Introduction
Proteolytic enzymes are ubiquitous in all organisms and constitute 2–4% of the encoded gene products. They are critical for diverse biological processes such as digestion, blood clotting, host defense, pathogenic infection, viral replication, wound healing, and disease progression, to name a few. Because proteases trigger an irreversible event - the cleavage of a protein - their activity must be tightly controlled. Dysregulated proteolytic activity causes a disruption in the homeostatic balance of a biological system and can result in any number of poor biological outcomes. As a result, nature has developed a number of strategies to control proteolysis, including spatial and temporal regulation, zymogen activation and protease degradation, and through the inhibition of proteases by macromolecular inhibitors. Somewhat surprisingly, relatively few design principles underlie the mechanisms of inhibition of a myriad range of macromolecular protease inhibitors. Significant engineering efforts have gone into modifying and improving inhibitor potency and specificity, and to a large extent, the same design principles that work well for naturally occurring protease inhibitors have proved valuable for inhibitors developed in the laboratory.
This review aims to survey the mechanisms by which macromolecular protease inhibitors function. To do this, inhibitors have been divided into categories based on their mechanism in order to illustrate that a relatively small number of design principles can be combined to develop new and effective protease inhibitors. These divisions are not strict, and many inhibitors could be grouped in a number of classes. The list of mechanisms presented here is not exhaustive in its treatment of all inhibitors, but aims to be illustrative of the many ways proteases can be inhibited. For more information on genome-wide protease mining,[1] protease mechanism,[2] pre-clinical inhibition,[3] and drug discovery efforts,[4] the reader is directed to excellent reviews that have been written in recent years. Figure 1 provides an overview of basic substrate and protease nomenclature that will be used in this review.
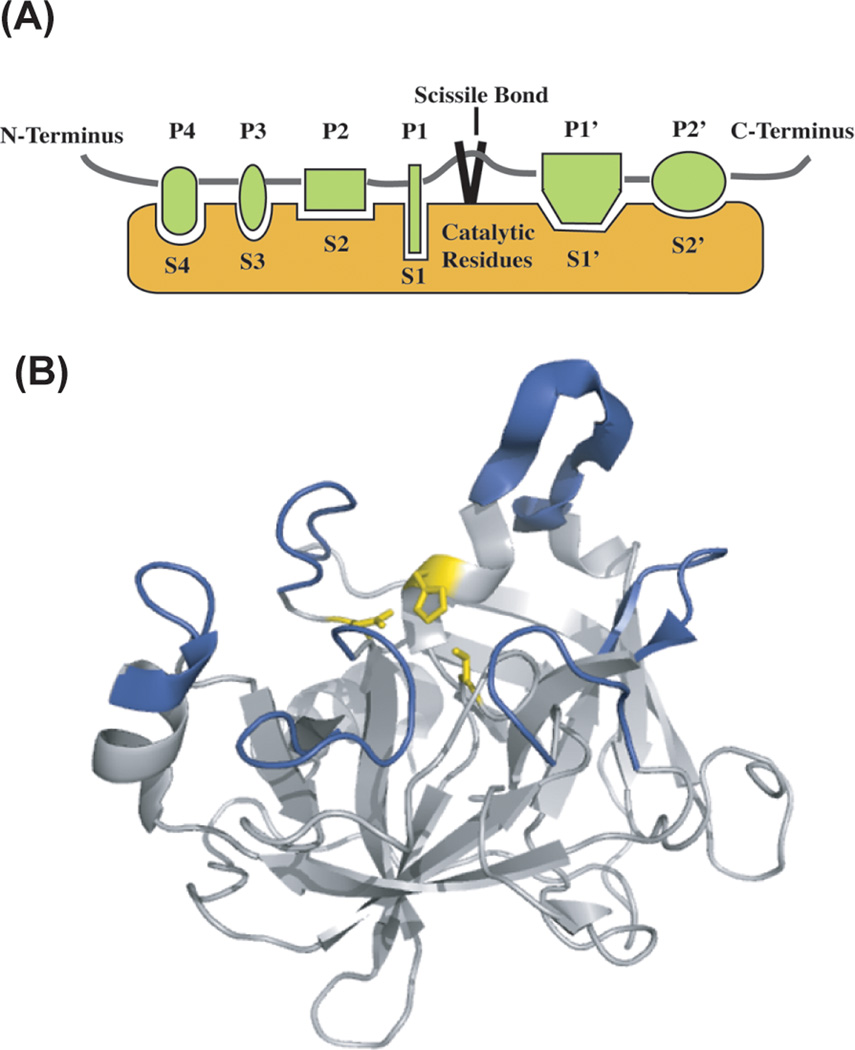
Figure 1.
(A) Diagram of a protease active site. A protease cleaves a peptide at the scissile bond, and has a number of specificity subsites, which determine protease specificity. Substrates bind to a protease with their non-prime residues on the N-terminal side of the scissile bond and their prime-side residues C-terminal to the scissile bond. The catalytic residues determine the class of protease. Serine, cysteine, and threonine proteases hydrolyze a peptide bond via a covalent acyl-enzyme intermediate, and aspartic, glutamic and metalloproteases activate a water molecule to hydrolyze the peptide bond in a non-covalent manner. (B) A serine protease (matriptase/MT-SP1, 1EAX.pdb) with the catalytic triad in yellow and the surface loops that surround the active site colored in blue. While the catalytic architecture of proteases is remarkably conserved, the surface loops are areas of high sequential and structural diversity.
