Abstract
Objective
To analyze the GNRHR in patients with normosmic isolated hypogonadotropic hypogonadism (IHH) and constitutional delay of growth and puberty (CDGP).
Design
Molecular analysis and in vitro experiments correlated with phenotype.
Setting
Academic medical center.
Patient(s)
110 individuals with normosmic IHH (74 males) and 50 with CGDP.
Intervention(s)
GNRHR coding region was amplified and sequenced.
Main Outcome Measure(s)
Novel variants were submitted to in vitro analysis. Frequency of mutations and genotype-phenotype correlation were analyzed. Microsatellite markers flanking GNRHR were examined in patients carrying the same mutation to investigate a possible founder effect.
Result(s)
Eleven IHH patients (10%) carried biallelic GNRHR mutations. In vitro analysis of novel variants (p.Y283H and p.V134G) demonstrated complete inactivation. The founder effect study revealed that Brazilian patients carrying the p.R139H mutation shared the same haplotype. Phenotypic spectrum in patients with GNRHR mutations varied from complete GnRH deficiency to partial and reversible IHH, with a relatively good genotype-phenotype correlation. One boy with CDGP was heterozygous for the p.Q106R variant, which was not considered pathogenic.
Conclusion(s)
GNRHR mutations are a frequent cause of congenital normosmic IHH and should be the first candidate gene for genetic screening in this condition, especially in autosomal recessive familial cases. The founder effect study suggested that the p.R139H mutation arises from a common ancestor in the Brazilian population. Finally, mutations in GNRHR do not appear to be involved in the pathogenesis of CDGP.
Keywords: GnRH receptor, hypogonadotropic hypogonadism, pubertal delay, mutation, founder effect
INTRODUCTION
Normal pubertal development and reproductive function depends on the intact release and action of hypothalamic gonadotropin-releasing hormone (GnRH), the fundamental regulator of the gonadotropic axis. The beginning of puberty is characterized by reactivation of pulsatile GnRH secretion, after a period of quiescence during childhood (1).
Delay in the normal timing of pubertal onset is the underlying feature of constitutional delay of growth and puberty (CDGP) (2). CDGP is considered to be an extreme of the normal spectrum in pubertal timing, with individuals spontaneously starting and attaining complete pubertal development 2.5 SD later than the mean age (2). Although CDGP represents the single most common cause of delayed puberty within the general population, it can be diagnosed only after other underlying conditions have been ruled out. The main differential diagnosis of CDGP is congenital normosmic isolated hypogonadotropic hypogonadism (IHH). Congenital IHH is characterized by failure of gonadal function secondary to deficient GnRH-induced gonadotropin secretion (3). IHH is a clinically and genetically heterogeneous condition, which can be sporadic or inherited as an autosomal trait (4). The phenotypic spectrum of patients with normosmic IHH varies from complete lack of secondary sexual characteristics and absent gonadotropin secretion to incomplete pubertal development (4). When IHH is associated with olfactory abnormalities, it characterizes Kallmann syndrome, associated with defects in the migration of olfactory and GnRH neurons during fetal life.
GnRH receptor (GNRHR) inactivating mutations were the first recognized monogenic cause of normosmic IHH (5). Since the first description by de Roux et al. (1997) (5), more than 20 mutations distributed throughout the coding sequence of the receptor have been reported in patients with sporadic or familial forms of normosmic IHH, in a classical autosomal recessive mode of inheritance (5–14). In addition, a number of other genes have been implicated in the pathogenesis of IHH (15).
In contrast to IHH, no specific genes have been identified in association with the molecular pathogenesis of CDGP to date. Nevertheless, genetic variation is known to influence the normal spectrum of pubertal timing. CDGP frequently aggregates in families, with a reported incidence of 50 to 75% of patients with a family history of delayed puberty, suggesting that it may be at least in part genetically determined (2, 16). Mutations in genes underlying IHH have been reported in families with a wide phenotypic spectrum, varying from pubertal delay to severe IHH. CDGP may represent a mild phenotype of IHH and defects in GNRHR could contribute to this phenotype.
In the present study, we examined the frequency and phenotype-genotype correlation of GNRHR mutations in a large cohort of patients with pubertal delay, including normosmic IHH and constitutional delay of growth and puberty. We also performed a founder effect study of a recurrent GNRHR mutation.
MATERIAL AND METHODS
Patients
The protocol was approved by the Ethical Committee of Hospital das Clinicas, Sao Paulo University. Written informed consent was obtained from all patients or their parents. 160 patients with pubertal delay were studied: 50 with CGDP (41 (82%) boys and 9 (18%) girls) and 110 with normosmic IHH (74 (67.3%) males and 36 (32.7%) females). Twenty patients (18%) had familial IHH. GNRHR had been previously sequenced in 15 patients from this cohort (8). Other genes associated to the IHH phenotype, including TAC3, TACR3, KISS1, KISS1R, GNRH1, FGF8, FGFR1, PROK2, PROKR2 and WDR11, have been previously sequenced in the IHH group (17–24). The control population was composed of 150 healthy Brazilian adults of both sexes, with normal pubertal development.
The diagnosis of CDGP was based on lack of breast development (Tanner stage 2) by the age of 13 and absent menarche by the age of 15 years in girls, and testicular volume < 4.0 mL by the age of 14 years in boys, delayed bone age, absence of other identifiable causes of delayed puberty, as well as spontaneous and complete achievement of puberty development by age 18 years during follow-up. The diagnosis of normosmic IHH was based on the following criteria: incomplete or absent pubertal development after 18 yrs in males and 16 yrs in females, prepubertal or low testosterone (T) or estradiol (E2) levels for age, and low or normal basal gonadotropin levels, otherwise normal pituitary function, normal hypothalamic-pituitary imaging and normal olfaction confirmed by a formal olfactory test (Smell Identification Test Philadelphia, PA or Alcohol Sniff Test, San Diego, CA) (25).
Hormone assays
Serum LH and FSH concentrations were measured by radioimmunoassay (RIA) up to 1991 or immunofluorometric assays (IFMA, AutoDELFIA hLH Spec and AutoDELFIA hFSH, Wallac Oy, Turku, Finland) after 1991. The sensitivity for IFMA was set at 0.1 IU/L for LH and 1.0 IU/L for FSH. Serum T and E2 concentrations were measured by commercial solid phase fluoroimmunoassay (FIA) (AutoDELFIA Testosterone, PerkinElmer, Turku, Finland). The interassay coefficient of variation was 5% or less for all assays. For the acute GnRH stimulation test, serum LH and FSH were measured at −15, 0, 15, 30, 45, and 60 min after intravenous (i.v.) administration of 100 μg GnRH.
