi: 10.1371/journal.pone.0084023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ren H, Xiao F, Zhan K, Kim Y-P, Xie H, Xia Z, Rao J. A Biocompatible Condensation Reaction for the Labeling of Terminal Cysteine Residues on Proteins. Angew Chem Int Ed. 2009;48:9658–9662. doi: 10.1002/anie.200903627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hardy RY, Resh MD. Identification of N-terminal Residues of Sonic Hedgehog Important for Palmitoylation by Hedgehog Acyltransferase. J Biol Chem. 2012;287:42881–42889. doi: 10.1074/jbc.M112.426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oballa RM, Truchon J-F, Bayly CI, Chauret N, Day S, Crane S, Berthelette C. A generally applicable method for assessing the electrophilicity and reactivity of diverse nitrile-containing compounds. Bioorg Med Chem Lett. 2007;17:998–1002. doi: 10.1016/j.bmcl.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 51.Cao C-Y, Shen Y-Y, Wang J-D, Li L, Liang G-L. Controlled intracellular self-assembly of gadolinium nanoparticles as smart molecular MR contrast agents. Sci Rep. 2013;3:1024. doi: 10.1038/srep01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye D, Shuhendler AJ, Cui L, Tong L, Tee SS, Tikhomirov G, Felsher DW, Rao J. Bioorthogonal cyclization-mediated in situ self-assembly of small-molecule probes for imaging caspase activity in vivo. Nat Chem. 2014;6:519–526. doi: 10.1038/nchem.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maguire CA, Bovenberg MS, Crommentuijn MH, Niers JM, Kerami M, Teng J, Sena-Esteves M, Badr CE, Tannous BA. Triple bioluminescence imaging for in vivo monitoring of cellular processes. Mol Ther Nucleic Acids. 2013;2:e99. doi: 10.1038/mtna.2013.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang KH, Xiang H, Dunaway-Mariano D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonas sp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry. 1997;36:15650–15659. doi: 10.1021/bi971262p. [DOI] [PubMed] [Google Scholar]
- 55.Sundlov JA, Fontaine DM, Southworth TL, Branchini BR, Gulick AM. Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism. Biochemistry. 2012;51:6493–6495. doi: 10.1021/bi300934s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oba Y, Ojika M, Inouye S. Firefly luciferase is a bifunctional enzyme: ATP-dependent monooxygenase and a long chain fatty acyl-CoA synthetase. FEBS Lett. 2003;540:251–254. doi: 10.1016/s0014-5793(03)00272-2. [DOI] [PubMed] [Google Scholar]
- 57.Oba Y, Ojika M, Inouye S. Characterization of CG6178 gene product with high sequence similarity to firefly luciferase in Drosophila melanogaster. Gene. 2004;329:137–145. doi: 10.1016/j.gene.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 58.Oba Y, Sato M, Ojika M, Inouye S. Enzymatic and Genetic Characterization of Firefly Luciferase and Drosophila CG6178 as a Fatty Acyl-CoA Synthetase. Biosci Biotechnol Biochem. 2005;69:819–828. doi: 10.1271/bbb.69.819. [DOI] [PubMed] [Google Scholar]
- 59*.Mofford DM, Reddy GR, Miller SC. Latent luciferase activity in the fruit fly revealed by a synthetic luciferin. Proc Natl Acad Sci US A. 2014;111:4443–4448. doi: 10.1073/pnas.1319300111. This work demonstrated that the fatty acyl-CoA synthetase CG6178 from Drosophila melanogaster is a latent luciferase that emits light with the synthetic luciferin CycLuc2, but not with D-luciferin. [DOI] [PMC free article] [PubMed] [Google Scholar]