Abstract
The interleukin-1 (IL-1) gene polymorphisms have been implicated in chronic obstructive pulmonary disease (COPD) risk, but results are controversial. We aimed to conduct a meta-analysis to address this issue. Odds ratio (OR) and 95% confidence interval (CI) were used to investigate the strength of the association. The meta-analysis revealed no association between the IL1B (−511), (−31), (+3954) polymorphisms and COPD risk. However, stratification by ethnicity indicated that the T allele carriers of the IL1B (−511) polymorphism and the C allele carriers of the IL1B (−31) variant were associated with an increased risk for developing COPD in East Asians (OR = 1.61, 95% CI: 1.13–2.31, Pz = 0.009 and OR = 1.55, 95% CI: 1.14–2.11, Pz = 0.006, respectively). The meta-analysis revealed a significant association between the IL1RN (VNTR) polymorphism and COPD risk in all study subjects and East Asians under homozygote model (22 vs. LL: OR = 3.16, 95% CI: 1.23–8.13, Pz = 0.017 and OR = 3.20, 95% CI: 1.13–9.12, Pz = 0.029, respectively). Our meta-analysis suggests that the IL1B (−511), (−31) and IL1RN (VNTR) polymorphisms are associated with COPD risk in East Asians. There is no association between the IL1B (+3954) polymorphism and COPD risk. Further studies should be performed in other ethnic groups besides East Asians.
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory disease characterized by the gradual progression of irreversible airflow obstruction and increased inflammation in the airways and lung parenchyma1. It is a leading cause of chronic morbidity and mortality worldwide. Although cigarette smoking has been showed to be a major environmental risk factor for COPD, only 10–20% of smokers develop COPD, implying that apart from environmental features, additional risk factors such as genetic variation contributes to COPD susceptibility2. Genetic linkage studies, candidate gene association studies and genome-wide association studies (GWAS) have identified genes that may have roles in the pathogenesis of COPD, including microsomal epoxide hydrolase (EPHX1), glutathione S-transferase (GST), interleukin-6 (IL-6), iron-responsive element binding protein 2 (IREB2), matrix metallopeptidase 9 (MMP9) and transforming growth factor-β (TGF-β)3,4. A better understanding of the genetic architecture of the disease will be important in helping to unravel the pathogenetic mechanism and develop novel therapeutic strategies for COPD.
IL-1 is a pro-inflammatory cytokine and key contributor to immune responses. IL-1 occurs in two forms, IL-1A and IL-1B, both of which bind to the IL-1 receptor5. The IL-1 receptor antagonist (IL-1RA) does not transmit any signal and functions as a cell bound inhibitor to both IL-1A and IL-1B5. The loci for human IL1A, IL1B, and IL1RN gene are found as a cluster on chromosome 2q12 to 2q146. The genes are arranged with IL1A situated 5-prime, and then IL1B and finally IL1RN 3-prime. Several common polymorphisms within the IL-1 gene complex have been described, including single nucleotide polymorphisms (SNPs) in IL1B at position −511 (rs16944) and −31 (rs1143627) in the promoter region and at position +3954 (rs1143634) in exon 5, and a penta-allelic polymorphism representing a variable number of an 86-bp tandem repeat in intron 2 (rs2234663) of the IL1RN gene7,8. Many genetic association studies have been conducted to investigate the relationship of these variants with COPD risk; however, small sample size and varying population characteristics led to conflicting results. To validate the potential association between the IL1B, IL1RN polymorphisms and COPD risk, we performed a meta-analysis of data reported in 12 case-control studies from 11 publications.