DNA analysis
Genomic DNA was extracted from all patients and the three exons of GNRHR (NM_000406.2) were amplified by polymerase chain reaction (PCR) using specific intronic primer pairs (8). PCR products were purified and automatically sequenced in an ABI Prism Genetic Analyzer 3100 (Perkin- Elmer, Foster City, CA, USA) (26).
In silico analysis
Mutation Taster (http://www.mutationtaster.org/), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/) and SIFT - Sorting Intolerant from Tolerant Human Protein (http://sift.jcvi.org/www/SIFT_enst_submit.html) programs were used to predict the putative impact of an amino acid substitution on protein structure and function and to predict the pathogenicity of the missense variants (27).
In vitro analysis
Two distinct GnRHR mutants were generated by site-directed mutagenesis with the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies, Wilmington, DE), using pcDNA3 containing the HA-tagged wild type (WT) human GnRHR as the template. The mutations were verified by direct DNA sequencing (Dana Farber/Harvard Cancer Center Sequencing Facility, Boston, MA).
COS-7 cells were grown and maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum, 100 U/ml of penicillin and 100 μg/ml of streptomycin sulfate in 5% CO2 humidified air at 37°C. The cells were transiently transfected using GenePorter transfection reagent (Gelantis, San Diego, CA) in six-well plates with 100 ng/well of WT, p.V134G, or p.Y283H GnRHR, or empty vector (pcDNA3) as a negative control. To evaluate transfection efficiency, COS-7 cells transfected with WT, V134G, or Y283H GnRHR were co-transfected with a GFP expression vector and DNA extracted from transfected cells were submitted PCR using primers designed based on the GnRHR cDNA sequence (Supplement). Inositol phosphate (IP) assays were performed as previously described (8). In brief, 24 hours after transfection, media on the cells was changed to 1 ml/well inositol-free DMEM containing 1 μCi myo-[2-3H]-inositol (PerkinElmer, Waltham, MA), and the cells were cultured overnight. The next day, cells were pretreated with 10 mM LiCl for 15 minutes and then stimulated with increasing concentrations of GnRH (Sigma-Aldrich, St. Louis, MO) (10−10 to 10−7 M) for 1 hour. After stimulation, the cells were lysed in ice-cold 20 mM formic acid. The lysates were collected and neutralized to pH 7.5 with 300 μl of 7.5mM HEPES containing 150 mM KOH. 3H-IP products were isolated through prepared AG 1-X8 resin (Bio-Rad, Hercules, CA) anion exchange columns by washing with 5 ml H2O, and then 5 ml of solution containing 5mM Borax and 60 mM sodium formate, followed by elution with 3 ml of solution containing 0.9 M ammonium formate and 0.1 M formic acid. The radioactivity in the eluates was measured using a scintillation counter (Beckman, Danvers MA). All assay points were performed in triplicate, and each experiment was repeated at least three times. Statistic analysis was performed by two way ANOVA.
Haplotype mapping
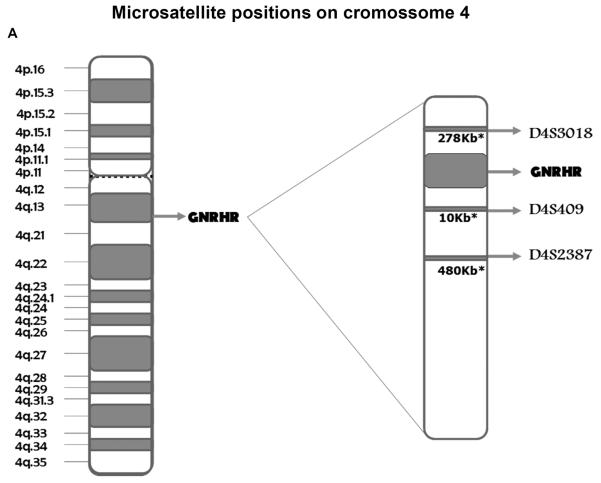
GNRHR haplotype mapping was performed in five unrelated normosmic IHH cases harboring the p.R139H mutation, belonging to four Brazilian pedigrees and one Polish family (8, 12). Family members with available genomic DNA and a control group composed of 50 individuals with normal pubertal development were also studied. However, as the DNA of parents was not available in the Brazilian families, haplotype phasing was not performed. Three microsatellite markers flanking the GNRHR gene were genotyped: D4S3018 (202/316, 278 kb centromeric to the GNRHR gene), D4S409 (278–294, 10 kb telomeric to the GNRHR gene), and D4S2387 (256/259, 480 kb telomeric to the GNRHR gene). The three microsatellite markers were genotyped by Gene Scan technology.
RESULTS
Normosmic Isolated Hypogonadotropic Hypogonadism
Mutational analysis
Nine GNRHR mutations were identified in eleven unrelated individuals with normosmic IHH in either a homozygous or compound heterozygous state (p.N10K, p.Q11K, p.Q106R, p.V134G, p.R139H, p.C200Y, p.R262Q, p.Y283H and p.Y284C) (Table 1). Only one variant, p.Q106R, was identified in the heterozygous state in a male IHH patient. Except for the novel p.V134G and p.Y283H variants, all other mutants have been reported previously and functionally characterized in vitro, leading to varying degrees of GnRH receptor function impairment (5, 6, 8, 9, 11, 28).
Table 1.
