ABSTRACT
Desmosomes are prominent adhesive junctions found in various epithelial tissues. The cytoplasmic domains of desmosomal cadherins interact with a host of desmosomal plaque proteins, including plakophilins, plakoglobin and desmoplakin, which, in turn, recruit the intermediate filament cytoskeleton to sites of cell–cell contact. Although the individual components of the desmosome are known, mechanisms regulating the assembly of this junction are poorly understood. Protein palmitoylation is a posttranslational lipid modification that plays an important role in protein trafficking and function. Here, we demonstrate that multiple desmosomal components are palmitoylated in vivo. Pharmacologic inhibition of palmitoylation disrupts desmosome assembly at cell–cell borders. We mapped the site of plakophilin palmitoylation to a conserved cysteine residue present in the armadillo repeat domain. Mutation of this single cysteine residue prevents palmitoylation, disrupts plakophilin incorporation into the desmosomal plaque and prevents plakophilin-dependent desmosome assembly. Finally, plakophilin mutants unable to become palmitoylated act in a dominant-negative manner to disrupt proper localization of endogenous desmosome components and decrease desmosomal adhesion. Taken together, these data demonstrate that palmitoylation of desmosomal components is important for desmosome assembly and adhesion.
KEY WORDS: Desmosome, Plakophilin, Palmitoylation
INTRODUCTION
Desmosomes are prominent cell–cell adhesive junctions found in epithelial and cardiac tissues, and they are widely thought to provide these tissues the ability to withstand mechanical stress (Brooke et al., 2012). Structurally, these adhesive structures link the intermediate filament cytoskeletal systems between adjacent cells and allow the cells of the tissue to function coordinately. Desmosomal cadherins (i.e. desmogleins and desmocollins) form the transmembrane core of the desmosome, and these transmembrane proteins interact with a set of desmosomal plaque proteins to recruit the intermediate filament cytoskeleton to sites of cell–cell contact. Desmosomal plaque proteins include plakoglobin, desmoplakin and the plakophilins. Disruption of the expression or function of plaque proteins can have devastating effects on desmosome integrity and function (Gerull et al., 2004; McGrath et al., 1997; Ruiz et al., 1996).
Cells possess the ability to modulate adhesive strength between neighboring cells in response to extracellular cues. For example, keratinocytes acquire a motile phenotype during the re-epithelialization phase of epidermal wound healing. During this process, cells decrease their adhesiveness with neighboring cells and migrate into the wound bed until the epithelial barrier is re-established (Thomason et al., 2012). When adhesive strength is altered or disrupted during tumorigenesis, tumor cells are believed to acquire the ability to detach from their normal neighbors and to migrate inappropriately. Altered desmosome assembly has been observed in several tumor types (Davies et al., 1999; Harada et al., 1996; Thomas and Speight, 2001). Additionally, it is likely that desmosomes are also responsive to extracellular cues, and these signals should result in altered desmosome dynamics (Roberts et al., 2011). The mechanisms regulating desmosome assembly, stability and turnover during normal migratory events and tumorigenesis are poorly understood.
Protein palmitoylation is a reversible posttranslational modification whereby a 16-carbon fatty acid (palmitate) is linked to specific cysteine residues through a labile thioester linkage (reviewed in Aicart-Ramos et al., 2011). Palmitoylation regulates diverse protein features including protein localization, activity and stability (Greaves and Chamberlain, 2011). Palmitoylation is thought to increase the association of substrate proteins with cellular membranes. Recent studies in yeast have identified the Asp-His-His-Cys (DHHC) family of proteins as palmitoyl acyltransferases (PATs) (Ohno et al., 2012). There are 23 evolutionarily conserved DHHC genes in mammals. The enzymes responsible for deacylation of protein targets are acyl protein thioesterases. Three cytosolic thioesterases (LYPLA1, LYPLA2 and LYPLAL1) have been identified to date (Sugimoto et al., 1996; Tomatis et al., 2010; Toyoda et al., 1999).
In the current study, we have demonstrated that multiple desmosomal components are indeed palmitoylated in cultured cells. Additionally, we have identified a single conserved cysteine residue in plakophilin-2 and plakophilin-3 that is the site of palmitoylation. Mutation of this cysteine residue results in decreased membrane targeting of plakophilin-2 and plakophilin-3 and the inability of mutant plakophilins to support de novo desmosome assembly in A431D cells. Finally, expression of mutant plakophilin-2 and plakophilin-3 in A431 weakens desmosomal adhesion compared with that of cells expressing wild-type plakophilin-2 and plakophilin-3, suggesting the mutants act in a dominant-negative manner with respect to cell adhesion. The data presented here suggest that palmitoylation of desmosomal components, and specifically plakophilins, plays an important role in the regulation of junction assembly and adhesive strength.
RESULTS
Desmosomal components are palmitoylated
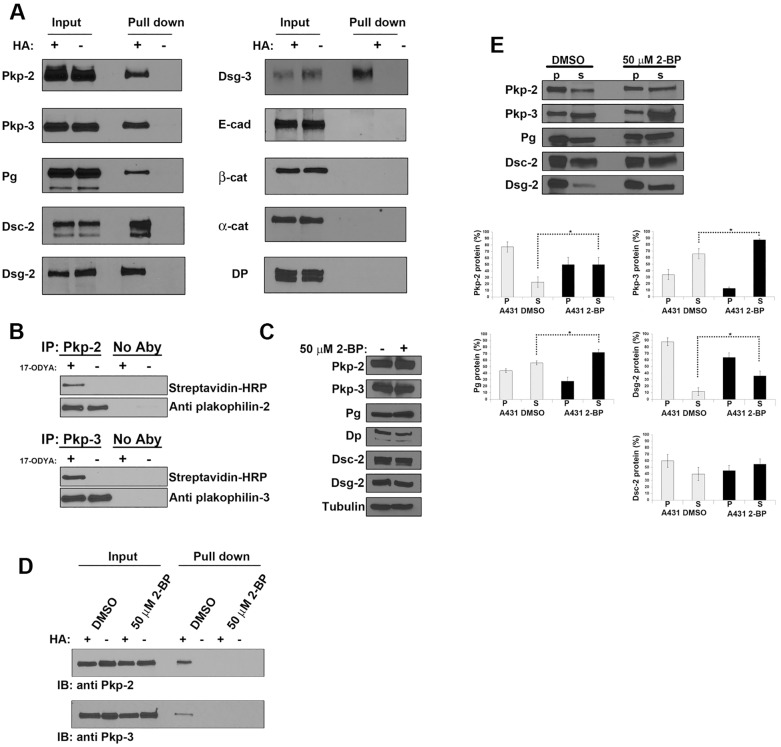
Recently, several studies have utilized large-scale proteomic screening strategies in an attempt to identify the landscape of palmitoylated proteins in cultured cells (Martin and Cravatt, 2009; Yang et al., 2010). These efforts have identified several desmosomal components as potential palmitoylated proteins (Yang et al., 2010). We set out to show that desmosomal proteins were indeed palmitoylated and to determine the effect of palmitoylation on desmosome assembly and function. We first chose to identify palmitoylated desmosomal components using an acyl-biotin exchange assay. Acyl-biotin exchange (ABE) is performed by selectively hydrolyzing thioester bonds followed by covalent tagging of the previously palmitoylated cysteine residues with biotin. Biotin-labeled proteins are captured by streptavidin pulldown and individual proteins are detected with specific antibodies (Wan et al., 2007). Streptavidin pulldown revealed that the desmosomal plaque proteins (plakophilin-2, plakophilin-3 and plakoglobin), as well as desmosomal cadherins (desmogleins-2, desmoglein-3 and desmocollin-2), were indeed palmitoylated in A431 cells (Fig. 1A). Because two bands were detected in the blot for desmocollin-2, it is likely both desmocollin-2a and desmocollin-2b (Collins et al., 1991; Koch et al., 1991) are palmitoylated. In addition, we detected palmitoylation of plakophilin-2 using ABE in murine heart tissue (data not shown). The adherens junction components E-cadherin, β-catenin and α-catenin were found to not be palmitoylated and served as negative controls. Proteomics analysis (Yang et al., 2010) had previously suggested that desmoplakin was also palmitoylated; however, we were unable to detect desmoplakin palmitoylation in A431 cells. We chose to further investigate the role of palmitoylation of plakophilin on desmosome assembly and adhesion.
Fig. 1.
Desmosomal components are palmitoylated. (A) A431 cell lysates were prepared, and ABE was performed to identify palmitoylated proteins. Lysates were split into two samples and processed with (+) and without (−) hydroxylamine (HA). After the final streptavidin–agarose pull-down, the captured proteins were resolved by SDS-PAGE, and individual desmosome and adherens junction components were detected by immunoblot analysis. Pkp, plakophilin; Pg, plakoglobin; DP, desmoplakin; Dsc-2, desmocollin-2; Dsg, desmoglein; E-cad, E-cadherin; α-cat, α-catenin; β-cat, β-catenin. (B) A431 cells were grown in medium with or without 100 µM 17-ODYA for 48 hours. Plakophilin-2 and plakophilin-3 were immunoprecipitated (IP) from cell lysates, and HRP–biotin was added to label cells using the Cu-catalyzed click reaction. Immunoprecipitations with no immunoprecipitating antibody (No Aby) served as negative controls. Immunoprecipitates were resolved by SDS-PAGE, and labeled proteins were detected by HRP–streptavidin or isoform-specific anti-plakophilin antibodies. (C) Cell lysates were prepared from A431 cells grown in the presence or absence of 50 µM 2-BP for 18 hours, and immunoblot analysis was performed to examine the levels of plakophilin-2, plakophilin-3, plakoglobin, desmoplakin, desmocollin-2 and desmoglein-2. β-tubulin was used as a loading control. (D) ABE demonstrated that plakophilin-2 and plakophilin-3 are palmitoylated in the absence of 2-BP, and palmitoylation is decreased in A431 cells grown in medium containing 50 µM 2-BP. IB, immunoblot. (E) Triton-X-100-insoluble (p) and -soluble (s) fractions were prepared from control A431 cultures (DMSO) or from A431 cells grown in medium containing 50 µM 2-BP. Immunoblot analysis determined that there was a significant increase in the soluble fraction of plakophilin-2, plakophilin-3, plakoglobin and desmoglein-2 in 2-BP-treated cultures. The Triton X-100 solubility of desmocollin-2 was unchanged following 2-BP treatment. Data show the mean±s.d. (n = 3 experiments); *P<0.05.