Methods
Search strategy and study identification
We conducted electronic searches, not limited to the English language, of PubMed, Embase, Scopus, Cochrane database, China National Knowledge Infrastructure (CNKI) and Wanfang databases to identify relevant studies reporting on the association between the IL1B, IL1RN polymorphisms and COPD risk in the medical literature from January 1990 up to the end of June 2014. Search terms included “chronic obstructive pulmonary disease”, “COPD”, “interleukin-1”, “IL-1”, “polymorphism”, “genetics”, and “association”. References from retrieved papers were also considered. Association studies were included in this meta-analysis if they met the following criteria: 1) studies on human subjects; 2) studies written in English or Chinese; 3) case-control design; 4) sufficient data for examining an odds ratio (OR) with 95% confidence interval (CI). The major exclusion criteria were as follows: 1) studies on animal populations; 2) no usable data reported; 3) duplicate data; 4) studies presenting deviation from Hardy-Weinberg equilibrium (HWE) in the control group. All relevant publications identified through the search were scanned on the basis of title, keywords and abstract, and were rejected in the initial screening if the paper clearly did not meet the inclusion criteria. Where a title/abstract could not be rejected with certainty, the full text of the publications was obtained for evaluation.
Data extraction
Two independent investigators extracted data from the published studies and entered them in a customized database. Disagreement was resolved by the evaluation of a third reviewer and discussion until a consensus was reached. For each study, the following data were collected: first author's name, country, year of publication, ethnicity, total number of cases and controls, characteristics of the control group, and genotypic frequencies of the IL-1 polymorphisms.
Quality score assessment
The quality of selected studies was evaluated by scoring according to Newcastle Ottawa Scale (NOS) (www.ohri.ca/programs/clinical_epidemiology/oxford.asp). This scale consists of three parts relating to selection, comparability and ascertainment of exposure. Quality scores ranged from 0 to 9 and studies were regarded as “high quality” if the score was ≥ 5. Otherwise, studies was regarded as “low quality”.
Data analysis
Data were entered and analyzed with the Stata version 11.0. We used raw data of genotypic distribution, without adjustment for calculation of the study-specific estimates of OR and 95% CI. Dominant, recessive and homozygote models were evaluated. Z-test was used for assessing the significance of the pooled OR, with P<0.05 considered statistically significant. Between-study heterogeneity was evaluated with Cochran's Q-test statistics among the studies, with significance level set at 0.10. The DerSimonian Laird random-effects model rather than the Mantel-Haenszel fixed-effect model was employed to calculate the pooled OR and 95% CI9,10. For the IL1RN (VNTR) polymorphism, L signifies any long allele embracing allele 1, 3, 4, or 5. We created a funnel plot to assess potential publication bias by plotting natural logarithm of individual study effect size against the standard error of the natural logarithm of individual study effect size. We also assessed publication bias using the Begg's test and Egger's test. The HWE law states that if two alleles, T and C, with frequencies p and q = 1-p, are in equilibrium in a population, then the proportion of people with genotypes TT, TC and CC will be p2, 2pq and q2. HWE in the control group for each study was tested by using a web-based programme (http://www.oege.org/software/hwe-mr-calc.html). Since we only utilized previously published data, we did not obtain approval of an ethics committee or written informed consent.
Results
Included studies
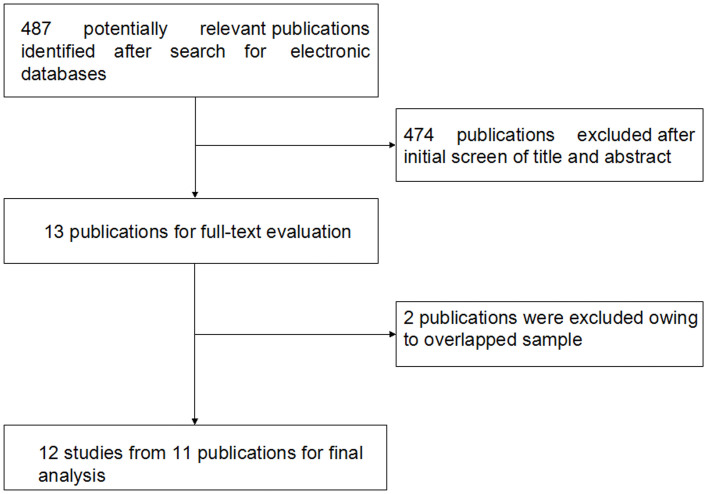
Figure 1 showed the process of identifying eligible studies. 487 publications were identified from the initial keywords search. After review, 474 were excluded. Overall, 12 studies from 11 publications met our inclusion criteria with a total of 1530 COPD patients and 1524 controls11,12,13,14,15,16,17,18,19,20,21. Table 1 summarized the characteristics of the studies included in the meta-analysis. In terms of ethnicity, eight studies including 1055 cases and 1199 controls were performed in East Asians11,12,14,15,16,17,19,20, two studies containing 169 cases and 97 controls were undertaken in Arabians12,21, one study with 102 cases and 20 controls was conducted in Europeans13, and one study including 204 cases and 208 controls was performed in South Asians18. The publication of Hegab et al. contained an Egyptian study and a Japanese study, respectively12. It was noteworthy that we did not extract data for the IL1B (−511), (−31) polymorphisms from the Liu et al. study17, since they further evaluated these two polymorphisms using larger sample numbers in another study19. In addition, the data for the IL1B (−31) SNP from the Liu et al. study19 and the data for the IL1RN (VNTR) polymorphism from the Hegab et al. study12, the Lee et al. study16, and the Shukla et al. study18 presented deviations from HWE in controls, respectively, so we did not include them in the final analysis.