Phenotypic and genotypic characterization of normosmic IHH patients carrying GNRHR mutations
| Sex | Age | Phenotype | Familial history | Breast (Tanner) | Testes (cm) | Ovarian (ml) | Penile Length | LH (U/L) | FSH (U/L) | T (ng/dl) | E (pg/ml) | Mutation | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||||||||
| Basal | Peak | Basal | Peak | ||||||||||||
|
| |||||||||||||||
| 1 * | F | 18 | Complete IHH | Affected cousins | I | - | 3 | - | 3.6 | 5.1 | 2.6 | 2.9 | - | 11 | p.[R139H; R139H] |
| 2 | M | 19 | Complete IHH | Consanguineous family, one 4 yr- old nephew with micropenis |
I | 1.0×1.0 | - | 4.0 × 1.0 | <0.6 | <0.6 | <1 | <1 | 33 | - | p.[R139H;R139H] |
| 3 | M | 38 | Complete IHH | Consanguineous parents, 2 affected brothers |
I | 1.0×0.5 | - | 7.5×1.5 | 0.3 | - | - | 1.5 | 30 | - | p.[R139H; R139H] |
| 4 | M | 18 | Partial IHH Reversal |
no | I | 1.5×0.9 | - | 3.5×1.5 | 0.9 | - | 2.6 | - | 30 | - | p.[R139H]; [Q106R] |
| 5 | F | 16 | Complete IHH | Consanguineous parents, one affected sister |
I | - | 1.5 | - | <0.6 | 1.7 | <1 | 2.4 | - | <13 | p.[C200Y; C200Y] |
| 6 * | F | 19 | Partial IHH | Two affected brothers and one affected sister with partial IHH |
III | - | 1.3×1.0 ×0.9 |
- | 9 | 46 | 8.5 | 27 | - | <50 | p.[N10K;Q11K]; [Q106R] |
| 7 | M | 16 | Complete IHH | no | I | 1.5×1.0 | - | 7×2 | 0.17 | - | 0.7 | - | 20 | - | p.[N10K;Q11K]; [Y283H] |
| 8 | M | 17 | Partial IHH | no | III | 2.6×1.5 | - | 8×2 | 2.5 | - | <1 | - | 58 | - | p.[V134G]; [R262Q] |
| 9 | M | 17 | Complete IHH | no | I | 1.5×1.0 | - | 5 | 0.3 | - | 0.7 | - | 110 | - | p.[V134G]; [Q106R] |
| 10 | M | 26 | Partial IHH Reversal |
no, Consanguineous parents |
I | 2.8×1.6 | - | 4 | 0.95 | - | 1.86 | - | - | - | p.[Q106R;Q106R] |
| 11 | M | 18 | Complete IHH | One sister with complete IHH, Consanguineous parents |
I | 2.0×1.5 | - | 10×2.7 | - | - | - | - | - | - | p.[Y284C;Y284C] |
Patients previously described in Costa et al. (10)
F: female; M: male; LH: Luteinizing hormone; FSH: Follicle-stimulating hormone; T: testosterone; E2: estradiol; -: not available. The immunofluorometric method was used in all hormonal analyses except for patients 1 and 6, in whom radioimmunoassays were used. Complete IHH refers to the complete absence of sexual development, whereas incomplete IHH indicates partial pubertal development. Testis size is given when available (normal, 15–25 ml). Breast development is given when available—Tanner 1 indicates no breast buds.
In silico and in vitro analysis of p.V134G and p.Y283H variants
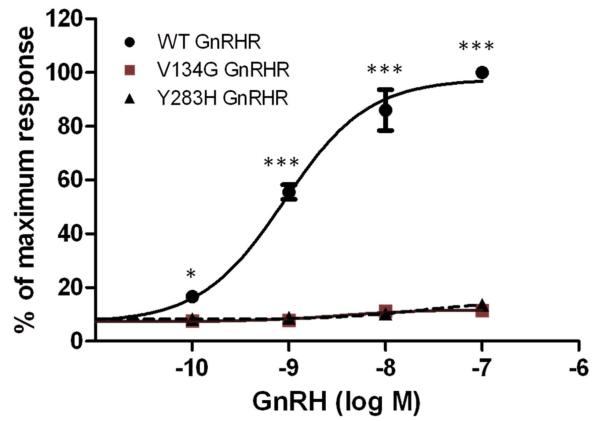
The novel variants, p.V134G and p.Y283H, were absent in the control group. Both affect well-conserved residues and in silico analysis using three different bioinformatic tools suggested that they were deleterious. In order to confirm the effects of the p.V134G and p.Y283H mutations on GnRHR function, in vitro signaling analysis was performed. IP production was measured in COS-7 cells transiently transfected with WT or mutant (p.V134G or p.Y283H) GNRHR or empty vector, and the response to stimulation with increasing concentrations (10−10–10−7 M) of GnRH was measured. The wild type GnRH receptor responded to GnRH stimulation in a dose dependent manner, with an EC50 of 0.75 nM. Dose–response curves did not elicit any response with the mutant receptors, showing complete loss of activity of both the p.Y283 and p. V134G mutants (Figure 1). Cells transfected with empty vector (pcDNA3) as a negative control failed to show any IP induction in response to GnRH treatment (not shown). Levels of GFP expression were similar in cells co-transfected with WT, V134G, or Y283H GnRHR, or empty vector (Suppl. Figure 1). Amplified GnRHR PCR products were detected in cells transfected with WT, V134G or Y283H GnRHR, but not in cells transfected with pcDNA3 (Suppl. Figure 1). These data indicate equivalent transfection efficiency in all cells, excluding variability in transfection efficiency as a factor contributing to the lack of response to GnRH in cells transfected with V134G or Y283H GnRHR.
Figure 1. GnRH stimulation of inositol phosphate (IP) accumulation by wild type (WT), V134G, and Y283H mutant GnRHR.
COS-7 cells were transiently transfected with WT, V134G, or Y283H GnRHR as indicated. GnRH-stimulated IP accumulation was measured following stimulation with increasing concentrations of GnRH. Data points represent the mean ± SEM of triplicate samples and are presented as the percentage of the maximum response. *, P<0.05; ***, P<0.001 compared to WT GnRHR. The results shown are from a representative of three independent experiments.