In order to further confirm the palmitoylation of plakophilins, we metabolically labeled A431 cells using the palmitic acid analog 17-ODYA (Martin and Cravatt, 2009). Proteins were immunoprecipitated from lysates of A431 cells that had been grown in medium containing 100 µM 17-ODYA or in control medium lacking 17-ODYA (DMSO). Biotin azide was covalently added to 17-ODYA by the Staudinger ligation and Cu(I)-catalyzed azide-alkyne cycloaddition (click chemistry). Immunoprecipitated proteins were separated by SDS-PAGE and biotin-labeled proteins were detected by horseradish peroxidase (HRP)–streptavidin blotting (Fig. 1B). Plakophilin-2 and plakophilin-3 were specifically labeled by 17-ODYA, whereas no biotin-labeled plakophilin-2 or plakophilin-3 was detected in cell lysate prepared from cells grown in control medium. Because A431 cells endogenously express plakophilin-2 and plakophilin-3 but not plakophilin-1, our studies here focused on plakophilin-2 and plakophilin-3.
Inhibition of palmitoylation disrupts desmosome assembly
Next, we sought to investigate the effect of inhibiting global palmitoylation on desmosome assembly in A431 cells. 2-bromopalmitate (2-BP) is an irreversible inhibitor of protein acyltransferases, which are responsible for palmitoylation of substrate proteins, and, as a consequence, 2-BP treatment inhibits global palmitoylation in a non-selective manner (Davda et al., 2013). Immunoblot analysis of cell lysates prepared from A431 cells grown in 50 µM 2-BP demonstrates that there is no substantial reduction in the expression of desmosomal components in 2-BP-treated cells compared with that of control cells (Fig. 1C). ABE was used to demonstrate that 2-BP treatment resulted in a reduction in the palmitoylation of plakophilin-2 and plakophilin-3 (Fig. 1D). Additionally, we examined the solubility of desmosomal components in buffer containing 0.5% Triton X-100. In cells treated with 2-BP, there was a significant shift from the insoluble pool to the soluble pool for plakophilin-2, plakophilin-3, plakoglobin and desmoglein-2. The solubility of desmocollin-2 was not significantly altered by 2-BP treatment (Fig. 1E).
Next, we examined the localization of desmosomal components in the presence of 2-BP. A431 cells expressing plakophilin-3–GFP (Roberts et al., 2011) were grown in the presence of 2-BP for 18 hours, and the localization of plakophilin-3–GFP and other desmosomal components was examined. Immunofluorescence microscopy revealed a disruption in the localization of desmosomal components in A431 cells grown in medium containing 2-BP compared with that of control cells. As expected, plakophilin-3–GFP and endogenous desmoglein-2 were found to colocalize in a linear punctate staining pattern at cell–cell contact sites in DMSO-treated control cells (Fig. 2A–C). In cells grown in medium containing 2-BP, plakophilin-3–GFP and desmoglein-2 colocalized; however, the pattern of desmosome localization was disrupted compared with that of control cells (Fig. 2D–F). The linear punctate array present in untreated cells was replaced in the 2-BP-treated cells with larger aggregates of desmosomal components with increased space between punctate structures and some intracellular accumulation of signal. Cytoplasmic aggregates containing desmosomal components are observed throughout the 2-BP-treated cultures. The nature of these structures is unclear; however, they do not colocalize with markers of the Golgi, lysosomes or the endocytic recycling compartment (data not shown). A similar staining pattern was observed for other desmosomal components, plakophilin-2 (Fig. 2M–R) and desmoplakin (Fig. 2S–X). E-cadherin localization (Fig. 2G–L) was not altered in cells grown in medium containing 2-BP, demonstrating that palmitoylation impacts on the assembly of desmosome junctions, whereas adherens junction assembly is not substantially affected by treatment with 2-BP.
Fig. 2.
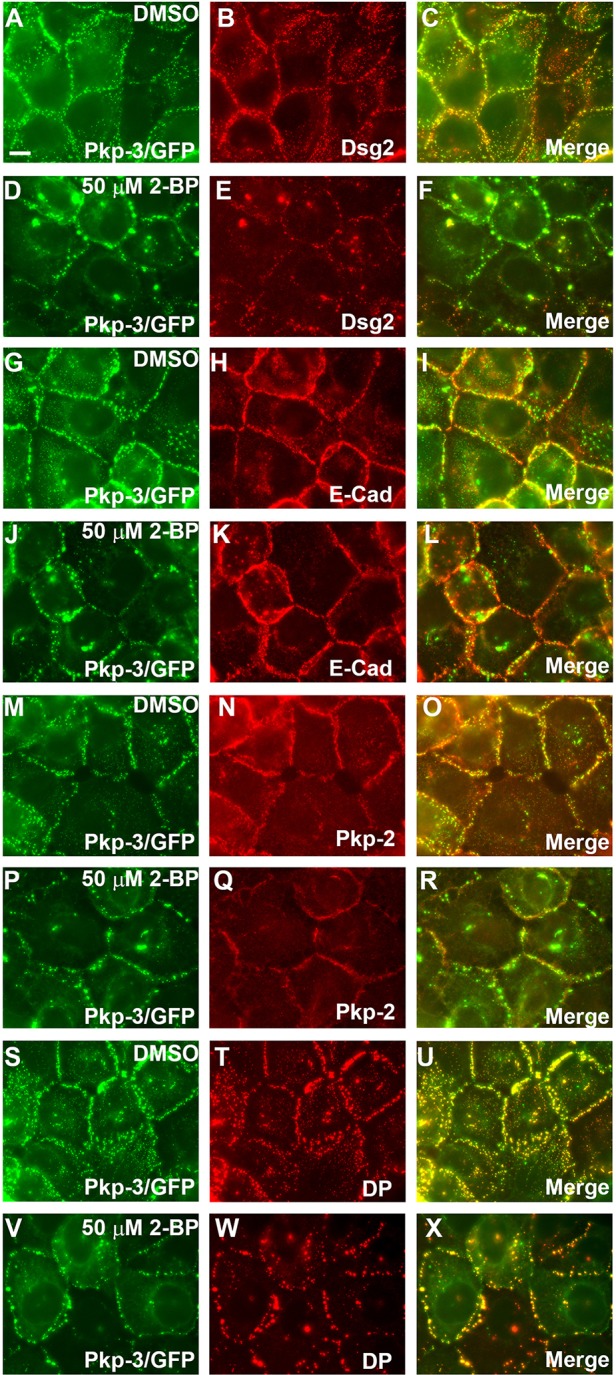
Inhibition of palmitoylation by 2-BP alters the localization of desmosomal components at cell borders. A431 cells expressing plakophilin-3–GFP (Pkp-3/GFP) were grown on glass coverslips in medium containing 50 µM 2-BP or DMSO as a negative control (18 hours). Cells were fixed and immunostained with antibodies specific for desmoglein-2 (Dsg2) (A–F), E-cadherin (E-Cad) (G–L), plakophilin-2 (Pkp-2) (M–R) and desmoplakin (DP) (S–X). Inhibition of palmitoylation has little effect on the localization of E-cadherin, whereas the localization of desmosomal components is disrupted at cell–cell borders. Scale bar: 10 µm.
Inhibition of palmitoylation delays desmosome assembly in HaCat keratinocytes
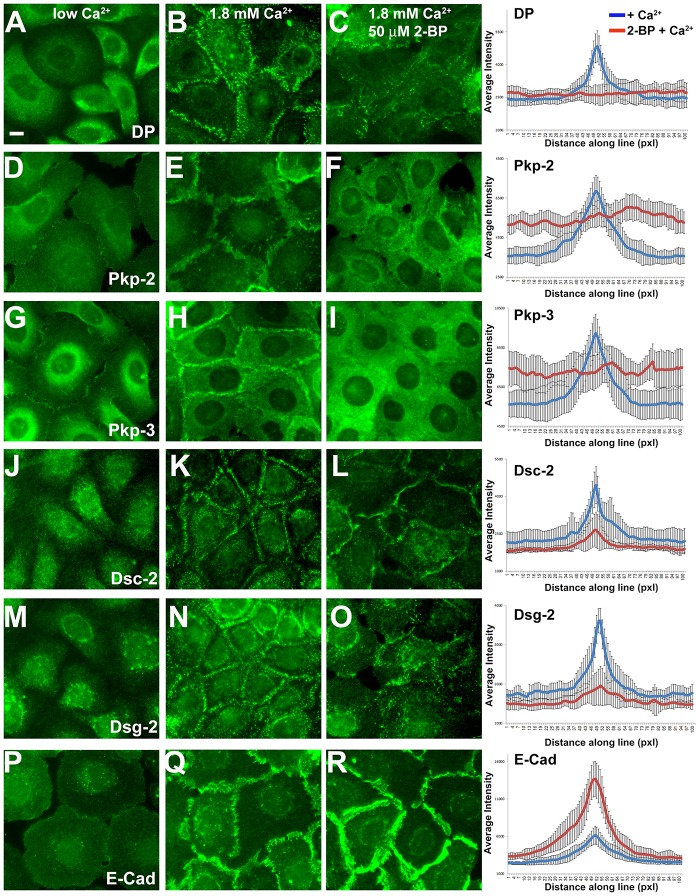
In order to investigate the role of palmitoylation during desmosome assembly, we performed a calcium-switch experiment using HaCat keratinocytes in the presence or absence of 2-BP. Cells were seeded on glass coverslips and grown in medium containing a low concentration of Ca2+ (0.050 mM Ca2+) until the culture reached ∼75–80% confluence. Cells were grown overnight in low-Ca2+-containing medium with or without 50 mM 2-BP. Ca2+ was added to the medium to a final concentration of 1.8 mM, and cells were processed for immunofluorescence microcopy after 2.5 hours under normal Ca2+ conditions.