Figure 1. Flowchart of study selection process.
Table 1. Characteristics of the studies evaluating IL-1 gene polymorphisms and COPD risk.
| Number | Characteristics of controls | |||||||
|---|---|---|---|---|---|---|---|---|
| First author | Country or Area | Ethnicity | Year | Cases | Controls | IL-1 polymorphisms | Score | |
| Ishii | Japan | East Asians | 2000 | 53 | 65 | Sex- and smoking history-matched healthy subjects with normal pulmonary function | IL1B (−511), IL1B (+3954) and IL1RN (VNTR) | 6 |
| Hegab | Egypt and Japan | Arabians and East Asians | 2005 | 88 (East Asians) 106 (Arabians) | 61 (East Asians) 72 (Arabians) | Age- and smoking history-matched healthy subjects with normal pulmonary function | IL1B (−511), IL1B (−31), IL1B (+3954) and IL1RN (VNTR) | 8 |
| Broekhuizen | Netherlands | Europeans | 2005 | 102 | 20 | Age- and sex-matched healthy volunteers | IL1B (−511) | 9 |
| Shi | China | East Asians | 2006 | 88 | 96 | Age- and sex-matched healthy subjects with smoking history | IL1A (VNTR), IL1B (−511) and IL1RN (VNTR) | 7 |
| Hsieh | Taiwan | East Asians | 2008 | 30 | 115 | Age- and sex-matched healthy subjects | IL1B (−31) and IL1RN (VNTR) | 8 |
| Lee | Korea | East Asians | 2008 | 311 | 386 | Sex- but not age-matched healthy subjects | IL1B (−3737), IL1B (−1464), IL1B (−511), IL1B (−31), and IL1RN (VNTR) | 7 |
| Liu | China | East Asians | 2012 | 162 | 162 | Age- and sex-matched healthy subjects | IL1B (−511), IL1B (−31), IL1B (+3954) | 7 |
| Shukla | India | South Asians | 2012 | 204 | 208 | Healthy age-and sex- matched subjects | IL1B (−511) and IL1RN (VNTR) | 7 |
| Liu | China | East Asians | 2013 | 260 | 260 | Healthy age-and sex- matched subjects | IL1B (−511) and IL1B (−31) | 6 |
| Sun | China | East Asians | 2013 | 63 | 54 | Age-and sex- matched healthy smokers | IL1B (−511) | 7 |
| Issac | Egypt | Arabians | 2014 | 63 | 25 | Age- matched smokers with no clinical suspicion of COPD and a normal spirometry | IL1B (−511) and IL1RN (VNTR) | 7 |
IL-1, interleukin-1; COPD, chronic obstructive pulmonary disease.
Individual polymorphism meta-analysis
Table 2 showed genotypic distribution of the four polymorphisms. The genotype-wise meta-analytic results for each of the four polymorphisms were showed in Table 3.