Founder effect study
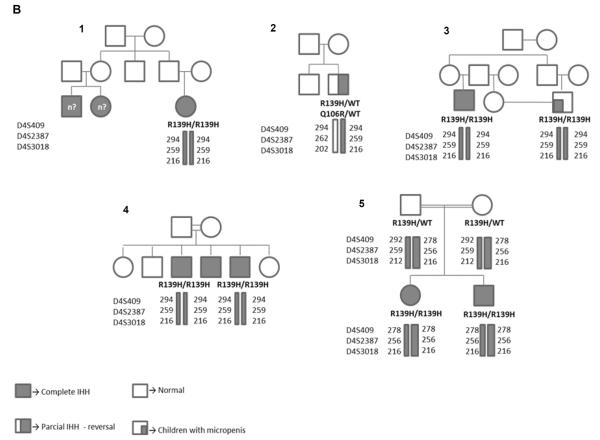
Three microsatellite markers (D4S409, D4S2387, D4S3018) were analyzed in six IHH patients carrying the GNRHR variant encoding the p.R139H mutation, belonging to four families, and in their unaffected relatives. One consanguineous Polish family, with two siblings homozygous for the p.R139H mutation, was also studied (Figure 2). Although haplotype phasing was not performed duet to unavailability of the trios (parents and child), the genotypes detected are consistent with the same haplotype (294, 259, 216) being present in all of the affected Brazilian patients harboring the p.R139H mutation, especially considering that all of them, except one, were homozygous for the same genotypes. In the control population comprised of 150 Brazilian subjects, this haplotype was found in only one subject. On the other hand, the affected Polish family presented with a different haplotype (278,256,216) (Table 2).
Figure 2.
(A) Schematic representation of the location of the microsatellite markers in relation to the GNRHR gene on chromosome 4. *Distance from the marker to GNRHR. (B) Pedigrees of the families carrying the p.R139H mutation and showing haplotypes obtained by microsatellite analysis. Pedigrees A, B, C and D: Brazilian families; Pedigree E: Polish family.
TABLE 2.
Haplotypes of the patients carrying the p.R139H GNRHR mutation
| Origin | Brazil | Brazil | Brazil | Brazil | Poland |
|---|---|---|---|---|---|
|
| |||||
| Family | 1 | 2 | 3 | 4 | 5 |
| Genotype | R139H/R139H | R139H/+ | R139H/R139H | R139H/R139H | R139H/R139H |
| D4S409 | 294/294 | 294/294 | 294/294 | 294/294 | 278/278 |
| D4S2387 | 259/259 | 259/262 | 259/259 | 259/259 | 256/278 |
| D4S3018 | 216/216 | 216/202 | 216/216 | 216/216 | 216/216 |
Clinical characterization and genotype/phenotype correlation
Considering all affected IHH patients, with and without mutations, the mean age at diagnosis was 21.5 yrs. Spontaneous pubarche was observed in 51% of the affected patients. All males had micropenis before treatment (mean penile length: 5.7 ± 1.9 cm (−5.4 ± 1.2 SD)). Unilateral or bilateral cryptorchidism was present in 23%. Median hormone levels were T = 32 ng/dL (range < 14–239), E2 < 13 pg/mL (range < 13–40.2) and LH < 0.6 U/L (< 0.6–3.82). All females had hypoplastic uteri (<3 0 mL) and ovaries (< 3 mL), but 34% had spontaneous thelarche. No phenotypic differences were observed between patients with and without GNRHR mutations. On the other hand, a relatively good genotype-phenotype correlation was observed in the patients with GNRHR mutations (Table 1). The p.R139H and p.C200Y mutations had been shown previously to cause complete loss of function of the receptor in vitro and the p.Y284C severely affects the receptor signaling capacity, with minimal activity retained, whereas the [p.N10K;p.Q11K], p.Q106R, and p.R262Q are partially inactivating mutations (5, 6, 8, 9, 11, 28). Functional analysis of the novel p.Y283H and p.V134G mutations demonstrated that both are completely inactivating. All individuals homozygous for loss-of-function mutations (p.R139H, p.C200Y and p.Y284C) had a phenotype of complete IHH. Patients bearing two partially inactivating mutations had partial IHH, including four siblings (two males and two females) compound heterozygous for the [p.N10K; p.Q11K] and p.Q106R mutations and a male homozygous for the p.Q106R. However in the case of completely inactivating mutations (p.R139H, p.Y283H and p.V134G) identified in the compound heterozygous state together with partially inactivating mutations (p.Q106R, p.R262Q and p.[N10K; Q11K]) the severity of the hypogonadism varied from complete GnRH deficiency to partial IHH with sustained reversal of hypogonadism.
Hypogonadism reversal
Two males with biallelic GNRHR mutations and partial IHH had reversal of hypogonadotropic hypogonadism. IHH reversal was defined as the presence of normal adult testosterone levels after hormone replacement was discontinued for an appropriate washout period (3 to 6 months for testosterone injections), and no symptoms of hypogonadism after cessation of treatment (29, 30). The male patient compound heterozygous for the p.R139H/p.Q106R mutations was on testosterone replacement for 3 years. During routine examination, an increase in testicular volume was noted (3.5 × 2.0 cm), therapy was discontinued, and he had a sustained reversal of hypogonadism, which has been maintained for 4 years. The male homozygous for p.Q106R had spontaneous fertility during testosterone replacement. Further evaluation showed an increase in testicular size (4.0 × 2.5 cm bilaterally) and a sperm count of 17×106/ml. He maintained normal testosterone levels after treatment discontinuation, but hypogonadism relapsed after one year.
Constitutional Delay of Growth and Puberty
Of the 50 CDGP patients studied, 82% were males, 31% had family history of delayed puberty, 94% had short stature (mean height SDS, - 2.9) and none of them had cryptorchidism or micropenis. All had delayed bone age (mean −2.8 yrs) and the pubertal development stage was compatible with the bone age. The p.Q106R GNRHR mutation was identified in the heterozygous state in a boy with CDGP. This partially inactivating mutation has been described previously in patients with autosomal recessive normosmic IHH, either in a homozygous state or in a compound heterozygous state with other variants (5). This boy was first evaluated at 14 years of age due to short stature (height SDS, −2.8) and pubertal delay, with no other associated conditions. LH and T levels were pre-pubertal and bone age was delayed (12.6 yrs). He was born from non-consanguineous parents and denied similar cases in the family. The patient was not treated and attained spontaneous complete pubertal development.
DISCUSSION
In the normosmic IHH group, we described two novel GNRHR mutations, p.V134G and p.Y283H, both completely inactivating, as demonstrated by in vitro functional analysis. The p.V134G was identified in the compound heterozygous state with p.Q106R and p.R262Q in two males with sporadic complete and partial IHH, respectively. The p.Y283H was identified in the compound heterozygous state with the [p.N10K; p.Q11K] mutations in a patient with partial IHH. This is the third time that the p.N10K and p.Q11K mutations have been described together (8, 11). In the case described here, familial segregation showed that the patient's father, with normal sexual development, harbored the [p.N10K; p.Q11K] mutations in the heterozygous state, confirming that these variants were inherited on the same allele, and are not sufficient to cause the IHH phenotype in the heterozygous state. This finding might be ascribed to a founder effect or to the existence of a hotspot region involving these two codons.