Competitive Inhibitors
The vast majority of protease inhibitors are competitive inhibitors. Despite divergent targets and different mechanisms of inhibition, most protease inhibitors bind a critical portion of the inhibitor in the active site in a substrate-like manner (Figure 2). This is an effective paradigm for potent inhibition, but because related proteases often show a high degree of homology in the active site, substrate-like binding often leads to inhibitors that can potently inhibit more than one target protease. This inhibitor promiscuity is evidenced by the fact that there are 115 annotated human protease inhibitors responsible for regulating the activity of the 612 known human proteases. Though these numbers will change as more refinement of protease and inhibitor families are achieved, the ratio of approximately one protease inhibitor to five proteases is likely to remain constant.[5]
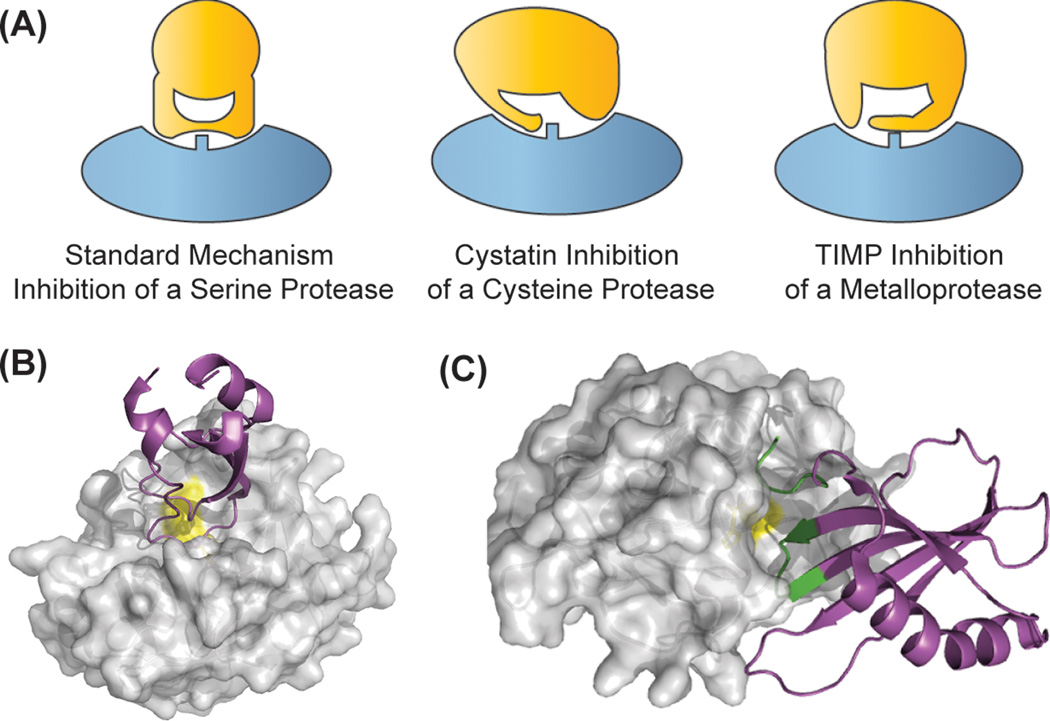
Figure 2.
Competitive, active site inhibitors of proteases. (A) Inhibitors bind in the active site, but not in a substrate-like manner. Peptide extensions bind in specificity subsites, and can interact with the catalytic residues (rectangle). Crystal structures of (B) a serine protease (matriptase/MT-SP1, 1EAW.pdb) in complex with the standard mechanism inhibitor aprotinin, and (C) the cystatin stefin A in complex with a cysteine protease (cathepsin H, 1NB5.pdb). The portion of stefin A that interacts with the protease is colored in green. Both inhibitors bind in the active site groove of their targets.
The most thoroughly studied mechanism of protein protease inhibitors is that of the standard mechanism (or Canonical, or Laskowski mechanism) inhibitors of serine proteases.[6] These inhibitors include the Kazal, Kunitz, and Bowman-Birk family of inhibitors and bind in a lock-and-key fashion. Standard mechanism inhibitors insert a reactive loop into the active site of the protease, which is complementary to the substrate specificity of the target protease and binds in an extended β-sheet with the enzyme in a substrate-like manner. While bound to the protease, the “scissile bond” of standard mechanism inhibitors is hydrolyzed very slowly, but products are not released, and the amide bond can be re-ligated.[7,8] The standard mechanism is an efficient way to inhibit serine proteases, and is thus used by many structurally disparate protein scaffolds to create potent inhibitors. However, the majority of standard mechanism protease inhibitors tend to have relatively broad specificity within sub-classes of serine proteases. For example, bovine pancreatic trypsin inhibitor (BPTI) efficiently inhibits almost all trypsin-fold serine proteases with P1-Arg specificity with sub-nanomolar potency, and can also potently inhibit chymotrypsin (Phe P1 specificity) with a KI of 10 nM[9] (Figure 2B).
The majority of protease inhibitors bind in and block access to the active site of their target protease, but do not bind in a strictly substrate-like manner. Instead they interact with the protease subsites and catalytic residues in a non-catalytically competent manner. This differentiates them from standard mechanism inhibitors, but like standard mechanism inhibitors, they get most of their potency from interactions within the protease active site, and tend to potently inhibit many related proteases.