Table 2. Genotypic distribution of the IL-1 polymorphisms in cases and controls.
| Polymorphisms | Cases | Controls | HWE in controls | ||||
|---|---|---|---|---|---|---|---|
| IL1B (-511) | CC | TC | TT | CC | TC | TT | |
| Ishii et al | 14 | 29 | 10 | 16 | 27 | 22 | Yes |
| Hegab et ala | 20 | 52 | 16 | 21 | 31 | 8 | Yes |
| Hebab et alb | 49 | 45 | 11 | 26 | 29 | 16 | Yes |
| Broekhuizen et al | 54 | 39 | 5 | 8 | 9 | 3 | Yes |
| Shi et al | 14 | 48 | 26 | 36 | 44 | 16 | Yes |
| Lee et al | 62 | 174 | 75 | 107 | 175 | 104 | Yes |
| Shukla et al | 31 | 93 | 80 | 23 | 101 | 84 | Yes |
| Liu et al | 44 | 164 | 52 | 45 | 158 | 57 | Yes |
| Sun et al | 12 | 32 | 19 | 21 | 26 | 7 | Yes |
| IL1B (-31) | TT | CT | CC | TT | CT | CC | |
| Hegab et ala | 20 | 52 | 16 | 21 | 31 | 8 | Yes |
| Hegab et alb | 49 | 45 | 11 | 26 | 29 | 16 | Yes |
| Hsieh et al | 6 | 18 | 6 | 28 | 64 | 23 | Yes |
| Lee et al | 58 | 179 | 74 | 100 | 177 | 109 | Yes |
| Liu et al | 54 | 151 | 55 | 50 | 153 | 57 | No |
| IL1B (+3954) | CC | TC | TT | CC | TC | TT | |
| Ishii et al | 49 | 4 | 0 | 58 | 7 | 0 | Yes |
| Hegab et ala | 78 | 10 | 0 | 55 | 6 | 0 | Yes |
| Hegab et alb | 50 | 45 | 8 | 37 | 29 | 5 | Yes |
| Liu et al | 141 | 21 | 0 | 146 | 15 | 1 | Yes |
| IL1RN (VNTR) | LL | 2L | 22 | LL | 2L | 22 | |
| Ishii et al | 49 | 3 | 1 | 58 | 6 | 1 | Yes |
| Hegab et ala | 80 | 8 | 0 | 55 | 6 | 0 | Yes |
| Hegab et alb | 83 | 18 | 5 | 50 | 15 | 7 | No |
| Shi et al | 45 | 32 | 11 | 71 | 21 | 4 | Yes |
| Hsieh et al | 24 | 6 | 0 | 99 | 15 | 1 | Yes |
| Lee et al | 296 | 15 | 0 | 347 | 35 | 4 | No |
| Shukla et al | 104 | 86 | 14 | 110 | 74 | 24 | No |
| Issac et al | 38 | 19 | 6 | 19 | 5 | 1 | Yes |
IL-1, interleukin-1; HWE, Hardy–Weinberg equilibrium.
aHegab et al, a study containing Japanese subjects.
bHegab et al, a study containing Egyptian subjects.
Table 3. Meta-analysis of the relationship of the IL-1 polymorphisms with COPD risk.