Junctional proteins are largely cytosolic when HaCat keratinocytes are grown in medium containing low Ca2+ (Fig. 3, left panels). Adding Ca2+ to the culture medium induced the redistribution of adhesive junction components to the plasma membrane, the assembly of desmosomes and the recruitment of E-cadherin (Fig. 3, middle panels). Cells pretreated with 2-BP failed to efficiently recruit desmosomal components to the plasma membrane and, as a result, failed to assemble desmosomes after 2.5 hours (Fig. 3, right panels). By contrast, E-cadherin was recruited to sites of cell–cell contact in the presence of 2-BP. In fact, E-cadherin localization at cell borders was enhanced in HaCat cells treated with 2-BP. Overall, 2-BP treatment inhibited desmosome assembly and this inhibition was not due to a disruption of E-cadherin recruitment to the plasma membrane but rather was specific to events directing desmosome assembly.
Fig. 3.
Inhibition of palmitoylation disrupts Ca2+-induced desmosome assembly in HaCat keratinocytes. HaCat keratinocytes were grown on glass coverslips in medium containing low Ca2+ (left panels) in the absence (middle panels) or the presence of 50 µM 2-BP (right panels) overnight. Ca2+ was added (1.8 mM final concentration) to the medium for 2.5 hours and cells were processed for immunofluorescence microscopy using the antibodies indicated. Scale bar: 10 µm. Average fluorescence intensity was determined using Slidebook5 image software by measuring a segment of equal length and width across 30 individual cell borders for each treatment. Blue lines, the average fluorescence intensity for control cultures; red lines, fluorescence intensity in 2-BP-treated cell cultures. DP, desmoplakin; Pkp, plakophilin; Dsc-2, desmocollin-2; Dsg-2, desmoglein-2; E-cad, E-cadherin; Pxl, pixels. Data show the mean±s.d. (n = 30 cell border measurements).
A conserved cysteine in the plakophilin armadillo repeat domain is palmitoylated
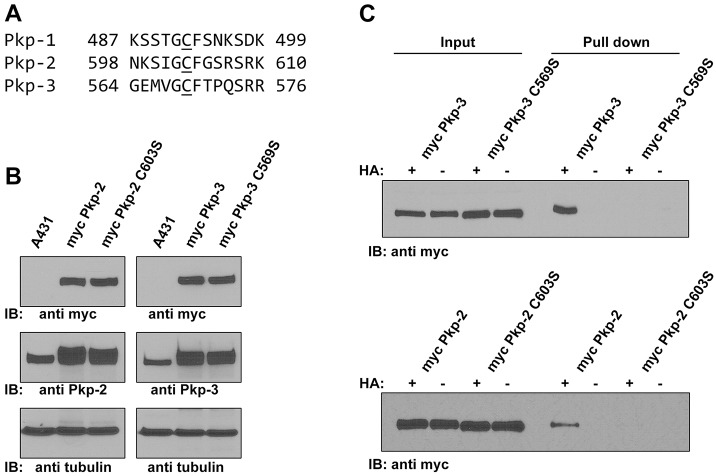
Examination of the amino acid sequence of the plakophilins revealed a conserved cysteine residue present in all three plakophilin isoforms (Fig. 4A), residing in the unstructured ‘loop’ sequence between armadillo repeats 5 and 6 (Choi and Weis, 2005). Additionally, a recent proteomics dataset generated by Yang et al. (Yang et al., 2010) identified this conserved cysteine as a potentially palmitoylated residue. In addition, based on a web-based algorithm, these conserved cysteine residues are predicted to be palmitoylated (CCS Palm; Ren et al., 2008). To test these predictions, we generated Myc-tagged plakophilin-2 with a serine residue in place of the conserved cysteine (C603S) and stably expressed this plakophilin-2 mutant protein in A431 cells. Additionally, we generated the corresponding mutant in plakophilin-3 (C569S). Mutation of these cysteine residues results in the loss of palmitoylation of the tagged plakophilin construct, whereas Myc-tagged wild-type plakophilin-2 and plakophilin-3 were palmitoylated (Fig. 4C). These data show that plakophilins are palmitoylated on a single conserved cysteine residue present in an unstructured loop in the armadillo repeat domain.
Fig. 4.
A conserved cysteine in plakophilins is palmitoylated. (A) Sequence alignment of plakophilin-1 (NP_001005337), plakophilin-2 (NP_001005242) and plakophilin-3 (NP_009114) (Pkp-1, Pkp-2 and Pkp-3, respectively) shows similarity in the sequences surrounding the conserved cysteine residue. (B) Immunoblot (IB) analysis of cell lysates prepared from control A431 cells, A431 cells expressing Myc-tagged wild-type plakophilin-2 (myc Pkp-2), Myc-tagged plakophilin-2 C603S (myc Pkp-2 C603S), Myc-tagged wild-type plakophilin-3 (myc Pkp-3) and Myc-tagged plakophilin-3 C569S (myc Pkp-3 C569S). Lysates were blotted with antibodies against Myc, plakophilin-2 or plakophilin-3. Exogenous proteins were expressed at levels similar to those of the endogenous plakophilins, and the Myc-tagged proteins were all expressed at similar levels to one another. Tubulin is shown as a loading control. (C) ABE demonstrates that plakophilin-2 C603S and plakophilin-3 C569S are not palmitoylated, whereas wild-type plakophilin-2 and plakophilin-3 are palmitoylated. HA, hydroxylamine.
Because A431 cells endogenously assemble desmosomes, we determined the localization of the Myc-tagged plakophilin constructs in these cells. Exogenous wild-type plakophilin-2 (Fig. 5A–C) and plakophilin-3 (supplementary material Fig. S1) localized to cell borders and colocalized extensively with endogenous desmoplakin. By contrast, mutant plakophilin-2 (C603S) (Fig. 5D–F) and plakophilin-3 (C569S) (supplementary material Fig. S1) did not efficiently localize to sites of cell–cell contact and are noticeably more cytoplasmic in their distribution. Interestingly, the expression of mutant plakophilins in A431 cells results in noticeable disruption in the localization of endogenously expressed desmosomal components. For example, desmoplakin is partially redistributed to cytosolic punctate structures in cells expressing plakophilin-2 C603S and plakophilin-3 C569S (compare Fig. 5B,E; supplementary material Fig. S1B,E). Additionally, the localization of the desmosomal cadherins was also disrupted in cells expressing mutant plakophilins compared with cells expressing wild-type plakophilin-2 or plakophilin-3 (Fig. 5; supplementary material Fig. S1G–R). These data suggest that plakophilin-2 C603S and plakophilin-3 C569S act as dominant-negative mutants with respect to the localization of desmosomal components at sites of cell–cell contact. The mechanism of this dominant-negative effect is currently under investigation.
Fig. 5.
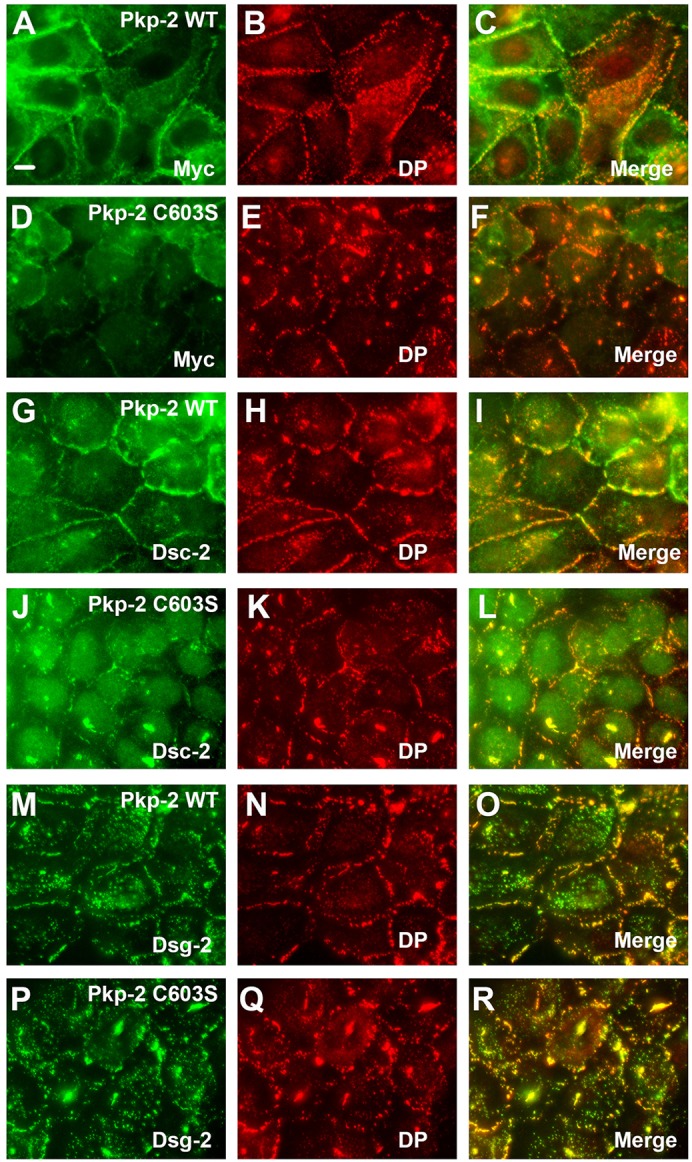
Palmitoylation-defective plakophilin-2 disrupts the localization of endogenous desmosomal components. A431 cells expressing wild-type plakophilin-2 (Pkp-2 WT; A–C, G–I and M–O) or plakophilin-2 C603S (Pkp-2 C603S; D–F, J–L and P–R) were grown on glass coverslips, fixed and immunostained using antibodies recognizing the Myc epitope tag (A,D), desmocollin-2 (Dsc-2; G,J) and desmoglein-2 (Dsg-2; M,P). Desmoplakin (DP) colocalization is shown in panels B, E, H, K, N and Q. Scale bar: 10 µm.