The cystatins, a superfamily of proteins that inhibit papain-like cysteine proteases, are a classic example of these inhibitors (Figure 2A, 2C). The cystatins insert a wedge-like face of the inhibitor consisting of the protein N-terminus and two hairpin loops into the V-shaped active site of a cysteine protease. The N-terminal residues bind in the S3-S1 pockets in a substrate-like manner, but the peptide then turns away from the catalytic residues and out of the active site. The two hairpin loops bind to the prime-side of the active site, which provides the majority of the binding energy for the interaction. Thus, both the prime and non-prime sides of the active site are occupied, but no interactions are actually made with the catalytic machinery of the enzyme.[10]
The four human tissue inhibitors of metalloproteases (TIMPs) are responsible for inhibition of dozens of extracellular metalloproteases (Figure 2B). They bind to their target enzymes in a two-step mechanism similar to that of cystatins. While the N-terminal residues of cystatins bind to the non-prime side of cysteine proteases, TIMPs N-termini bind in the P1–P3’ pockets of the protease, coordinate the catalytic Zn2+ ion, and exclude a catalytic water molecule from the active site. Meanwhile a second loop of the TIMP binds both in the P3 and P2 pockets, and binds to the N-terminus of the MMP. Despite the similarities in mechanistic architecture between TIMPs and cystatins (hairpin loops and N-terminal residues in substrate binding pockets), TIMPs interfere with the catalytic machinery of MMPs by chelating the catalytic Zn2+.[11]
Competitive Inhibition with Exosite Binding
A number of protease inhibitors are competitive, and bind in the protease active site, but also have secondary binding sites outside the active site, which are critical to inhibition. Exosite binding provides two major benefits; it increases the surface area of the protein-protein interaction, leading to a greater affinity, and it can have a significant effect on the specificity of the inhibitor.
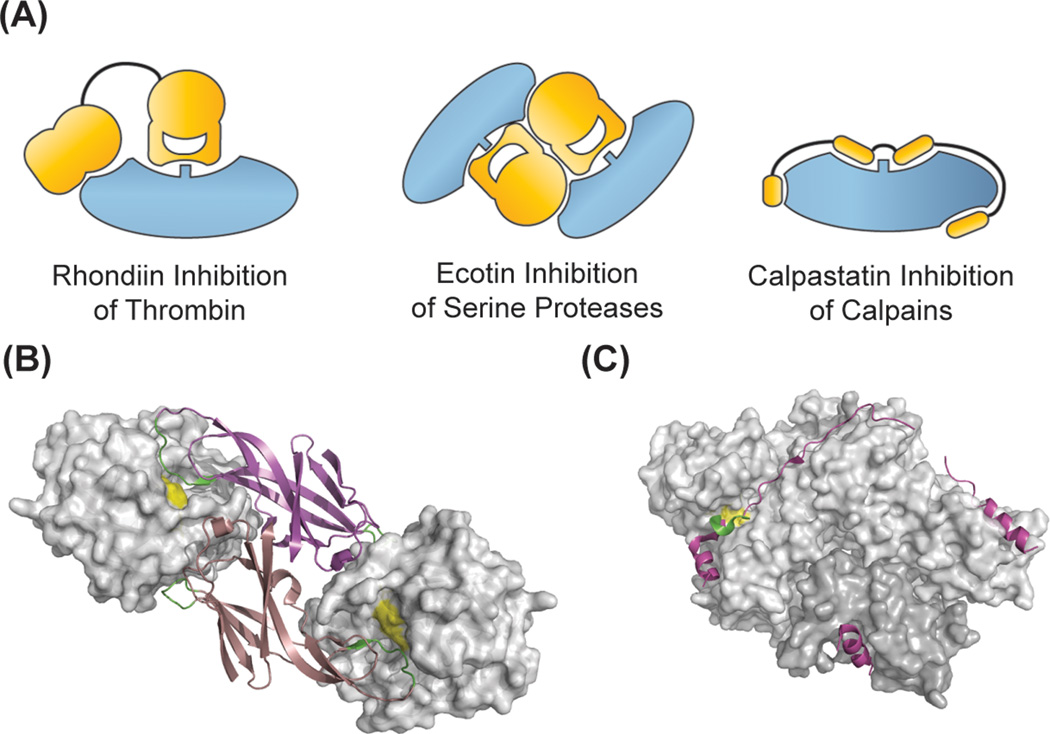
Many blood-meal parasites have evolved protease inhibitors that take advantage of exosites to prevent host blood clotting. These inhibitors often use similar competitive inhibitory mechanisms as described above, but have domains that bind to protease exosites and provide a high degree of target specificity. Rhodniin, a thrombin inhibitor from the assassin bug Rhodnius prolixus has two Kazal-type inhibitory domains, a common standard mechanism serine protease inhibitor domain (Figure 3A). While the N-terminal domain binds and inhibits via the standard mechanism, the second Kazal-type domain has evolved to bind to exosite I on thrombin. The binding affinities of the individual domains are roughly additive, and the resultant inhibitor has a KI of 0.2 pM and exquisite specificity for thrombin.[12] Therefore, the inhibitor gains both potency and specificity from exosite binding.
Figure 3.