| No. of studies | Dominant | Recessive | Homozygote | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Polymorphism | OR (95% CI) | Phet | Pz | OR (95% CI) | Phet | Pz | OR (95% CI) | Phet | Pz | |
| IL1B (-511)a | ||||||||||
| Total | 9 | 1.22 (0.84-1.75) | 0.002 | 0.293 | 0.94 (0.67-1.32) | 0.010 | 0.730 | 1.10 (0.65-1.85) | <0.001 | 0.734 |
| East Asians | 6 | 1.61 (1.13-2.31) | 0.061 | 0.009 | 1.13 (0.74-1.73) | 0.017 | 0.574 | 1.62 (0.91-2.88) | 0.005 | 0.105 |
| IL1B (-31)b | ||||||||||
| Total | 4 | 1.25 (0.79-1.96) | 0.101 | 0.340 | 0.80 (0.52-1.21) | 0.225 | 0.287 | 1.00 (0.52-1.95) | 0.065 | 0.993 |
| East Asians | 3 | 1.55 (1.14-2.11) | 0.842 | 0.006 | 0.87 (0.64-1.18) | 0.469 | 0.356 | 1.27 (0.87-1.87) | 0.599 | 0.221 |
| IL1B (+3954)c | ||||||||||
| Total | 4 | 1.16 (0.78-1.73) | 0.830 | 0.464 | 0.97 (0.32-2.88) | 0.486 | 0.951 | 1.02 (0.33-3.12) | 0.480 | 0.974 |
| East Asians | 3 | 1.16 (0.75-1.78) | 0.645 | 0.503 | 0.33 (0.01-8.19) | NA | 0.500 | 0.35 (0.01-8.54) | NA | 0.516 |
| IL1RN (VNTR)d | ||||||||||
| Total | 5 | 1.64 (0.99-2.73) | 0.241 | 0.056 | 2.59 (1.02-6.58) | 0.891 | 0.046 | 3.16 (1.23-8.13) | 0.800 | 0.017 |
| East Asians | 4 | 1.48 (0.77-2.83) | 0.146 | 0.241 | 2.60 (0.93-7.31) | 0.732 | 0.069 | 3.20 (1.13-9.12) | 0.605 | 0.029 |
CI, confidence interval; COPD, chronic obstructive pulmonary disease; IL-1, interleukin-1; NA, not available; OR, odds ratio; Phet, P-value for heterogeneity; Pz, P-value for overall effect.
aFor IL1B (-511) polymorphism: dominant (TT + TC vs CC), recessive (TT vs TC + CC) and homozygote (TT vs CC).
bFor IL1B (-31) polymorphism: dominant (CC + CT vs TT), recessive (CC vs CT + TT) and homozygote (CC vs TT).
cFor IL1B (+3954) polymorphism: dominant (TT + TC vs CC), recessive (TT vs TC + CC) and homozygote (TT vs CC).
dFor IL1RN (VNTR) polymorphism: dominant (22 + 2L vs LL), recessive (22 vs 2L + LL) and homozygote (22 vs LL).
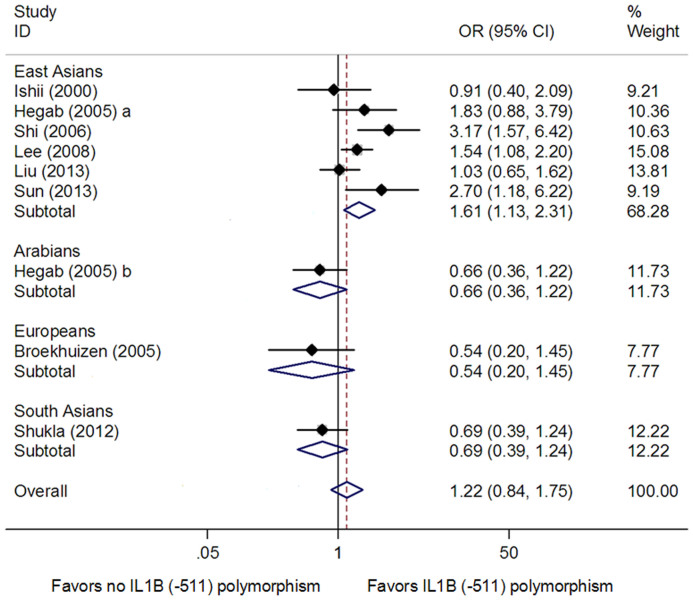
For the IL1B (−511) polymorphism, nine studies from eight publications with 1270 cases and 1220 controls were included in the meta-analysis11,12,13,14,16,18,19,20. There was no significant association between the IL1B (−511) polymorphism and COPD risk in all study subjects under dominant model (OR = 1.22, 95% CI: 0.84–1.75, Ph = 0.002, Pz = 0.293) (Table 3 and Fig. 2), recessive model (OR = 0.94, 95% CI: 0.67–1.32, Ph = 0.010, Pz = 0.730) (Table 3), and homozygote model (OR = 1.10, 95% CI: 0.65–1.85, Ph<0.001, Pz = 0.734) (Table 3). In subgroup analysis stratified by ethnicity, we found that the IL1B (−511) polymorphism was associated with COPD risk in East Asians under dominant model (OR = 1.61, 95% CI: 1.13–2.31, Ph = 0.061, Pz = 0.009) (Table 3 and Fig. 2), but not under recessive model (OR = 1.13, 95% CI: 0.74–1.73, Ph = 0.017, Pz = 0.574) (Table 3) and homozygote model (OR = 1.62, 95% CI: 0.91–2.88, Ph = 0.005, Pz = 0.105) (Table 3). Since there was only one study performed in Arabians, Europeans and South Asians, respectively, we did not conduct subgroup analysis in these ethnic groups. Between-study heterogeneity for the genotype-wise ORs was found in dominant model (P = 0.002) (Table 3), recessive model (P = 0.010) (Table 3), and homozygote model (P<0.001) (Table 3).