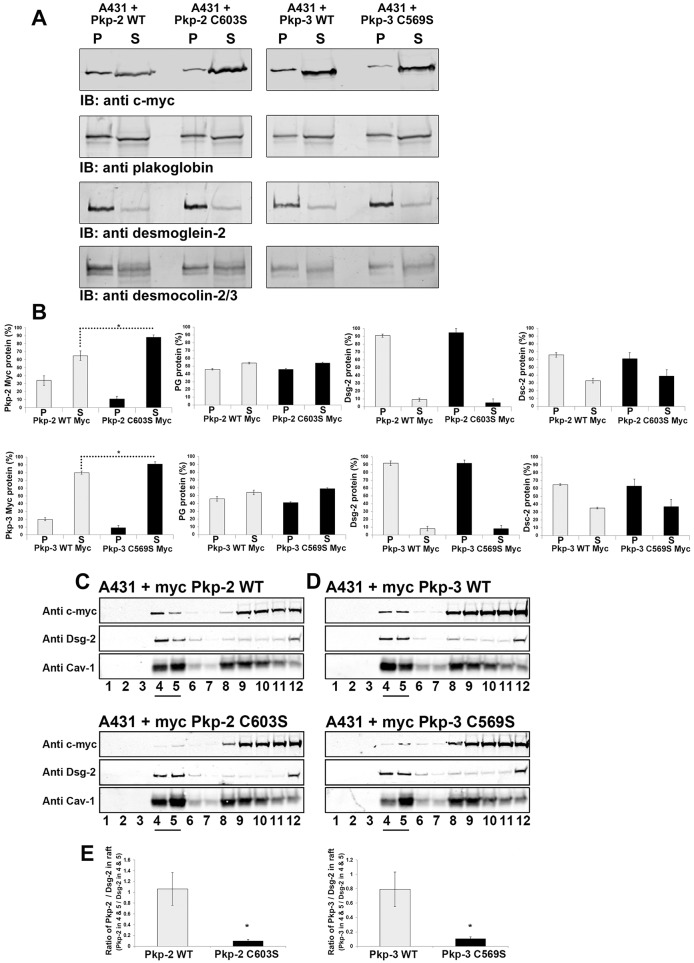
Additionally plakophilin-2 C603S and plakophilin-3 C569S were found to be more soluble in lysis buffer containing Triton X-100 compared with wild-type plakophilins expressed in A431 cells (Fig. 6A,B). Although the localization of endogenous desmosomal components was altered in cells expressing mutant plakophilins, the Triton X-100 solubility of these proteins was unaltered. These data suggest that the mutant plakophilins affect desmosome localization rather than solubility of the desmosomal components. The nature of the intracellular clusters is not understood.
Fig. 6.
Plakophilin mutants display altered Triton X-100 solubility and lipid raft association compared with that of wild-type plakophilin. (A) Triton-X-100-insoluble (P) and soluble fractions (S) were prepared from A431 cells expressing wild-type plakophilin-2 (Pkp-2 WT), plakophilin-2 C603S (Pkp-2 C603S), wild-type plakophilin-3 (Pkp-3 WT) or plakophilin-3 C569S (Pkp-3 C569S). Immunoblot (IB) analysis was performed using antibodies against the Myc epitope tag, plakoglobin, desmoglein-2 and desmocollin-2/3. (B) Immunoblot analysis was performed, and the signal was quantified using a Li-Cor Odyssey Imaging system. Myc-tagged plakophilin-2 C603S and Myc-tagged plakophilin-3 C569S were found to be more soluble compared with Myc-tagged wild-type plakophilin-2 and plakophilin-3. The Triton X-100 solubility of endogenous plakoglobin (PG), desmoglein-2 (Dsg-2) and desmocollin-2/3 (Dsc-2) was not altered by the expression of mutant plakophilin-2 or plakophilin-3. (C,D) Cell lysates were prepared from A431 cells expressing Myc-tagged wild-type plakophilin-2 and Myc-tagged plakophilin-2 C603S (C) and Myc-tagged wild-type plakophilin-3 and Myc-tagged plakophilin-3 C569S (D). Lysates were subjected to sucrose-density centrifugation, 1-ml fractions were collected and immunoblot analysis was performed with the indicated antibodies. Fractions 4 and 5 (underlined) are enriched in the lipid raft component caveolin-1 (Cav-1), endogenous desmoglein-2 and wild-type plakophilin-2 and wild-type plakophilin-3. (E) The ratio of Myc signal to the desmoglein-2 signal present in fractions 4 and 5 is shown graphically. The ratios of plakophilin-2 C603S and plakophilin-3 C569S to the corresponding desmoglein-2 signal are reduced in fractions 4 and 5 compared with that of the wild-type plakophilins. For B,E, data show the mean±s.d. [n = 3 (B,E)]; *P<0.05.
Palmitoylation of the plakophilins enhances their association with lipid raft domains
Palmitoylation of cysteine residues is proposed to increase the association of modified proteins with lipid raft domains. Recent studies have demonstrated that desmosomal components are indeed associated with membrane microdomains enriched in lipid rafts components (Brennan et al., 2012; Resnik et al., 2011), and the disruption of lipid rafts has effects on desmosome dynamics (Stahley et al., 2014). We examined the ability of Myc-tagged wild-type plakophilins and plakophilin mutants to associate with lipid raft components by sucrose-gradient centrifugation. Cell lysates prepared from A431 cells expressing wild-type and mutant plakophilins were separated by sucrose-gradient centrifugation and the co-sedimentation of the Myc-tagged plakophilins with desmoglein-2 and caveolin-1 was examined by immunoblot analysis. Desmoglein-2 was shown previously to associate with lipid rafts, and, in our lysates, desmoglein-2 partitioned with the lipid raft component caveolin-1 in fractions 4 and 5 of our preparations (Fig. 6C–E). Wild-type plakophilins were found to be present in fractions 4 and 5, indicating an association with lipid raft components. Plakophilin mutants (Pkp-2 C603S and Pkp-3 C569S) showed a decreased association with the lipid raft fractions compared with that of the wild-type plakophilins. These data suggest that lack of palmitoylation of the plakophilins results in decreased association with lipid raft domains, but other interactions are likely to be involved in recruitment to lipid raft domains, possibly direct interactions with other endogenous desmosomal components present in A431 lipid rafts (e.g. desmosomal cadherins).
Plakophilin mutants fail to support desmosome assembly in A431D cells
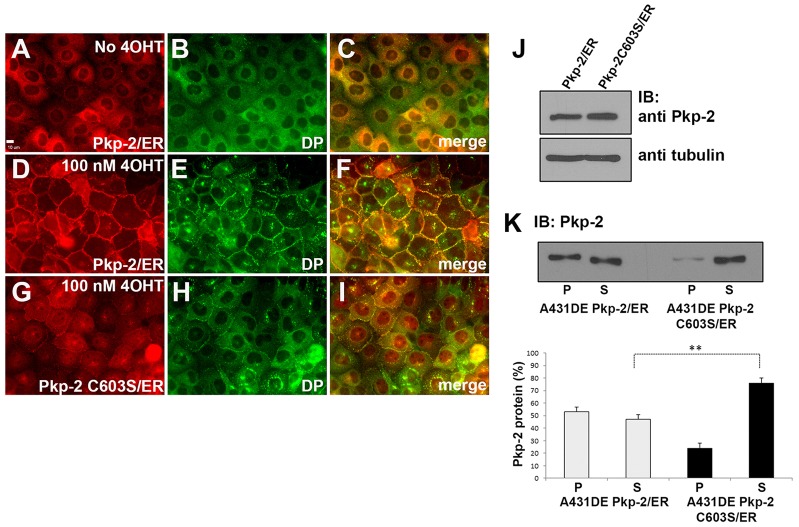
Previously, our laboratory characterized a cell culture system that allowed us to examine plakophilin-dependent assembly of desmosomes (Lewis et al., 1997; Wahl, 2005). A431D cells expressing E-cadherin (A431DE cells) assemble adherens junctions but lack the expression of an endogenous plakophilin and do not assemble desmosomal components at sites of cell–cell contact. Expression of plakophilin-2 fused at its carboxyl terminus to the estrogen receptor ligand-binding domain resulted in a plakophilin-2–ER fusion protein that can be ‘activated’ by the addition of 4-hydroxytamoxifen (4OHT) to the culture medium. In the absence of 4OHT, plakophilin-2–ER was localized in the cytoplasm (Fig. 7A) and desmoplakin was cytosolic and not recruited to the plasma membrane (Fig. 7B). Addition of 4OHT to the culture medium resulted in the localization of plakophilin-2–ER to the plasma membrane (Fig. 7D) and the recruitment of desmoplakin to punctate structures at sites of cell–cell contact (Fig. 7E). In contrast to the wild-type protein, plakophilin-2-C603S–ER failed to localize efficiently to sites of cell–cell contact, and the recruitment of desmoplakin to the plasma membrane was impaired (Fig. 7G–I). Wild-type plakophilin-2–ER fusion protein becomes incorporated into a Triton-X-100-insoluble fraction (pellet fraction) following the addition of 4OHT. However, the mutant plakophilin–ER fusion protein remained in the Triton-X-100-soluble fraction, indicating the inability of the mutant fusion protein to initiate desmosome assembly in A431DE cells (Fig. 7K).