In total, nine GNRHR mutations were identified in eleven unrelated individuals with normosmic IHH, resulting in a frequency of 10% of all affected patients. Considering only the familial cases, the frequency of GNRHR mutations increased to 30%. Mutations in other genes associated with the IHH phenotype, including TAC3, TACR3, KISS1, KISS1R, GNRH1, FGF8, FGFR1, PROK2, PROKR2 and WDR11, were present in 17.3% of all cases altogether (data presented elsewhere and not within the scope of the current study) (6). The GNRHR was the most affected gene in the population studied, in accordance with the literature (9, 13, 39). Only one heterozygous p.Q106R GnRHR mutation was identified in a male IHH patient in the present study. Heterozygous GnRHR mutations have been identified in rare sporadic IHH patients, but it is not known if these mutations alone are responsible for the reproductive phenotype (13, 36, 40). In fact, no phenotype has been observed in heterozygote parents or siblings of nIHH patients bearing biallelic inactivating GnRHR mutations (13). To date, all cases of loss-of-function GnRHR mutations reported in the literature, confirmed to cause normosmic IHH, were transmitted as an autosomic recessive trait (13). This recessive transmission confirms the genetic model observed in Gnrhr knockout mice (41). Taken together, these findings suggest that heterozygous GnRHR mutations are not sufficient to cause a reproductive phenotype and it is likely that additional variants in other genes contribute to disease pathogenesis in IHH patients heterozygous for GnRHR mutations. Indeed, mutations in more than one gene accounting for the IHH phenotype have been reported in 11% of patients, characterizing a digenic or oligogenic inheritance (36, 42). However, no digenic defects were identified in our cohort of normosmic IHH patients.
Examining the severity of GnRHR functional impairment caused by the mutations and the spectrum of reproductive phenotypes of the affected individuals, a relatively good genotype-phenotype correlation was observed. All individuals homozygous for complete loss-of-function mutations (p.R139H and p.C200Y) or with severe impairment of receptor function (p.Y284C) had a phenotype of complete normosmic IHH. Patients bearing two partially inactivating mutations had a milder GnRH deficiency phenotype. However, this correlation was not so clear in individuals bearing a combination of a complete and a partially inactivating mutation. These patients presented with a variable phenotypic spectrum, ranging from complete GnRH deficiency to partial and reversible IHH, suggesting that the phenotype depends on the interaction between the mutant receptors (43). Other authors have observed a similar correlation in individuals with biallelic GnRHR mutations (13, 44).
It is noteworthy that two males harboring the p.Q106R, one homozygous and the other compound heterozygous with the inactivating p.R139H, presented with reversal of GnRH deficiency, denoted by an increase in testicular volume during androgen replacement and maintenance of normal gonadotropins and T values after interruption of treatment. The homozygous p.Q106R patient presented with spontaneous fertility, but later had a relapse of hypogonadism. Interestingly, Pitteloud et al. (31) described a 26-yr-old male homozygous for the p.Q106R mutation, with a mild phenotypic form of IHH, the fertile eunuch syndrome (IHH in the presence of normal testicular size and some degree of spermatogenesis). Reversibility has been described in 10% of the IHH cases after sex steroid replacement, especially in patients with the partial IHH phenotype. However, in some patients the reversal was not sustained indefinitely after discontinuation of therapy (29, 30, 45, 46). As the reported incidence of reversal is relatively high, we suggest temporary discontinuation of hormonal therapy to assess reversibility of hypogonadism, particularly if spontaneous testicular growth occurred during androgen therapy. Nevertheless, even in cases of reversal, follow-up needs to be continued due to the possibility of relapse. The mechanism of reversal is not known, and it has been suggested that androgen exposure may up-regulate the neuroendocrine regulation of GnRH secretion (29, 45).
The p.R139H mutation is particularly common in the Brazilian population. It was first described in 2001 (8) and in five other patients belonging to three unrelated Brazilian families in the present study. A Polish family carrying the p.R139H was included in this study, given that there was considerable Polish immigration to Brazil in 19th century and early in the 20th century. For the haplotype mapping analysis three microsatellite markers flanking the GNRHR gene were genotyped. Although a larger number of markers could be more informative, these were selected for being the closest to the GNRHR gene, decreasing the possibility of recombination, and the results were consistent with all affected Brazilian patients with the p.R139H mutation sharing the same haplotype, suggesting inheritance from a common ancestor. Although these families were not known to be related, a detailed interview revealed that all of them had family members originating from the same region in the Northeast of Brazil (Itabaiana-SE), consistent with the hypothesis of a common ancestor. Conversely, the affected Polish family had a different haplotype (278, 256, 216), with only the closest microsatellite marker to the GNRHR (D4S3018 - 216) in common with the Brazilian patients. Indeed, most of the Polish population settled in Southern Brazil, and not in the Northeast area. Nevertheless, the possibility of a more remote common ancestry cannot be totally discarded. The p.R139H has also been reported in five other unrelated cases of different geographic origins worldwide, which were not evaluated here, and further studies would be necessary to determine if they too have a common ancestor (12, 30, 44, 47, 48). On the other hand, another mutation affecting the same codon (p.R139C) was described in two cases, raising the possibility that this may be a hotspot region (48, 49).
Genes involved in the pathogenesis of IHH, including GNRHR, have been associated with a broad phenotypic spectrum, from complete IHH to more subtle phenotypes, such as the fertile eunuch syndrome, IHH reversal and hypothalamic amenorrhea (31–35). In the hypothesis that CDGP might represent a mild phenotypic variation of normosmic IHH, we investigated the presence of GNRHR rare variants in this group of patients and identified a boy heterozygous for the p.Q106R mutation. Vaaralahti et al. in 2011 (33) studied 146 individuals with CDGP and also identified one female heterozygous for the same variant. This partially inactivating mutation has been described previously and in the present study in patients with normosmic IHH in the homozygous or compound heterozygous state (5, 33). It has also been identified in heterozygous state in unaffected patients, parents, and siblings, and in control individuals with a similar frequency as in CDGP and IHH patients (13, 33, 36). Considering these results, it is very unlikely that the p.Q106R mutation is involved in the pathogenesis of CDGP. Other authors have screened CDGP patients for GnRHR variants, but no consistent alteration was found (37, 38). Overall, these data suggest a distinct molecular etiology between normosmic IHH and CDGP.