Figure 2. Meta-analysis for the association between the IL1B (-511) polymorphism and COPD risk in dominant model.
Each study is shown by the point estimate of the odds ratio, and a horizontal line denotes the 95% confidence interval. The pooled odds ratio is represented by a diamond. The area of the grey squares reflects the weight of the study in the meta-analysis.
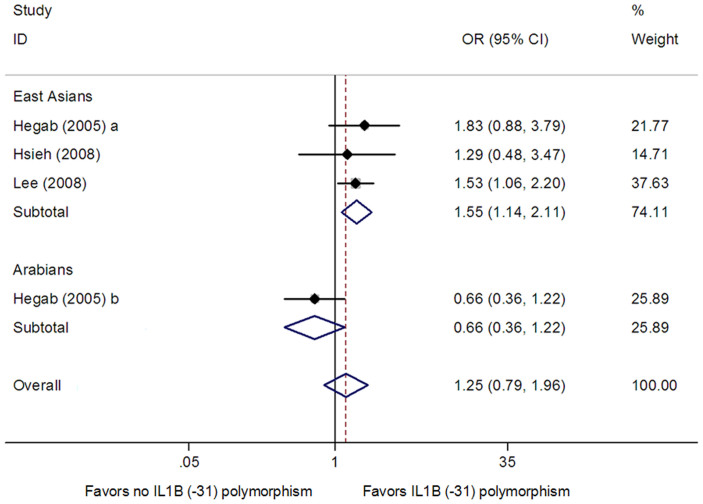
For the IL1B (−31) polymorphism, four studies from three publications with 534 cases and 632 controls were included in the meta-analysis12,15,16. Pooling data provided no evidence of a relationship between this polymorphism and COPD risk in all study subjects in dominant model (OR = 1.25, 95% CI: 0.79–1.96, Ph = 0.101, Pz = 0.340) (Table 3 and Fig. 3), recessive model (OR = 0.80, 95% CI: 0.52–1.21, Ph = 0.225, Pz = 0.287) (Table 3) and homozygote model (OR = 1.00, 95% CI: 0.52–1.95, Ph = 0.065, Pz = 0.993) (Table 3). However, in subgroup analysis based on ethnicity, we found a significant association between this SNP and COPD risk in East Asians in dominant model (OR = 1.55, 95% CI: 1.14–2.11, Ph = 0.842, Pz = 0.006) (Table 3 and Fig. 3), but not in recessive model (OR = 0.87, 95% CI: 0.64–1.18, Ph = 0.469, Pz = 0.356) (Table 3) and homozygote model (OR = 1.27, 95% CI: 0.87–1.87, Ph = 0.599, Pz = 0.221) (Table 3). Between-study heterogeneity was found for the genotype-wise OR in homozygote model (P = 0.065) (Table 3).
Figure 3. Meta-analysis for the association between the IL1B (-31) polymorphism and COPD risk in dominant model.
Each study is shown by the point estimate of the odds ratio, and a horizontal line denotes the 95% confidence interval. The pooled odds ratio is represented by a diamond. The area of the grey squares reflects the weight of the study in the meta-analysis.