Fig. 7.
Plakophilin-2 C603S fails to efficiently initiate desmosome assembly in A431DE cells. In the absence of 4OHT, neither plakophilin-2–ER (Pkp-2/ER) nor desmoplakin (DP) localize to cell borders (A–C). Addition of 100 nM 4OHT to the medium for 18 hours results in plakophilin-2–ER localization to cell borders and the recruitment of desmoplakin (D–F). In contrast to the wild-type protein, plakophilin-2-C603S–ER (Pkp-2 C603S/ER) did not efficiently localize to cell borders and desmoplakin was not efficiently recruited (G–I). Scale bar: 10 µm. (J) Immunoblot (IB) analysis of A431DE cells expressing plakophilin-2–ER or plakophilin-2-C603S–ER demonstrates equivalent expression of the plakophilin-2 fusion proteins. β-tubulin is included as a loading control (K) A431DE cells expressing plakophilin-2–ER or plakophilin-2-C603S–ER were grown in medium containing 100 nM 4OHT for 18 hours. Triton-X-100-soluble (S) and Triton-X-100-insoluble pellet (P) fractions were prepared and immunoblotted with antibodies against plakophilin-2. Data were collected using Li-Cor Odyssey near-infrared imaging. Data show the mean±s.d. (n = 3); **P<0.01.
Expression of mutant plakophilins disrupts cell–cell adhesion
We examined the relative intercellular adhesive strength of A431 cells expressing Myc-tagged plakophilin-2, Myc-tagged plakophilin-3 and Myc-tagged plakophilin mutants using the dispase adhesion assay (Calautti et al., 1998; Huen et al., 2002). In this assay, a decrease in the number of cell fragments following mild mechanical stress indicates increased intercellular adhesion. As expected, A431 cells expressing wild-type Myc-tagged plakophilin-2 and plakophilin-3 conferred increased intercellular adhesion compared with that of parental A431 cells (Fig. 8). Others have previously shown that exogenous expression of plakophilin isoforms can increase intercellular adhesion (Roberts et al., 2013; Setzer et al., 2004; Wolf et al., 2013). By contrast, expression of the Myc-tagged plakophilin palmitoylation mutants failed to increase intercellular adhesion in these cultures. In fact, expression of the plakophilin palmitoylation mutants in A431 cells resulted in decreased adhesion compared with that of parental A431 cells. Taken together with the observation that desmoplakin localization is also disrupted in A431 cells expressing plakophilin mutants (Fig. 5; supplementary material Fig. S1), these data suggest that the plakophilin isoforms that are unable to be palmitoylated function as dominant-negative proteins with respect to strength of desmosomal adhesion.
Fig. 8.
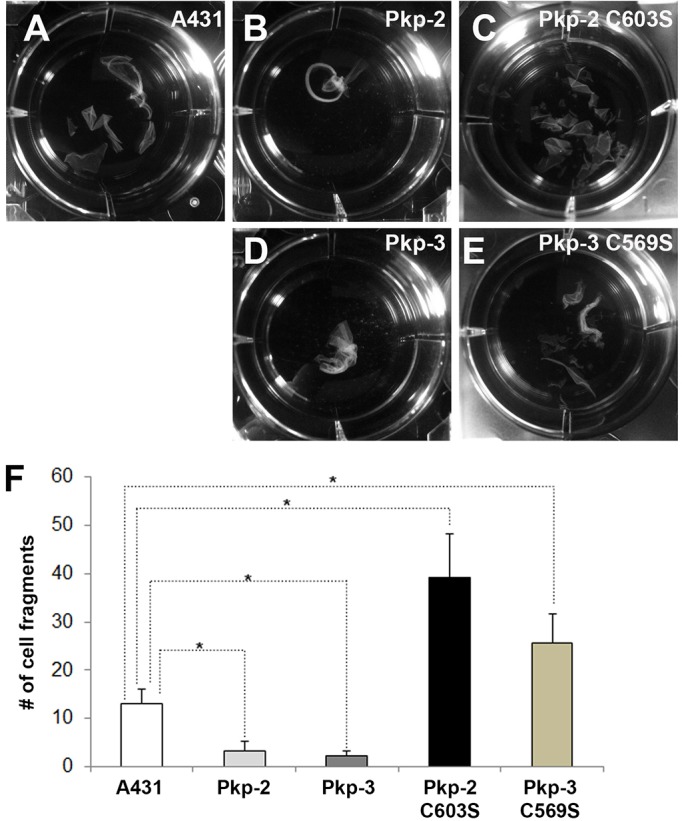
Expression of plakophilin palmitoylation mutants disrupts desmosomal adhesion. Dispase adhesion assays were performed to examine the relative strength of desmosomal adhesion in parental A431 cells (A) and in A431 cells expressing Myc-tagged plakophilin-2 (Pkp-2; B), Myc-tagged plakophilin-2 C603S (C), Myc-tagged plakophilin-3 (Pkp-3; D) and Myc-tagged plakophilin-3 C569S (E). A–E depict representative examples of cell sheet fragments counted in the dispase assay. (F) Quantification of the dispase assays. Data show the mean±s.d. (n = 3); *P<0.05.
DISCUSSION
In the present study, we have demonstrated that several desmosomal components are palmitoylated in vivo, uncovering a novel posttranslational modification as well as a potential mechanism of regulating junction dynamics. Previously, several desmosomal components were identified as potential palmitoylated proteins in a large-scale proteomics screen of palmitoylated proteins in DU145 prostate cells (Yang et al., 2010). Given the potential for a high incidence of ‘false positives’ associated with large proteomic data sets, we set out to show that desmosomal proteins are palmitoylated and to determine the role of palmitoylation in desmosome dynamics.
We demonstrated that both the desmosomal cadherins (desmoglein-2, desmoglein-3 and desmocollin-2) and desmosomal plaque proteins are palmitoylated, whereas the adherens junction constituents E-cadherin, α-catenin, β-catenin and desmoplakin are not palmitoylated. Plakophilin-2 and plakophilin-3 each contain eight cysteines. We mapped the site of palmitoylation in plakophilin-2 and plakophilin-3 to a conserved cysteine present in the armadillo repeat domain between armadillo repeats 5 and 6 (Choi and Weis, 2005). Recent studies have demonstrated that another p120 catenin family member is also palmitoylated. Delta-catenin has been shown to be palmitoylated in cultured hippocampal neurons, and disruption of the palmitoylation of delta-catenin decreases its association with N-cadherin. Interestingly, these authors identified two cysteines (C960 and C961) that are palmitoylated by the PAT DHHC5 (Brigidi et al., 2014). These cysteines are distinct from the conserved cysteine that we have identified in the plakophilins.
Mutation of the conserved cysteine residue in plakophilin-2 and plakophilin-3 resulted in the inability of the plakophilins to become palmitoylated, disrupted their localization to the plasma membrane and interfered with their ability to initiate desmosome assembly in A431D cells. Preventing global palmitoylation by 2-BP resulted in an inhibition of desmosome assembly in a calcium-switch assay. E-cadherin localization and adherens junction assembly were not disrupted following 2-BP incubation in HaCat cells, suggesting that 2-BP does not simply disrupt global cell–cell adhesion. Taken together, these data indicate that palmitoylation of the plakophilins is of particular importance during desmosome assembly, but do not address the possibility that palmitoylation of desmosomal proteins also plays a role in desmosome maturation, stability or disassembly. Further studies will be required to address these possibilities.
Palmitoylation is a reversible posttranslational modification resulting in the addition of 16-carbon palmitate to specific cysteine side chains through a thioester linkage. A family of PATs containing the conserved Cys-rich DHHC domain (Fukata et al., 2004; Roth et al., 2002) are responsible for the addition of palmitate to substrate proteins, whereas the removal of palmitate is catalyzed by cytosolic thioesterases (LYPLA1, LYPLA2 and LYPLA1L) (Tian et al., 2012; Veit and Schmidt, 2001; Zeidman et al., 2009). The reversible nature of this particular modulation makes it an attractive mechanism that is capable of regulating desmosome assembly, stability or even adhesive strength in epithelial cells. Currently, efforts are underway in our laboratory to identify the PATs responsible for palmitoylation of the plakophilins as well as the other desmosomal components. Identification of the PAT responsible for modification of the desmosomal components will be necessary to understand the sequence of events in palmitoylation-dependent desmosome assembly.
Several groups have demonstrated that the palmitoylation of substrate proteins results in increased association with cellular membranes, including the plasma membrane (Resh, 2006). Furthermore, palmitoylation is thought to play a role in the targeting of proteins to membrane microdomains. Recent studies have demonstrated that desmosomal components are associated with lipid rafts. (Brennan et al., 2012; Resnik et al., 2011; Stahley et al., 2014). Here, we have shown that plakophilin mutants that are unable to become palmitoylated show decreased association with lipid raft markers in sucrose-gradient centrifugation. The association of desmoglein-2 with raft fractions in this assay was not affected by plakophilin palmitoylation. It is likely that palmitoylation of the desmosomal components increases the incorporation of these proteins into lipid rafts and, ultimately, into desmosomal plaques. Palmitoylation might assist in the packing of desmosomal components in the plane of the membrane to generate the characteristic electron-dense plaque seen by electron microscopy. Characterization of the palmitoylation of additional desmosomal components should shed light on the role of lipid rafts on desmosome dynamics.