Inhibitors that take advantage of exosite binding. Most exosite inhibitors are competitive inhibitors that prevent substrate binding at the active site. In the case of (B) ecotin (bound to trypsin, 1EZU.pdb), the exosites provide binding energy and allow for broad specificity, while (C) calpastatin gains binding energy and specificity by forming critical interactions across the calpain protease surface (3BOW.pdb).
In contrast, the E. coli serine protease inhibitor ecotin uses its exosites to provide binding energy and actually broaden the inhibitor promiscuity and protect the bacteria from host proteases (Figure 3A, 3B). Ecotin is a dimeric protein that inhibits trypsin fold serine proteases regardless of their primary specificity. It functions as a dimer, and inhibits serine proteases through a standard mechanism at a primary binding site, but it also has a secondary binding site that can contribute up to 5 kcal/mol of binding energy to the very tight enzyme-inhibitor complex. Surprisingly, the individual binding energies of the two binding sites are not additive; the effect of the secondary binding site on affinity was found to be inversely proportional to the strength of binding at the primary site. The secondary binding site seems to provide compensatory effects that can overcome sub-optimal binding at the primary binding site; if binding at the primary site is not optimal, the secondary binding interaction tends to be stronger. In this way, the exosite actually makes the inhibitor less specific, or more capable of inhibiting a broad range of proteases, and allows one bacterial inhibitor to protect against a number of host proteases.[13]
The recent structures of the calcium-dependent protease calpain 2 in complex with the inhibitory domain CAST4 of calpastatin reveal another unique use of binding sites outside the active site (Figure 3A, 3C).[14,15] Free calpastatin is intrinsically unstructured,[16] but upon binding to calpain, the polypeptide forms three helices, strings across the surface of the enzyme, and binds in the protease active site to act as a competitive inhibitor. Incredibly, CAST4 buries 2,800 Å2 of surface area on calpain, which is approximately 3 times the surface area standard mechanism protease inhibitors,[17] cystatins,[18] or TIMPs[19] bury when complexed with their respective substrates. The majority of competitive protease inhibitors do not show much induced fit upon binding, and thus do not require a lot of buried surface area. In contrast, CAST4 uses this large amount of buried surface area outside the protease active site to compensate for the entropic penalty of ordering the inhibitor upon binding, and still allows for a KI in the low nanomolar range. As discussed later, the use of exosites to increase the surface area of a protease inhibitor, and allow for structural arrangements to take place upon binding, is a theme that has been extremely useful in the design of novel protein protease inhibitors.
Irreversible Inhibition
Sometimes called suicide substrates, a handful of protease inhibitors require proteolytic activation by the enzymes they inhibit, which leads to covalent modification of the enzyme. This sort of activity dependent inhibition is powerful and fundamentally different than the competitive mechanisms outlined above; the inhibitor acts as a substrate, then utilizes the enzymes’ catalytic machinery to trap and then inhibit the enzyme.
The inhibitor α-2-macroglobin (α2M) and its relatives are responsible for clearing excess proteases from plasma. Less an inhibitor than a “protease sponge”, α2M is a large protein, a tetramer of about 600 kD that has four bait loops on its surface. When a protease cleaves one of these reactive loops, it triggers a conformational change, and the protease becomes cross-linked to the inhibitor through surface lysines and arginines. The enzyme is still active; small molecule substrates can be hydrolyzed by proteases complexed with α2M, but protein substrates are occluded from the active site, and the complex is quickly cleared from the blood.[20]
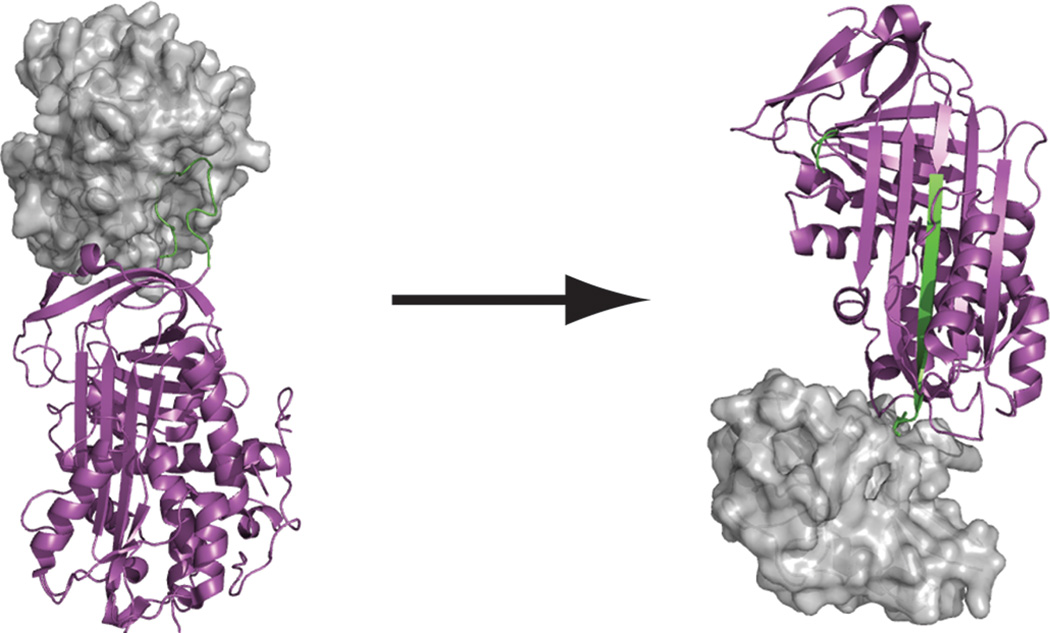
The serpins are a family of inhibitors that covalently and irreversibly inhibit primarily serine proteases[21] (the serpin Crm1 inhibits cysteine proteases). Serpins have a large reactive center loop (RCL) that is presented to a protease for proteolytic processing. Upon productive cleavage of the RCL, the N-terminal half of the RCL, still attached to the protease as an acyl-enzyme intermediate, is inserted into a β-sheet in the body of the inhibitor. The resulting free-energy change is enough to translocate the protease (still covalently attached to the RCL) to the distal side of the inhibitor, and the resulting steric collisions completely deform the protease active site, thus leaving the protease tethered to the serpin and completely inactive (Figure 4). The serpin inhibitory mechanism is completely irreversible. Because of the drastic nature and irreversibility of this mechanism, serpins function as protease scavengers, protecting cells and tissues from unwanted proteolytic activity.