In conclusion, although there are no particular phenotypic characteristics associated with GnRHR defects, GNRHR is one of the most commonly affected genes in normosmic IHH and should be the first candidate for genetic analysis, especially in autosomal recessive familial cases. GNRHR mutations are associated with a broad phenotypic spectrum of normosmic IHH, from complete GnRH deficiency to a partial and reversible phenotype, with a relatively good genotype-phenotype correlation. However, GNRHR defects do not seem to be involved in the pathogenesis of CDGP, and this appears to be a condition with a distinct molecular mechanism, rather than a milder form of IHH.
Supplementary Material
Acknowledgments
This work was supported by FAPESP grant 2011/15530-0 to D.B., by the Eunice Kennedy Shriver National Institute of Child Health and Human Development through cooperative agreement U54HD028138 as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research and R01HD019938 to U.B.K., and by T32 DK007529 and K08 HD070957 to L.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors have no conflict of interest
REFERENCES
- 1.Palmert MR, Boepple PA. Variation in the timing of puberty: clinical spectrum and genetic investigation. J Clin Endocrinol Metab. 2001;86(6):2364–8. doi: 10.1210/jcem.86.6.7603. [DOI] [PubMed] [Google Scholar]
- 2.Palmert MR, Dunkel L. Clinical practice. Delayed puberty. N Engl J Med. 2012;366(5):443–53. doi: 10.1056/NEJMcp1109290. Epub 2012/02/03. [DOI] [PubMed] [Google Scholar]
- 3.Seminara SB, Hayes FJ, Crowley WF., Jr. Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann's syndrome): pathophysiological and genetic considerations. Endocr Rev. 1998;19(5):521–39. doi: 10.1210/edrv.19.5.0344. [DOI] [PubMed] [Google Scholar]
- 4.Quinton R, Duke VM, Robertson A, Kirk JM, Matfin G, de Zoysa PA, et al. Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin Endocrinol (Oxf) 2001;55(2):163–74. doi: 10.1046/j.1365-2265.2001.01277.x. [DOI] [PubMed] [Google Scholar]
- 5.de Roux N, Young J, Misrahi M, Genet R, Chanson P, Schaison G, et al. A family with hypogonadotropic hypogonadism and mutations in the gonadotropin-releasing hormone receptor. N Engl J Med. 1997;337(22):1597–602. doi: 10.1056/NEJM199711273372205. [DOI] [PubMed] [Google Scholar]
- 6.Layman LC, Cohen DP, Jin M, Xie J, Li Z, Reindollar RH, et al. Mutations in gonadotropin-releasing hormone receptor gene cause hypogonadotropic hypogonadism. Nat Genet. 1998;18(1):14–5. doi: 10.1038/ng0198-14. [DOI] [PubMed] [Google Scholar]
- 7.Kottler ML, Chauvin S, Lahlou N, Harris CE, Johnston CJ, Lagarde JP, et al. A new compound heterozygous mutation of the gonadotropin-releasing hormone receptor (L314X, Q106R) in a woman with complete hypogonadotropic hypogonadism: chronic estrogen administration amplifies the gonadotropin defect. J Clin Endocrinol Metab. 2000;85(9):3002–8. doi: 10.1210/jcem.85.9.6783. [DOI] [PubMed] [Google Scholar]
- 8.Costa EM, Bedecarrats GY, Mendonca BB, Arnhold IJ, Kaiser UB, Latronico AC. Two novel mutations in the gonadotropin-releasing hormone receptor gene in Brazilian patients with hypogonadotropic hypogonadism and normal olfaction. J Clin Endocrinol Metab. 2001;86(6):2680–6. doi: 10.1210/jcem.86.6.7551. [DOI] [PubMed] [Google Scholar]
- 9.Beranova M, Oliveira LM, Bedecarrats GY, Schipani E, Vallejo M, Ammini AC, et al. Prevalence, phenotypic spectrum, and modes of inheritance of gonadotropin-releasing hormone receptor mutations in idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2001;86(4):1580–8. doi: 10.1210/jcem.86.4.7395. [DOI] [PubMed] [Google Scholar]
- 10.Silveira LF, Stewart PM, Thomas M, Clark DA, Bouloux PM, MacColl GS. Novel homozygous splice acceptor site GnRH receptor (GnRHR) mutation: human GnRHR “knockout”. J Clin Endocrinol Metab. 2002;87(6):2973–7. doi: 10.1210/jcem.87.6.8535. Epub 2002/06/07. [DOI] [PubMed] [Google Scholar]
- 11.Meysing AU, Kanasaki H, Bedecarrats GY, Acierno JS, Jr., Conn PM, Martin KA, et al. GNRHR mutations in a woman with idiopathic hypogonadotropic hypogonadism highlight the differential sensitivity of luteinizing hormone and follicle-stimulating hormone to gonadotropin-releasing hormone. J Clin Endocrinol Metab. 2004;89(7):3189–98. doi: 10.1210/jc.2003-031808. [DOI] [PubMed] [Google Scholar]
- 12.Fichna P, Fichna M, Zurawek M, Nowak J. Hypogonadotropic hypogonadism due to GnRH receptor mutation in a sibling. Endokrynol Pol. 2011;62(3):264–7. Epub 2011/07/01. [PubMed] [Google Scholar]
- 13.Chevrier L, Guimiot F, de Roux N. GnRH receptor mutations in isolated gonadotropic deficiency. Mol Cell Endocrinol. 2011;346(1–2):21–8. doi: 10.1016/j.mce.2011.04.018. Epub 2011/06/08. [DOI] [PubMed] [Google Scholar]
- 14.Tello JA, Newton CL, Bouligand J, Guiochon-Mantel A, Millar RP, Young J. Congenital hypogonadotropic hypogonadism due to GnRH receptor mutations in three brothers reveal sites affecting conformation and coupling. PLoS One. 2012;7(6):e38456. doi: 10.1371/journal.pone.0038456. Epub 2012/06/09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silveira LF, Trarbach EB, Latronico AC. Genetics basis for GnRH-dependent pubertal disorders in humans. Mol Cell Endocrinol. 2010;324(1–2):30–8. doi: 10.1016/j.mce.2010.02.023. Epub 2010/03/02. [DOI] [PubMed] [Google Scholar]