Based on the data presented here, we propose that palmitoylation of the plakophilins (and possibly other desmosomal components) affects the ability of these proteins to interact with the membrane in an ordered manner and affects junction assembly. Our studies were unable to examine the effect of palmitoylation on desmosome disassembly or degradation of desmosomal components. The effect of palmitoylation on these processes should be addressed in the future. Inhibition of global palmitoylation (by 2-BP treatment) prevents desmosome assembly following Ca2+ addition in HaCat cells, whereas E-cadherin localization is unaffected by inhibition of palmitoylation (Figs 2, 3). More specifically, plakophilin-2-C603S–ER is unable to initiate desmosome assembly in A431DE cells upon 4-OHT addition (Fig. 7). Recent studies have demonstrated that lipid raft association is required for desmosome assembly (Brennan et al., 2012; Resnik et al., 2011; Stahley et al., 2014). Palmitoylation is likely to influence the association of desmosomal components with this highly ordered membrane microdomain and thereby affect desmosome assembly.
Several interesting questions remain to be addressed. The PATs and thioesterases responsible for the addition and removal of palmitate from desmosomal components must be identified and their activity characterized with respect to desmosomal dynamics. It will be important to know in which specific membrane compartment the PATs reside and at which subcellular compartment do the desmosomal components become palmitoylated.
Additionally, it is currently unknown how palmitoylation of the plakophilin-2 and plakophilin-3 armadillo repeat domains influences membrane association. Because no binding partners of the plakophilin armadillo repeat domain have been identified, it is unclear whether palmitoylation alters protein–protein interaction or simply strengthens the association with membrane. Previous analysis of plakophilin-3 has shown that the interaction of plakophilin-3 with desmosomal components is mediated by the N-terminal head domain. Interestingly, the expression of the plakophilin-3 N-terminal head domain alone is not sufficient for recruitment to the plasma membrane, and expression of the armadillo repeat domain of plakophilin-3 results in its recruitment to the plasma membrane (Bonné et al., 2003; Roberts et al., 2011). Taken together, these data suggest that sequences in the head domain as well as the armadillo repeats are needed for proper localization and insertion into desmosomal complexes. Here, we have demonstrated that a single cysteine-to-serine substitution in the armadillo repeat domain disrupts membrane localization in A431 cells. We recently characterized desmosome assembly in migrating epithelial cells and we determined that desmosomes initiate assembly in close proximity to actin filaments between cells near the leading edge (Roberts et al., 2011). Plakophilin-1 has previously been shown to localize to the actin cytoskeleton when it is overexpressed in HaCat keratinocytes (Hatzfeld et al., 2000). Like plakophilin-2 and plakophilin-3, plakophilin-1 is also palmitoylated in cultured cells (data not shown). Palmitoylation of the plakophilins might influence the ability of these desmosomal components to interact with the actin cytoskeleton and initiate desmosome assembly.
MATERIALS AND METHODS
Cell culture
The A431 cervical squamous cell carcinoma cell line was obtained from ATCC (Manassas, VA). A431 and HaCat keratinocytes (Boukamp et al., 1988) (a kind gift from Pamela Jensen, University of Pennsylvania, PA) were routinely grown in DMEM (Sigma, St Louis, MO) supplemented with 10% fetal bovine serum (FBS) (HyClone Laboratories, Logan, UT). A431D and A431DE cells were described previously (Lewis et al., 1997; Wahl, 2005). Generation of retroviral particles and retroviral infection has been described previously (Roberts et al., 2011; Sobolik-Delmaire et al., 2007). Retrovirally infected cell populations were routinely grown in DMEM containing 500 µg/ml G418 (Mediatech, Herndon, VA). For calcium-switch experiments, HaCat cells were grown in DMEM lacking Ca2+ supplemented with 10% dialyzed FBS for at least 48 hours prior to the addition of 1.8 mM CaCl2 and 50 µM 2-BP.
Antibodies
Monoclonal antibodies specific for plakophilin-3 (11F2) and plakophilin-2 (8H6) were generated as described previously (Hall et al., 2009; Johnson et al., 1993; Roberts et al., 2011; Wahl, 2002). Antibodies against desmoplakin (20B6), desmoglein-2 (6D8), E-cadherin (4A2) and desmocollin-2/3 (7G6) were as described previously (Bazzi et al., 2006; Nieman et al., 1999; Sobolik-Delmaire et al., 2007; Wahl et al., 1996). Rabbit anti-desmoplakin (NW6) was a kind gift of Kathleen Green (Northwestern University, Chicago, IL). Rabbit anti-caveolin-1 (N20) was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-β-tubulin (E7) hybridoma was obtained from The Developmental Studies Hybridoma Bank (The University of Iowa, Iowa City, IA) and hybridoma supernatant was generated. Rabbit antibody against estrogen receptor was obtained from Sigma (St Louis, MO; catalog number E0646). The generation of cell lysates, immunoblot analysis and immunofluorescence microscopy were performed as described previously (Roberts et al., 2011; Sobolik-Delmaire et al., 2007).
Detergent extraction
Cell lysates were prepared as described previously (Sobolik-Delmaire et al., 2006; Wahl et al., 2000). Triton-X-100-soluble (s) and -insoluble (p) fractions were isolated by first washing cell monolayers with 1× phosphate buffered saline. Cells grown in a T25 flask were scraped into 1 ml of Triton-X-100-containing buffer (10 mM Tris-HCl pH 8.0, 0.5% Triton X-100 and 2 mM EDTA) and incubated on ice with shaking for 15 minutes. The insoluble material was collected by centrifugation for 15 minutes at 14,000 g. The insoluble pellet was washed once with Triton X-100 lysis buffer prior to resuspension of the pellet in 1 ml of SDS-containing buffer (10 mM Tris-HCl pH 8.0, 2% SDS, 2 mM EDTA). Lysates were prepared in Laemmli sample buffer and resolved by SDS-PAGE.
Generation of cDNA constructs
Human plakophilin-2a (NP_001005242) and plakophilin-3 (NP_009114) cDNA were as described previously (Hall et al., 2009; Roberts et al., 2011). Plakophilin-2 (pkp-2-ER) fused to the estrogen receptor ligand-binding domain T2 variant (Feil et al., 1997) was generated by standard procedures to remove the stop codon from each cDNA and add an XhoI restriction site. Plakophilin point mutations (pkp-2 C603S and pkp-3 C569S) were generated using the QuikChange™ site-directed mutagenesis kit (Stratagene/Agilent Technologies, Santa Clara, CA). The modified cDNAs were completely sequenced and shown to have no unintended changes. Addition of a 2×c-Myc epitope tag to the N-terminus was achieved by subcloning plakophilin cDNAs downstream of the cDNA encoding the epitope tag in a modified pSPUTK vector (Falcone and Andrews, 1991). The plakophilin cDNAs together with the c-Myc epitope tag were subsequently subcloned into an LZRS retroviral expression vector. Plakophilin-3–GFP has been described previously (Roberts et al., 2011). cDNA constructs were cloned into vectors based upon LZRS (Ireton et al., 2002), which were transfected into Phoenix cells for the generation of retroviral particles.
Acyl-biotin exchange
A431 cells were grown to 80–90% confluence and lysates were prepared as described previously (Wan et al., 2007). Cells were harvested in lysis buffer containing 1% Triton X-100 or 0.5% Empigen BB (EMD Millipore, Darmstadt, Germany) (150 mM NaCl, 50 mM Tris-HCl pH 7.4, 1% Triton X-100 or 0.5% Empigen BB, 5 mM EDTA) containing 10 mM NEM (N-ethylmaleimide, Sigma), and were collected by scraping on ice and passed through a 25-gauge needle. Triton X-100 was used for most ABE experiments but Empigen BB was used to solubilize desmoplakin from A431 cell monolayers. Samples were chloroform-methanol precipitated and the pellet was allowed to air dry for 2–3 minutes. The pellet was resuspended in 300 µl of 4% SDS buffer (4% SDS, 50 mM Tris-HCl pH 7.4, 5 mM EDTA) and diluted fourfold in lysis buffer containing 10 mM NEM. The samples are incubated at 4°C overnight with gentle agitation. NEM was removed by performing three sequential chloroform-methanol precipitations and, following the last precipitation, the pellet was resuspended in 100 µl of 4% SDS buffer. The sample was divided in two – one half was diluted fivefold with buffer containing 0.7 M hydroxylamine (+HA) [0.7 M hydroxylamine, 1 mM HPDP–biotin, 0.2% Triton X-100, 1 mM phenylmethanesulfonyl fluoride (PMSF) and 1× protease inhibitor cocktail (Sigma)] and the other half was diluted fivefold with buffer lacking hydroxylamine (−HA). Samples were incubated at room temperature with gentle rocking for 1 hour. Samples were chloroform-methanol extracted three times and the final pellets were resuspended in 240 µl of 4% SDS buffer and diluted with 960 µl of low-HPDP–biotin buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.4, 5 mM EDTA, 0.2 mM HPDP–biotin, 0.2% Triton X-100, 1 mM PMSF, 1× protease inhibitor cocktail). Samples were incubated at room temperature with gentle agitation for 1 hour prior to three sequential chloroform-methanol precipitations. The final pellet was resuspended in 75 µl of 2% SDS buffer (2% SDS, 50 mM Tris-HCl pH 7.4, 5 mM EDTA) and samples were diluted to 0.1% SDS in lysis buffer containing 0.2% Triton X-100, 1× protease inhibitor cocktail and 1 mM PMSF. Protein concentrations were determined and equal amounts of protein were added to streptavidin–agarose. Biotin-labeled proteins were captured on streptavidin–agarose and non-specifically bound proteins were removed by washing the beads in lysis buffer containing 0.1% SDS and 0.2% Triton X-100. Captured proteins were boiled in 2× Laemmli sample buffer and resolved by SDS-PAGE.