Figure 4.
Serpins inhibit serine proteases by binding a reactive center loop in the active site, forming a covalent complex with the enzyme, undergoing a large conformational change, and irreversibly distorting the active site of the protease (2GD4 and 1EZX.pdb).
These types of inhibitors, which take advantage of the catalytic activity of a protease to trap and inhibit the enzyme, are effective and powerful inhibitors responsible for protecting the organism from aberrant proteolytic activity from a wide range of proteases. Thus they tend to be relatively non-specific. As reviewed by Powers et al,[3] many small molecule covalent and/or irreversible inhibitors have been developed that rely on the same fundamental mechanism of using enzyme activity to trap and inactivate a protease. These molecules use a reactive warhead that binds to the catalytic machinery of the enzyme, and specificity elements are often linked to the warheads to gain selectivity for one target. But due to the reactive nature of the warhead, and the mechanistic and structural similarities displayed by protease families, absolute specificity is difficult to achieve.
Inhibitors Developed via Protein Engineering
Protein engineering has allowed for the development of new protease inhibitors with increased potency and altered specificity, and diverse mechanisms of action. Because proteases tend to have relatively shallow active sites, a high degree of homology, and can have broad specificity, macromolecules are attractive inhibitors in that they can bury more surface area upon binding, and hopefully gain more potency and specificity than a small molecule. And from a functional point of view, the extracellular localization of many proteases make them amenable to regulation by proteins and other macromolecules in vivo. Structure-base modification of naturally occurring protease inhibitors[22] or other scaffolds[23] have been successful, but the evolution of phage display (and similar technologies) has revolutionized our ability to specifically inhibit individual proteases.
The combinatorial nature of phage display allows for the optimization of a number of individual positions on an inhibitor at once, and early protein engineering efforts focused on improving the specificity of naturally occurring protease inhibitors. In an early example of this, Dennis et al. used phage-displayed libraries to randomize residues that interact with the protease subsites, to modify and improve the specificity of the Kunitz-type serine protease inhibitor APPI. Using a series of competitive selection strategies, the authors were able to engineer potent inhibitors that were selective for the clotting enzyme factor VIIa (FVIIa), and lost inhibitory potency against homologous enzymes.[24,25] While modification of the residues that interact with the protease active site have drastic effects on inhibitor affinity, in naturally occurring inhibitors specificity tends to be gained through evolution of secondary interactions. As such the randomization of secondary binding sites of protease inhibitors can greatly increase the specificity of an inhibitor. The standard mechanism serine protease inhibitors ecotin[26] and eglin c[27] have both been refined at both their primary and secondary interaction sites, which has drastically improved their specificity for plasma kallikrein and kexin 2, respectively. Functioning with inhibitory mechanisms similar to that of rhodniin, the engineering of secondary sites on standard mechanism protein scaffolds results in potent and selective inhibitors that allow for modulation of a single protease in complex biological processes.[28]
Another strategy has been to develop polypeptide-based inhibitors of proteases. Typically consisting of 10–20 amino acids, and often containing disulfide bonds or chemical scaffolding components[29] to rigidify the inhibitors and decrease the entropic cost of binding, constrained peptides have been developed to inhibit aspartic, cysteine, serine, and threonine proteases. While peptides are not thought to be ideal drug molecules due to their susceptibility to proteolysis, the relatively small size of constrained peptides allows for the creation of extremely diverse libraries. Furthermore, they are amenable to the incorporation of non-natural or D-amino acids, thus greatly increasing potential diversity. The mechanisms of action of these inhibitors have sometimes mimicked known biological mechanisms, and other times have been completely novel. Constrained peptide phage display libraries have yielded standard mechanism inhibitors of the serine proteases chymotrypsin[30] and urokinase-type plasminogen activator (uPA)[31] with moderate potency and specificity. Cyclic peptides have also been shown to competitively inhibit the aspartic protease renin, and are also thought to bind to the enzyme in a substrate-like manner.[32]
Constrained peptides that mimic natural inhibitors are essentially a reduction of naturally occurring inhibitors to just their reactive elements. But a number of allosteric peptide inhibitors have been developed that have novel mechanisms of inhibition, which reveal information about enzyme function, and suggest new ways of regulating proteolysis. Constrained peptide libraries have yielded two potent exosite inhibitors of the clotting enzyme factor VIIa (FVIIa).[33,34] The two inhibitors bound to two different sites outside of the active site of the enzyme, and had unique mechanisms of inhibition. One inhibitor, A-183, functioned by forcing a loop near the active site into an inactive conformation, and occluding substrate binding to the enzyme. The other inhibitor, E-76 was a non-competitive inhibitor of FVIIa’s natural substrate, factor X, and it seems to work by locking the enzyme in a zymogen-like conformation. In another example of allosteric inhibition, an α-helical peptide was designed to disrupt - and thus prevent activation of - the dimerization of the protease from Kaposi’s sarcoma-associated herpesvirus (KSHV).