- 16.Gajdos ZK, Henderson KD, Hirschhorn JN, Palmert MR. Genetic determinants of pubertal timing in the general population. Mol Cell Endocrinol. 2010;324(1–2):21–9. doi: 10.1016/j.mce.2010.01.038. Epub 2010/02/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trarbach EB, Costa EM, Versiani B, de Castro M, Baptista MT, Garmes HM, et al. Novel fibroblast growth factor receptor 1 mutations in patients with congenital hypogonadotropic hypogonadism with and without anosmia. J Clin Endocrinol Metab. 2006;91(10):4006–12. doi: 10.1210/jc.2005-2793. [DOI] [PubMed] [Google Scholar]
- 18.Abreu AP, Trarbach EB, de Castro M, Frade Costa EM, Versiani B, Matias Baptista MT, et al. Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive Kallmann syndrome. J Clin Endocrinol Metab. 2008;93(10):4113–8. doi: 10.1210/jc.2008-0958. Epub 2008/08/07. [DOI] [PubMed] [Google Scholar]
- 19.Trarbach EB, Teles MG, Costa EM, Abreu AP, Garmes HM, Guerra G, Jr, et al. Screening of autosomal gene deletions in patients with hypogonadotropic hypogonadism using multiplex ligation-dependent probe amplification: detection of a hemizygosis for the fibroblast growth factor receptor 1. Clin Endocrinol (Oxf) 2009 doi: 10.1111/j.1365-2265.2009.03642.x. Epub 2009/06/06. [DOI] [PubMed] [Google Scholar]
- 20.Silveira LG, Noel SD, Silveira-Neto AP, Abreu AP, Brito VN, Santos MG, et al. Mutations of the KISS1 gene in disorders of puberty. J Clin Endocrinol Metab. 2010;95(5):2276–80. doi: 10.1210/jc.2009-2421. Epub 2010/03/20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teles M, Trarbach E, Sekoni N, Guerra-Junior G, Jorge A, Beneduzzi D, et al. A novel homozygous splice acceptor site mutation of KISS1R in two siblings with normosmic isolated hypogonadotropic hypogonadism. Eur J Endocrinol. 2010 doi: 10.1530/EJE-10-0012. Epub 2010/04/08. [DOI] [PubMed] [Google Scholar]
- 22.Trarbach EB, Abreu AP, Silveira LF, Garmes HM, Baptista MT, Teles MG, et al. Nonsense mutations in FGF8 gene causing different degrees of human gonadotropin-releasing deficiency. J Clin Endocrinol Metab. 2010;95(7):3491–6. doi: 10.1210/jc.2010-0176. Epub 2010/05/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gianetti E, Tusset C, Noel SD, Au MG, Dwyer AA, Hughes VA, et al. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab. 2010;95(6):2857–67. doi: 10.1210/jc.2009-2320. Epub 2010/03/25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tusset C, Noel SD, Trarbach EB, Silveira LF, Jorge AA, Brito VN, et al. Mutational analysis of TAC3 and TACR3 genes in patients with idiopathic central pubertal disorders. Arq Bras Endocrinol Metabol. 2012;56(9):646–52. doi: 10.1590/s0004-27302012000900008. Epub 2013/01/19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson TM, Freed C, Healy MP, Murphy C. Rapid clinical evaluation of anosmia in children: the Alcohol Sniff Test. Ann N Y Acad Sci. 1998;855:787–92. doi: 10.1111/j.1749-6632.1998.tb10659.x. Epub 1999/02/04. [DOI] [PubMed] [Google Scholar]
- 26.Beneduzzi D, Trarbach EB, Latronico AC, Mendonca BB, Silveira LF. Novel mutation in the gonadotropin-releasing hormone receptor (GNRHR) gene in a patient with normosmic isolated hypogonadotropic hypogonadism. Arq Bras Endocrinol Metabol. 2012;56(8):540–4. doi: 10.1590/s0004-27302012000800013. Epub 2013/01/09. [DOI] [PubMed] [Google Scholar]
- 27.Frousios K, Iliopoulos CS, Schlitt T, Simpson MA. Predicting the functional consequences of non-synonymous DNA sequence variants - evaluation of bioinformatics tools and development of a consensus strategy. Genomics. 2013 doi: 10.1016/j.ygeno.2013.06.005. Epub 2013/07/09. [DOI] [PubMed] [Google Scholar]
- 28.Bedecarrats GY, Linher KD, Janovick JA, Beranova M, Kada F, Seminara SB, et al. Four naturally occurring mutations in the human GnRH receptor affect ligand binding and receptor function. Mol Cell Endocrinol. 2003;205(1–2):51–64. doi: 10.1016/s0303-7207(03)00201-6. Epub 2003/08/02. [DOI] [PubMed] [Google Scholar]
- 29.Raivio T, Falardeau J, Dwyer A, Quinton R, Hayes FJ, Hughes VA, et al. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med. 2007;357(9):863–73. doi: 10.1056/NEJMoa066494. Epub 2007/09/01. [DOI] [PubMed] [Google Scholar]
- 30.Laitinen EM, Tommiska J, Sane T, Vaaralahti K, Toppari J, Raivio T. Reversible congenital hypogonadotropic hypogonadism in patients with CHD7, FGFR1 or GNRHR mutations. PLoS One. 2012;7(6):e39450. doi: 10.1371/journal.pone.0039450. Epub 2012/06/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitteloud N, Boepple PA, DeCruz S, Valkenburgh SB, Crowley WF, Jr., Hayes FJ. The fertile eunuch variant of idiopathic hypogonadotropic hypogonadism: spontaneous reversal associated with a homozygous mutation in the gonadotropin-releasing hormone receptor. J Clin Endocrinol Metab. 2001;86(6):2470–5. doi: 10.1210/jcem.86.6.7542. [DOI] [PubMed] [Google Scholar]
- 32.Lin L, Conway GS, Hill NR, Dattani MT, Hindmarsh PC, Achermann JC. A homozygous R262Q mutation in the gonadotropin-releasing hormone receptor presenting as constitutional delay of growth and puberty with subsequent borderline oligospermia. J Clin Endocrinol Metab. 2006;91(12):5117–21. doi: 10.1210/jc.2006-0807. Epub 2006/09/14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaaralahti K, Wehkalampi K, Tommiska J, Laitinen EM, Dunkel L, Raivio T. The role of gene defects underlying isolated hypogonadotropic hypogonadism in patients with constitutional delay of growth and puberty. Fertil Steril. 2011;95(8):2756–8. doi: 10.1016/j.fertnstert.2010.12.059. Epub 2011/02/05. [DOI] [PubMed] [Google Scholar]