Metabolic labeling and acyl-biotin exchange
For 17-ODYA metabolic labeling, A431 cells were grown to 80% confluence in 100-mm tissue culture dishes and were treated with 100 µM 17-ODYA (17-octadecynoic acid; Cayman Chemical Co.) or DMSO vehicle (Sigma) for 48 hours (Zoltewicz et al., 2012). To facilitate dissolution of 17-ODYA in the medium, 75 µl of 20 mM 17-ODYA stock in DMSO (or DMSO only) mixed with 150 µl 10% fatty-acid-free bovine serum albumin (Sigma) was added to 15 ml of DMEM, and the mixture was vortexed prior to being added to cells. Cells were lysed in extraction buffer containing 0.5% Empigen BB detergent (10 mM Tris-HCl pH 8.0, 0.5% Empigen BB, 2 mM EDTA), and insoluble proteins were pelleted by centrifugation (14,000 g for 15 min). Plakophilins were immunoprecipitated from Empigen-BB-soluble lysates and immune complexes were washed three times in TBST (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% Tween-20). Immunoprecipitated proteins were eluted from the beads in 47 µl of elution buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 2% SDS). The Cu-catalyzed click reaction was performed as described previously (Charron et al., 2009). Briefly, 47 µl of eluted protein was added to 0.25 µl of 10 mM biotin azide (Invitrogen), 0.5 µl of 50 mM TCEP [tris-(2-carboxyethyl)phosphine] hydrochloride (Sigma), 0.25 µl of 10 mM TBTA {tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine} (Sigma) and 0.5 µl of 50 mM CuSO4 (Sigma) for a total reaction volume of 50 µl, and the solution was incubated at room temperature for 1 hour. Samples were prepared in 1× Laemmli sample buffer containing 2-mercaptoethanol. Samples were not boiled prior to SDS-PAGE. Labeled proteins were separated by SDS-PAGE and biotin-labeled proteins were detected by blotting with HRP-labeled streptavidin (Jackson ImmunoResearch, West Grove, PA)
Lipid raft fraction analysis
Cell lysates were prepared from A431 cell lines expressing wild-type or mutant plakophilins and analyzed by sucrose-gradient centrifugation as described previously (McGuinn and Mahoney, 2014).
Dispase assays
Cells were grown to confluence in six-well dishes and, 24 hours post-confluence, the cell sheets were removed from the culture dish by incubating with dispase (Roche Applied Science, Indianapolis, IN). Cell sheets were carefully transferred to 15-ml conical tubes and subjected to mechanical stress by inversion. Cell sheet fragments were counted, assays were performed in triplicate and the data are expressed as the average number of fragments per well.
Statistical analysis
For all experiments, error bars represent standard deviation, and data were compared using two-tailed Student's t-tests assuming unequal variances. Statistical significance was assumed when P<0.05. *P<0.05 and **P<0.01.
Supplementary Material
Footnotes
Competing interests
K.R.J. and J.K.W. receive royalties from the sale of some of the antibodies used in these studies.
Author contributions
B.J.R. performed the microscopy. B.J.R. and K.E.J. performed acyl-biotin exchange experiments and metabolic labeling experiments. K.P.M. and M.G.M. performed the lipid raft analysis. J.S. and R.A.S. performed the site-directed mutagenesis and assembled the cDNA constructs for this study. The experiments were directed by K.R.J. and J.K.W., and the manuscript was primarily written by B.R.J., K.R.J. and J.K.W., with input from the other authors.
Funding
This project was supported by grants from the National Institute of General Medical Sciences [grant number P20GM103489 to J.K.W. and K.R.J.]; and National Institute of Arthritis and Musculoskeletal and Skin Diseases [grant number R01AR056067 to M.G.M.] from the National Institutes of Health. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.149849/-/DC1
References
- Aicart-Ramos C., Valero R. A., Rodriguez-Crespo I. (2011). Protein palmitoylation and subcellular trafficking. Biochim. Biophys. Acta 1808, 2981–2994 10.1016/j.bbamem.2011.07.009 [DOI] [PubMed] [Google Scholar]
- Bazzi H., Getz A., Mahoney M. G., Ishida-Yamamoto A., Langbein L., Wahl J. K., 3rd, Christiano A. M. (2006). Desmoglein 4 is expressed in highly differentiated keratinocytes and trichocytes in human epidermis and hair follicle. Differentiation 74, 129–140 10.1111/j.1432-0436.2006.00061.x [DOI] [PubMed] [Google Scholar]
- Bonné S., Gilbert B., Hatzfeld M., Chen X., Green K. J., van Roy F. (2003). Defining desmosomal plakophilin-3 interactions. J. Cell Biol. 161, 403–416 10.1083/jcb.200303036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukamp P., Petrussevska R. T., Breitkreutz D., Hornung J., Markham A., Fusenig N. E. (1988). Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J. Cell Biol. 106, 761–771 10.1083/jcb.106.3.761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan D., Peltonen S., Dowling A., Medhat W., Green K. J., Wahl J. K., 3rd, Del Galdo F., Mahoney M. G. (2012). A role for caveolin-1 in desmoglein binding and desmosome dynamics. Oncogene 31, 1636–1648 10.1038/onc.2011.346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigidi G. S., Sun Y., Beccano-Kelly D., Pitman K., Mobasser M., Borgland S. L., Milnerwood A. J., Bamji S. X. (2014). Palmitoylation of δ-catenin by DHHC5 mediates activity-induced synapse plasticity. Nat. Neurosci. 17, 522–532 10.1038/nn.3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke M. A., Nitoiu D., Kelsell D. P. (2012). Cell-cell connectivity: desmosomes and disease. J. Pathol. 226, 158–171 10.1002/path.3027 [DOI] [PubMed] [Google Scholar]
- Calautti E., Cabodi S., Stein P. L., Hatzfeld M., Kedersha N., Paolo Dotto G. (1998). Tyrosine phosphorylation and src family kinases control keratinocyte cell-cell adhesion. J. Cell Biol. 141, 1449–1465 10.1083/jcb.141.6.1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron G., Zhang M. M., Yount J. S., Wilson J., Raghavan A. S., Shamir E., Hang H. C. (2009). Robust fluorescent detection of protein fatty-acylation with chemical reporters. J. Am. Chem. Soc. 131, 4967–4975 10.1021/ja810122f [DOI] [PubMed] [Google Scholar]
- Choi H. J., Weis W. I. (2005). Structure of the armadillo repeat domain of plakophilin 1. J. Mol. Biol. 346, 367–376 10.1016/j.jmb.2004.11.048 [DOI] [PubMed] [Google Scholar]
- Collins J. E., Legan P. K., Kenny T. P., MacGarvie J., Holton J. L., Garrod D. R. (1991). Cloning and sequence analysis of desmosomal glycoproteins 2 and 3 (desmocollins): cadherin-like desmosomal adhesion molecules with heterogeneous cytoplasmic domains. J. Cell Biol. 113, 381–391 10.1083/jcb.113.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davda D., El Azzouny M. A., Tom C. T., Hernandez J. L., Majmudar J. D., Kennedy R. T., Martin B. R. (2013). Profiling targets of the irreversible palmitoylation inhibitor 2-bromopalmitate. ACS Chem. Biol. 8, 1912–1917 10.1021/cb400380s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies E. L., Gee J. M., Cochrane R. A., Jiang W. G., Sharma A. K., Nicholson R. I., Mansel R. E. (1999). The immunohistochemical expression of desmoplakin and its role in vivo in the progression and metastasis of breast cancer. Eur. J. Cancer 35, 902–907 10.1016/S0959-8049(99)00031-3 [DOI] [PubMed] [Google Scholar]
- Falcone D., Andrews D. W. (1991). Both the 5′ untranslated region and the sequences surrounding the start site contribute to efficient initiation of translation in vitro. Mol. Cell. Biol. 11, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R., Wagner J., Metzger D., Chambon P. (1997). Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem. Biophys. Res. Commun. 237, 752–757 10.1006/bbrc.1997.7124 [DOI] [PubMed] [Google Scholar]
- Fukata M., Fukata Y., Adesnik H., Nicoll R. A., Bredt D. S. (2004). Identification of PSD-95 palmitoylating enzymes. Neuron 44, 987–996 10.1016/j.neuron.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Gerull B., Heuser A., Wichter T., Paul M., Basson C. T., McDermott D. A., Lerman B. B., Markowitz S. M., Ellinor P. T., MacRae C. A. et al. (2004). Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 36, 1162–1164 10.1038/ng1461 [DOI] [PubMed] [Google Scholar]
- Greaves J., Chamberlain L. H. (2011). DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 10.1016/j.tibs.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Hall C., Li S., Li H., Creason V., Wahl J. K., 3rd (2009). Arrhythmogenic right ventricular cardiomyopathy plakophilin-2 mutations disrupt desmosome assembly and stability. Cell Commun. Adhes. 16, 15–27 10.1080/15419060903009329 [DOI] [PubMed] [Google Scholar]