[35] These allosteric peptide inhibitors lock their target enzymes in an inactive, closed, or zymogen-like state that prevents substrate binding and impairs catalytic activity, and thus they are usually mixed-type inhibitors. The allure of allosteric inhibition is founded in the idea that there are multiple sites on an enzyme that regulate activity, and because allosteric sites are less likely to show sequence conservation than the protease active site, allosteric inhibitors hopefully will show a higher degree of specificity. Nature utilizes a number of techniques to allosterically regulate proteases. Zymogen activation, dimerization, and the binding of proteins, co-factors or metal ions are all ways to more finely regulate protease, and localize proteolytic activity to a specific spatial or temporal location. But as discussed above, with very few exceptions (such as BIR3 domain inhibition of caspase-9[36]), naturally-occuring protease inhibitors do not take advantage of the allosteric movements in proteins, instead inhibiting the protease with a potent competitive or irreversible inhibitor that completely ablates proteolytic activity. Taking advantage of the chemical diversity of phage display libraries and screening libraries of peptides or antibodies against targets has allowed for the isolation of these allosteric inhibitors and the elucidation of mechanisms of allosteric regulation that have not been seen in nature and might not have been discovered using traditional high-throughput screening techniques.
A third approach has been to evolve specific protease inhibitors on other natural protein scaffolds. An enzyme-substrate interaction is really a protein-protein interaction, so any protein that can disrupt the protein-protein interaction can be an effective inhibitor. The antibody scaffold has been the most popular to date, but ankyrin repeats,[37] cysteine knots[38] and aptamers[39] have also been developed into protease inhibitors. Using either hybridoma or phage display technologies, antibody inhibitors have been raised against all four classes of proteases, and biochemical and biophysical characterization reveal they have familiar mechanisms of inhibition. The majority of antibody-based protease inhibitors work by binding to surface loops (see Figure 1) – thus gaining specificity and occluding macromolecular substrates from the protease active site.[40–43] The antibody inhibitors that have been shown to bind in the protease active site do so by binding antibody hypervariable loops in a non-substrate like manner in the substrate binding pockets.[44,45] In a manner analogous to the cystatins or calpastatin, they adopt conformations that avoid direct interaction with the catalytic nucleophile, thus avoiding hydrolysis. Antibodies directed to the dimer interface of HIV protease[46] or caspase 1[47] have been used to sequester monomeric forms of the protease, and thus, are able to inhibit their target protease with a mixed-type mechanism (both competitive and non-competitive inhibition). Antibodies are attractive inhibitors because they are exquisitely specific – antibodies have evolved to specifically bind to their antigen- and are useful biological tools for imaging and in vivo experiments. And that they have been able to incorporate many of the design aspects that make naturally occurring protease inhibitors effective suggests that specific inhibitors on alternate protein scaffolds can be developed for many proteases.
Conclusions and Outlook
Relatively few design principles underlie the mechanisms of inhibition of an awe-inspiring range of protease inhibitors. Protease inhibitors tend to compete with substrate binding, either through direct competition or deformation of the protease active site. While protein inhibitors can gain potency through the burial of a large surface area and specificity through contacts with specific exosites, small molecule inhibitors primarily gain potency through interactions with the catalytic machinery of the enzyme, and specificity through interactions with the substrate binding sites. While there are several examples of successful small molecule protease inhibitors in the clinic, selectivity and potency can be significant challenges when targeting particular protease family members. The search for novel modes of enzyme control, such as allosteric regulation, is therefore particularly exciting, with the hope that these regulatory sites will be more amenable to the design of specific and efficacious inhibitors.
Our evolving understanding of macromolecular protease inhibitor mechanisms has allowed us to use them as tools to dissect complex biological systems and to help determine the role of a single member or an entire family of proteases in either homeostasis or in disease states. Furthermore, protein protease inhibitors have begun to make their way into the clinic. The serpins α1-antitrypsin and antithrombin III[48] purified from human plasma are used as replacement therapies for patients deficient in those proteins. A recent illustration of how protein protease inhibitors can translate into the clinic is the example of Kalbitor (DX-88, Dyax Inc). The relatively unspecific standard mechanism Kuntiz domain of tissue factor pathway inhibitor 1 was matured via phage display to have a higher degree of selectivity for plasma kallikrein.[49] In 2009, it was approved for treatment of hereditary angioedema, and is the first example of an engineered protein therapeutic targeted against a protease. The extracellular location of many proteases, coupled with the increase in knowledge and techniques that allow for the development of more potent and specific inhibitors bodes well for the future of macromolecular therapeutic protease inhibitors.