- 34.Caronia LM, Martin C, Welt CK, Sykiotis GP, Quinton R, Thambundit A, et al. A genetic basis for functional hypothalamic amenorrhea. N Engl J Med. 2011;364(3):215–25. doi: 10.1056/NEJMoa0911064. Epub 2011/01/21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tommiska J, Jorgensen N, Christiansen P, Juul A, Raivio T. A homozygous R262Q mutation in the gonadotropin-releasing hormone receptor presenting as reversal of hypogonadotropic hypogonadism and late-onset hypogonadism. Clin Endocrinol (Oxf) 2013;78(2):316–7. doi: 10.1111/j.1365-2265.2012.04493.x. Epub 2012/07/14. [DOI] [PubMed] [Google Scholar]
- 36.Sykiotis GP, Plummer L, Hughes VA, Au M, Durrani S, Nayak-Young S, et al. Oligogenic basis of isolated gonadotropin-releasing hormone deficiency. Proc Natl Acad Sci U S A. 2010;107(34):15140–4. doi: 10.1073/pnas.1009622107. Epub 2010/08/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin L, Conway GS, Hill NR, Dattani MT, Hindmarsh PC, Achermann JC. A homozygous R262Q mutation in the gonadotropin-releasing hormone receptor (GNRHR) presenting as constitutional delay of growth and puberty with subsequent borderline oligospermia. J Clin Endocrinol Metab. 2006 doi: 10.1210/jc.2006-0807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sedlmeyer IL, Pearce CL, Trueman JA, Butler JL, Bersaglieri T, Read AP, et al. Determination of sequence variation and haplotype structure for the gonadotropin-releasing hormone (GnRH) and GnRH receptor genes: investigation of role in pubertal timing. J Clin Endocrinol Metab. 2005;90(2):1091–9. doi: 10.1210/jc.2004-0649. Epub 2004/11/18. [DOI] [PubMed] [Google Scholar]
- 39.Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009;5(10):569–76. doi: 10.1038/nrendo.2009.177. Epub 2009/08/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vagenakis GA, Sgourou A, Papachatzopoulou A, Kourounis G, Papavassiliou AG, Georgopoulos NA. The gonadotropin-releasing hormone (GnRH)-1 gene, the GnRH receptor gene, and their promoters in patients with idiopathic hypogonadotropic hypogonadism with or without resistance to GnRH action. Fertil Steril. 2005;84(6):1762–5. doi: 10.1016/j.fertnstert.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 41.Wu S, Wilson MD, Busby ER, Isaac ER, Sherwood NM. Disruption of the single copy gonadotropin-releasing hormone receptor in mice by gene trap: severe reduction of reproductive organs and functions in developing and adult mice. Endocrinology. 2010;151(3):1142–52. doi: 10.1210/en.2009-0598. Epub 2010/01/14. [DOI] [PubMed] [Google Scholar]
- 42.Quaynor SD, Kim HG, Cappello EM, Williams T, Chorich LP, Bick DP, et al. The prevalence of digenic mutations in patients with normosmic hypogonadotropic hypogonadism and Kallmann syndrome. Fertil Steril. 2011;96(6):1424–30. e6. doi: 10.1016/j.fertnstert.2011.09.046. Epub 2011/11/01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leanos-Miranda A, Ulloa-Aguirre A, Janovick JA, Conn PM. In vitro coexpression and pharmacological rescue of mutant gonadotropin-releasing hormone receptors causing hypogonadotropic hypogonadism in humans expressing compound heterozygous alleles. J Clin Endocrinol Metab. 2005;90(5):3001–8. doi: 10.1210/jc.2004-2071. Epub 2005/02/25. [DOI] [PubMed] [Google Scholar]
- 44.Gianetti E, Hall JE, Au MG, Kaiser UB, Quinton R, Stewart JA, et al. When genetic load does not correlate with phenotypic spectrum: lessons from the GnRH receptor (GNRHR) J Clin Endocrinol Metab. 2012;97(9):E1798–807. doi: 10.1210/jc.2012-1264. Epub 2012/06/30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sidhoum VF, Chan YM, Lippincott MF, Balasubramanian R, Quinton R, Plummer L, et al. Reversal and Relapse of Hypogonadotropic Hypogonadism: Resilience and Fragility of the Reproductive Neuroendocrine System. J Clin Endocrinol Metab. 2013:jc20132809. doi: 10.1210/jc.2013-2809. Epub 2014/01/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santhakumar A, Balasubramaniam R, Miller M, Quinton R. Reversal of Isolated Hypogonadotropic Hypogonadism: long-term integrity of hypothalamo-pituitary-testicular axis in two men is dependent on intermittent androgen exposure. Clin Endocrinol (Oxf) 2013 doi: 10.1111/cen.12347. Epub 2013/10/15. [DOI] [PubMed] [Google Scholar]
- 47.Wolczynski S, Laudanski P, Jarzabek K, Mittre H, Lagarde JP, Kottler ML. A case of complete hypogonadotropic hypogonadism with a mutation in the gonadotropin-releasing hormone receptor gene. Fertil Steril. 2003;79(2):442–4. doi: 10.1016/s0015-0282(02)04667-8. Epub 2003/02/06. [DOI] [PubMed] [Google Scholar]
- 48.Gurbuz F, Kotan LD, Mengen E, Siklar Z, Berberoglu M, Dokmetas S, et al. Distribution of gene mutations associated with familial normosmic idiopathic hypogonadotropic hypogonadism. J Clin Res Pediatr Endocrinol. 2012;4(3):121–6. doi: 10.4274/Jcrpe.725. Epub 2012/07/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Topaloglu AK, Lu ZL, Farooqi IS, Mungan NO, Yuksel B, O'Rahilly S, et al. Molecular genetic analysis of normosmic hypogonadotropic hypogonadism in a Turkish population: identification and detailed functional characterization of a novel mutation in the gonadotropin-releasing hormone receptor gene. Neuroendocrinology. 2006;84(5):301–8. doi: 10.1159/000098147. Epub 2006/12/21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.