- Harada H., Iwatsuki K., Ohtsuka M., Han G. W., Kaneko F. (1996). Abnormal desmoglein expression by squamous cell carcinoma cells. Acta Derm. Venereol. 76, 417–420 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M., Haffner C., Schulze K., Vinzens U. (2000). The function of plakophilin 1 in desmosome assembly and actin filament organization. J. Cell Biol. 149, 209–222 10.1083/jcb.149.1.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen A. C., Park J. K., Godsel L. M., Chen X., Bannon L. J., Amargo E. V., Hudson T. Y., Mongiu A. K., Leigh I. M., Kelsell D. P. et al. (2002). Intermediate filament-membrane attachments function synergistically with actin-dependent contacts to regulate intercellular adhesive strength. J. Cell Biol. 159, 1005–1017 10.1083/jcb.200206098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton R. C., Davis M. A., van Hengel J., Mariner D. J., Barnes K., Thoreson M. A., Anastasiadis P. Z., Matrisian L., Bundy L. M., Sealy L. et al. (2002). A novel role for p120 catenin in E-cadherin function. J. Cell Biol. 159, 465–476 10.1083/jcb.200205115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Lewis J. E., Li D., Wahl J., Soler A. P., Knudsen K. A., Wheelock M. J. (1993). P- and E-cadherin are in separate complexes in cells expressing both cadherins. Exp. Cell Res. 207, 252–260 10.1006/excr.1993.1191 [DOI] [PubMed] [Google Scholar]
- Koch P. J., Goldschmidt M. D., Walsh M. J., Zimbelmann R., Schmelz M., Franke W. W. (1991). Amino acid sequence of bovine muzzle epithelial desmocollin derived from cloned cDNA: a novel subtype of desmosomal cadherins. Differentiation 47, 29–36 10.1111/j.1432-0436.1991.tb00218.x [DOI] [PubMed] [Google Scholar]
- Lewis J. E., Wahl J. K., 3rd, Sass K. M., Jensen P. J., Johnson K. R., Wheelock M. J. (1997). Cross-talk between adherens junctions and desmosomes depends on plakoglobin. J. Cell Biol. 136, 919–934 10.1083/jcb.136.4.919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B. R., Cravatt B. F. (2009). Large-scale profiling of protein palmitoylation in mammalian cells. Nat. Methods 6, 135–138 10.1038/nmeth.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J. A., McMillan J. R., Shemanko C. S., Runswick S. K., Leigh I. M., Lane E. B., Garrod D. R., Eady R. A. (1997). Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nat. Genet. 17, 240–244 10.1038/ng1097-240 [DOI] [PubMed] [Google Scholar]
- McGuinn K. P., Mahoney M. G. (2014). Lipid Rafts and Detergent-Resistant Membranes in Epithelial Keratinocytes. Methods Mol. Biol 10.1007/7651_2014_71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman M. T., Kim J. B., Johnson K. R., Wheelock M. J. (1999). Mechanism of extracellular domain-deleted dominant negative cadherins. J. Cell Sci. 112, 1621–1632 [DOI] [PubMed] [Google Scholar]
- Wahl J. K., III, Nieset J. E., Sacco-Bubulya P. A., Sadler T. M., Johnson K. R., Wheelock M. J. (2000). The amino- and carboxyl-terminal tails of (beta)-catenin reduce its affinity for desmoglein 2. J. Cell Sci. 113, 1737–1745 [DOI] [PubMed] [Google Scholar]
- Ohno Y., Kashio A., Ogata R., Ishitomi A., Yamazaki Y., Kihara A. (2012). Analysis of substrate specificity of human DHHC protein acyltransferases using a yeast expression system. Mol. Biol. Cell 23, 4543–4551 10.1091/mbc.E12-05-0336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J., Wen L., Gao X., Jin C., Xue Y., Yao X. (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644 10.1093/protein/gzn039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resh M. D. (2006). Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006, re14 10.1126/stke.3592006re14 [DOI] [PubMed] [Google Scholar]
- Resnik N., Sepcic K., Plemenitas A., Windoffer R., Leube R., Veranic P. (2011). Desmosome assembly and cell-cell adhesion are membrane raft-dependent processes. J. Biol. Chem. 286, 1499–1507 10.1074/jbc.M110.189464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. J., Pashaj A., Johnson K. R., Wahl J. K., III (2011). Desmosome dynamics in migrating epithelial cells requires the actin cytoskeleton. Exp. Cell Res. 317, 2814–2822 10.1016/j.yexcr.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. J., Reddy R., Wahl J. K., III (2013). Stratifin (14-3-3 σ) limits plakophilin-3 exchange with the desmosomal plaque. PLoS ONE 8, e77012 10.1371/journal.pone.0077012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A. F., Feng Y., Chen L., Davis N. G. (2002). The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 10.1083/jcb.200206120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz P., Brinkmann V., Ledermann B., Behrend M., Grund C., Thalhammer C., Vogel F., Birchmeier C., Günthert U., Franke W. W. et al. (1996). Targeted mutation of plakoglobin in mice reveals essential functions of desmosomes in the embryonic heart. J. Cell Biol. 135, 215–225 10.1083/jcb.135.1.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setzer S. V., Calkins C. C., Garner J., Summers S., Green K. J., Kowalczyk A. P. (2004). Comparative analysis of armadillo family proteins in the regulation of a431 epithelial cell junction assembly, adhesion and migration. J. Invest. Dermatol. 123, 426–433 10.1111/j.0022-202X.2004.23319.x [DOI] [PubMed] [Google Scholar]
- Sobolik-Delmaire T., Katafiasz D., Wahl J. K., III (2006). Carboxyl terminus of Plakophilin-1 recruits it to plasma membrane, whereas amino terminus recruits desmoplakin and promotes desmosome assembly. J. Biol. Chem. 281, 16962–16970 10.1074/jbc.M600570200 [DOI] [PubMed] [Google Scholar]
- Sobolik-Delmaire T., Katafiasz D., Keim S. A., Mahoney M. G., Wahl J. K., III (2007). Decreased plakophilin-1 expression promotes increased motility in head and neck squamous cell carcinoma cells. Cell Commun. Adhes. 14, 99–109 10.1080/15419060701463082 [DOI] [PubMed] [Google Scholar]
- Stahley S. N., Saito M., Faundez V., Koval M., Mattheyses A. L., Kowalczyk A. P. (2014). Desmosome assembly and disassembly are membrane raft-dependent. PLoS ONE 9, e87809 10.1371/journal.pone.0087809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H., Hayashi H., Yamashita S. (1996). Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J. Biol. Chem. 271, 7705–7711 10.1074/jbc.271.13.7705 [DOI] [PubMed] [Google Scholar]
- Thomas G. J., Speight P. M. (2001). Cell adhesion molecules and oral cancer. Crit. Rev. Oral Biol. Med. 12, 479–498 10.1177/10454411010120060301 [DOI] [PubMed] [Google Scholar]
- Thomason H. A., Cooper N. H., Ansell D. M., Chiu M., Merrit A. J., Hardman M. J., Garrod D. R. (2012). Direct evidence that PKCα positively regulates wound re-epithelialization: correlation with changes in desmosomal adhesiveness. J. Pathol. 227, 346–356 10.1002/path.4016 [DOI] [PubMed] [Google Scholar]
- Tian L., McClafferty H., Knaus H. G., Ruth P., Shipston M. J. (2012). Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium channels. J. Biol. Chem. 287, 14718–14725 10.1074/jbc.M111.335547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomatis V. M., Trenchi A., Gomez G. A., Daniotti J. L. (2010). Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS ONE 5, e15045 10.1371/journal.pone.0015045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda T., Sugimoto H., Yamashita S. (1999). Sequence, expression in Escherichia coli, and characterization of lysophospholipase II. Biochim. Biophys. Acta 1437, 182–193 10.1016/S1388-1981(99)00007-4 [DOI] [PubMed] [Google Scholar]
- Veit M., Schmidt M. F. (2001). Enzymatic depalmitoylation of viral glycoproteins with acyl-protein thioesterase 1 in vitro. Virology 288, 89–95 10.1006/viro.2001.1063 [DOI] [PubMed] [Google Scholar]
- Wahl J. K., 3rd (2002). Generation of monoclonal antibodies specific for desmoglein family members. Hybrid. Hybridomics 21, 37–44 10.1089/15368590252917629 [DOI] [PubMed] [Google Scholar]
- Wahl J. K., 3rd (2005). A role for plakophilin-1 in the initiation of desmosome assembly. J. Cell. Biochem. 96, 390–403 10.1002/jcb.20514 [DOI] [PubMed] [Google Scholar]
- Wahl J. K., Sacco P. A., McGranahan-Sadler T. M., Sauppé L. M., Wheelock M. J., Johnson K. R. (1996). Plakoglobin domains that define its association with the desmosomal cadherins and the classical cadherins: identification of unique and shared domains. J. Cell Sci. 109, 1143–1154 [DOI] [PubMed] [Google Scholar]
- Wan J., Roth A. F., Bailey A. O., Davis N. G. (2007). Palmitoylated proteins: purification and identification. Nat. Protoc. 2, 1573–1584 10.1038/nprot.2007.225 [DOI] [PubMed] [Google Scholar]
- Wolf A., Rietscher K., Glaß M., Hüttelmaier S., Schutkowski M., Ihling C., Sinz A., Wingenfeld A., Mun A., Hatzfeld M. (2013). Insulin signaling via Akt2 switches plakophilin 1 function from stabilizing cell adhesion to promoting cell proliferation. J. Cell Sci. 126, 1832–1844 10.1242/jcs.118992 [DOI] [PubMed] [Google Scholar]
- Yang W., Di Vizio D., Kirchner M., Steen H., Freeman M. R. (2010). Proteome scale characterization of human S-acylated proteins in lipid raft-enriched and non-raft membranes. Mol. Cell. Proteomics 9, 54–70 10.1074/mcp.M800448-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidman R., Jackson C. S., Magee A. I. (2009). Protein acyl thioesterases. Mol. Membr. Biol. 26, 32–41 10.1080/09687680802629329 [DOI] [PubMed] [Google Scholar]
- Zoltewicz S. J., Lee S., Chittoor V. G., Freeland S. M., Rangaraju S., Zacharias D. A., Notterpek L. (2012). The palmitoylation state of PMP22 modulates epithelial cell morphology and migration. ASN Neuro 4, 409–421 10.1042/AN20120045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.