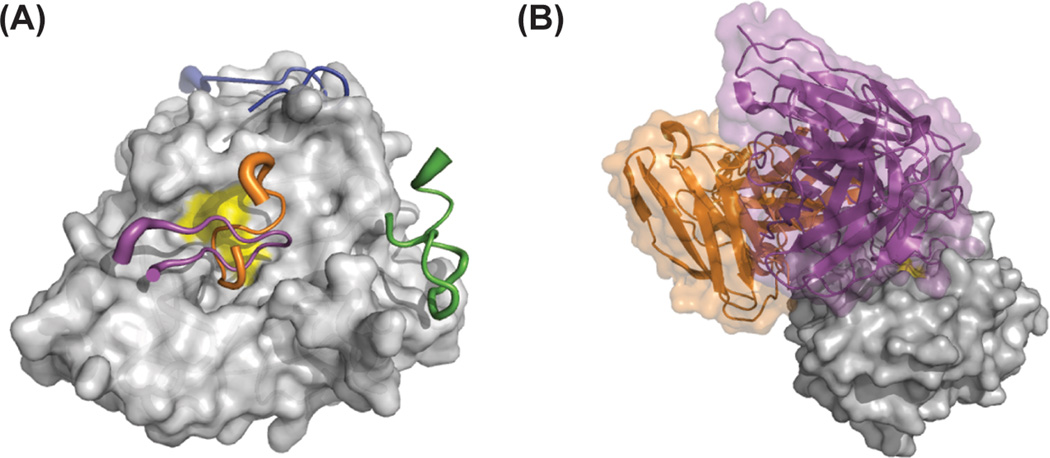
Figure 5.
(A) Inhibitory cyclic peptides bound to the surface of a serine protease. The cyclic peptide SFTI (magenta, 1SFI.pdb) inhibits trypsin in a canonical manner, upain (orange, 2NWN.pdb) binds to the prime side of uPA, and the fVIIa inhibitors E-76 (green, 1DVA.pdb) and A-183 (blue, 1JBU.pdb) bind to protease exosites. (B) Antibodies provide an alternative scaffold on which to build protease inhibitors with a high degree of specificity. The antibody E2 (magenta) binds in the protease active site (MT-SP1, 3BN9.pdb) in a non-canonical manner, while the Fab40 (orange, 3K2U.pdb) antibody inhibits HGFA while binding outside the protease active site through an allosteric mechanism.
References
- 1.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Nat Rev Genet. 2003;4:544–558. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AJ, Salvesen G. Research monographs in cell and tissue physiology. Vol. 12. New York: Elsevier, Amsterdam; 1986. p. xxii.p. 661. [Google Scholar]
- 3.Powers JC, Asgian JL, Ekici OD, James KE. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 4.Turk B. Nat Rev Drug Discov. 2006;5:785–799. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 5.Rawlings ND, Barrett AJ, Bateman A. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laskowski M, Jr, Kato I. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- 7.Luthy JA, Praissman M, Finkenstadt WR, Laskowski M., Jr J Biol Chem. 1973;248:1760–1771. [PubMed] [Google Scholar]
- 8.Zakharova E, Horvath MP, Goldenberg DP. Proc Natl Acad Sci U S A. 2009;106:11034–11039. doi: 10.1073/pnas.0902463106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castro MJ, Anderson S. Biochemistry. 1996;35:11435–11446. doi: 10.1021/bi960515w. [DOI] [PubMed] [Google Scholar]
- 10.Bode W, Huber R. Biochim Biophys Acta. 2000;1477:241–252. doi: 10.1016/s0167-4838(99)00276-9. [DOI] [PubMed] [Google Scholar]
- 11.Brew K, Dinakarpandian D, Nagase H. Biochim Biophys Acta. 2000;1477:267–283. doi: 10.1016/s0167-4838(99)00279-4. [DOI] [PubMed] [Google Scholar]
- 12.Roussel A, Mathieu M, Dobbs A, Luu B, Cambillau C, Kellenberger C. J Biol Chem. 2001;276:38893–38898. doi: 10.1074/jbc.M105707200. [DOI] [PubMed] [Google Scholar]
- 13.Eggers CT, Wang SX, Fletterick RJ, Craik CS. J Mol Biol. 2001;308:975–991. doi: 10.1006/jmbi.2001.4754. [DOI] [PubMed] [Google Scholar]
- 14.Hanna RA, Campbell RL, Davies PL. Nature. 2008;456:409–412. doi: 10.1038/nature07451. [DOI] [PubMed] [Google Scholar]
- 15.Moldoveanu T, Gehring K, Green DR. Nature. 2008;456:404–408. doi: 10.1038/nature07353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendt A, Thompson VF, Goll DE. Biol Chem. 2004;385:465–472. doi: 10.1515/BC.2004.054. [DOI] [PubMed] [Google Scholar]
- 17.Scheidig AJ, Hynes TR, Pelletier LA, Wells JA, Kossiakoff AA. Protein Sci. 1997;6:1806–1824. doi: 10.1002/pro.5560060902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jenko S, Dolenc I, Guncar G, Dobersek A, Podobnik M, Turk D. J Mol Biol. 2003;326:875–885. doi: 10.1016/s0022-2836(02)01432-8. [DOI] [PubMed] [Google Scholar]
- 19.Maskos K, Lang R, Tschesche H, Bode W. J Mol Biol. 2007;366:1222–1231. doi: 10.1016/j.jmb.2006.11.072. [DOI] [PubMed] [Google Scholar]
- 20.Kolodziej SJ, Wagenknecht T, Strickland DK, Stoops JK. J Biol Chem. 2002;277:28031–28037. doi: 10.1074/jbc.M202714200. [DOI] [PubMed] [Google Scholar]
- 21.Gettins PG. Chem Rev. 2002;102:4751–4804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 22.Izaguirre G, Rezaie AR, Olson ST. J Biol Chem. 2009;284:1550–1558. doi: 10.1074/jbc.M807340200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohfeldt E, Gohring W, Mayer U, Zweckstetter M, Holak TA, Chu ML, Timpl R. Eur J Biochem. 1996;238:333–340. doi: 10.1111/j.1432-1033.1996.0333z.x. [DOI] [PubMed] [Google Scholar]
- 24.Dennis MS, Lazarus RA. J Biol Chem. 1994;269:22137–22144. [PubMed] [Google Scholar]
- 25.Dennis MS, Lazarus RA. J Biol Chem. 1994;269:22129–22136. [PubMed] [Google Scholar]
- 26.Stoop AA, Craik CS. Nat Biotechnol. 2003;21:1063–1068. doi: 10.1038/nbt860. [DOI] [PubMed] [Google Scholar]
- 27.Komiyama T, VanderLugt B, Fugere M, Day R, Kaufman RJ, Fuller RS. Proc Natl Acad Sci U S A. 2003;100:8205–8210. doi: 10.1073/pnas.1032865100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lilla JN, Joshi RV, Craik CS, Werb Z. J Biol Chem. 2009;284:13792–13803. doi: 10.1074/jbc.M900508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinis C, Rutherford T, Freund S, Winter G. Nat Chem Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 30.Dumez E, Snaith JS, Jackson RF, McElroy AB, Overington J, Wythes MJ, Withka JM, McLellan TJ. J Org Chem. 2002;67:4882–4892. doi: 10.1021/jo025615o. [DOI] [PubMed] [Google Scholar]
- 31.Hansen M, et al. J Biol Chem. 2005;280:38424–38437. doi: 10.1074/jbc.M505933200. [DOI] [PubMed] [Google Scholar]
- 32.Nakaie CR, Oliveira MC, Juliano L, Paiva AC. Biochem J. 1982;205:43–47. doi: 10.1042/bj2050043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis MS, Eigenbrot C, Skelton NJ, Ultsch MH, Santell L, Dwyer MA, O'Connell MP, Lazarus RA. Nature. 2000;404:465–470. doi: 10.1038/35006574. [DOI] [PubMed] [Google Scholar]
- 34.Roberge M, Santell L, Dennis MS, Eigenbrot C, Dwyer MA, Lazarus RA. Biochemistry. 2001;40:9522–9531. doi: 10.1021/bi010592d. [DOI] [PubMed] [Google Scholar]
- 35.Shimba N, Nomura AM, Marnett AB, Craik CS. J Virol. 2004;78:6657–6665. doi: 10.1128/JVI.78.12.6657-6665.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shiozaki EN, et al. Mol Cell. 2003;11:519–527. doi: 10.1016/s1097-2765(03)00054-6. [DOI] [PubMed] [Google Scholar]
- 37.Schweizer A, et al. Structure. 2007;15:625–636. doi: 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Kolmar, H H. Febs J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- 39.Gopinath, S.C SC. Thromb Res. 2008;122:838–847. doi: 10.1016/j.thromres.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 40.Matias-Roman S, Galvez BG, Genis L, Yanez-Mo M, de la Rosa G, Sanchez-Mateos P, Sanchez-Madrid F, Arroyo AG. Blood. 2005;105:3956–3964. doi: 10.1182/blood-2004-06-2382. [DOI] [PubMed] [Google Scholar]
- 41.Obermajer N, Premzl A, Zavasnik Bergant T, Turk B, Kos J. Exp Cell Res. 2006;312:2515–2527. doi: 10.1016/j.yexcr.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 42.Petersen HH, Hansen M, Schousboe SL, Andreasen PA. Eur J Biochem. 2001;268:4430–4439. doi: 10.1046/j.1432-1327.2001.02365.x. [DOI] [PubMed] [Google Scholar]
- 43.Ganesan R, Eigenbrot C, Kirchhofer D. Biochem J. 2010;430:179–189. doi: 10.1042/BJ20100634. [DOI] [PubMed] [Google Scholar]
- 44.Farady CJ, Egea PF, Schneider EL, Darragh MR, Craik CS. J Mol Biol. 2008;380:351–360. doi: 10.1016/j.jmb.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu Y, et al. Proc Natl Acad Sci U S A. 2007;104:19784–19789. doi: 10.1073/pnas.0708251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rezacova P, Lescar J, Brynda J, Fabry M, Horejsi M, Sedlacek J, Bentley GA. Structure. 2001;9:887–895. doi: 10.1016/s0969-2126(01)00654-2. [DOI] [PubMed] [Google Scholar]
- 47.Gao J, Sidhu SS, Wells JA. Proc Natl Acad Sci U S A. 2009;106:3071–3076. doi: 10.1073/pnas.0812952106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leader B, Baca QJ, Golan DE. Nat Rev Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 49.Lehmann A. Curr Opin Investig Drugs. 2006;7:282–290. [PubMed] [Google Scholar]