For the IL1B (+3954) polymorphism, four studies from three publications with 406 cases and 359 controls were included in the meta-analysis11,12,17. Available data did not suggest an association between this polymorphism and COPD risk in all study subjects in dominant model (OR = 1.16, 95% CI: 0.78–1.73, Ph = 0.830, Pz = 0.464) (Table 3), recessive model (OR = 0.97, 95% CI: 0.32–2.88, Ph = 0.486, Pz = 0.951) (Table 3) and homozygote model (OR = 1.02, 95% CI: 0.33–3.12, Ph = 0.480, Pz = 0.974) (Table 3). In subgroup analysis according to ethnicity, we also did not find an association between this SNP and COPD risk in East Asians (Table 3). Because of limited availability of published results, we were unable to perform subgroup analysis in Arabians, Europeans and South Asians, respectively. Between-study heterogeneity for the genotype-wise ORs was not found (Table 3).
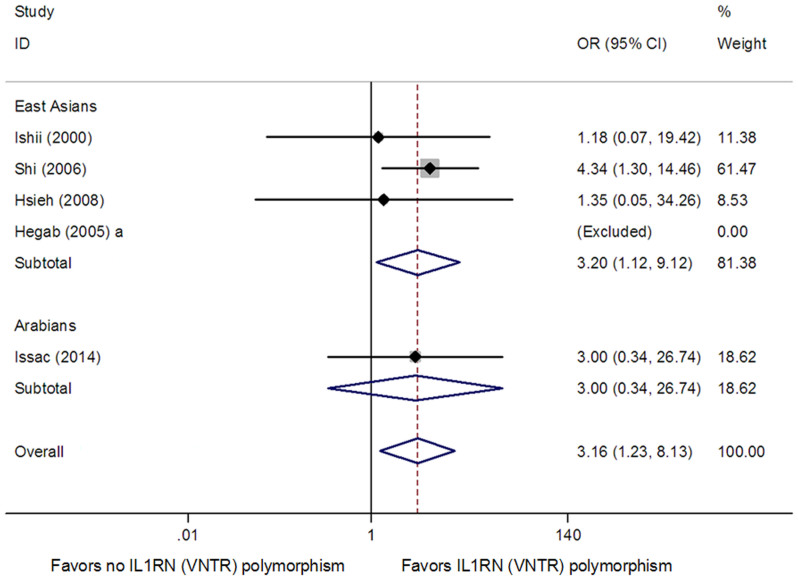
For the IL1RN (VNTR) polymorphism, five studies with 322 cases and 362 controls were included11,12,14,15,21. The pooled effect estimates among all studies suggested a significant association between the IL1RN (VNTR) polymorphism and COPD risk in recessive model (OR = 2.59, 95% CI: 1.02–6.58, Ph = 0.891, Pz = 0.046) (Table 3) and homozygote model (OR = 3.16, 95% CI: 1.23–8.13, Ph = 0.800, Pz = 0.017) (Table 3 and Fig. 4), but not in dominant model (OR = 1.64, 95% CI: 0.99–2.73, Ph = 0.241, Pz = 0.056) (Table 3). In subgroup analysis stratified by ethnicity, we found that the IL1RN (VNTR) polymorphism was associated with COPD risk in East Asians in homozygote model (OR = 3.20, 95% CI: 1.13–9.12, Ph = 0.605, Pz = 0.029) (Table 3 and Fig. 4), but not in dominant model (OR = 1.48, 95% CI: 0.77–2.83, Ph = 0.146, Pz = 0.241) (Table 3) and recessive model (OR = 2.60, 95% CI: 0.93–7.31, Ph = 0.732, Pz = 0.069) (Table 3). We did not perform subgroup analysis in Arabians, Europeans and South Asians because of limited availability of published results. Between-study heterogeneity for the genotype-wise OR was not indicated (Table 3).
Figure 4. Meta-analysis for the association between the IL1RN (VNTR) polymorphism and COPD risk in homozygote model.
Each study is shown by the point estimate of the odds ratio, and a horizontal line denotes the 95% confidence interval. The pooled odds ratio is represented by a diamond. The area of the grey squares reflects the weight of the study in the meta-analysis.
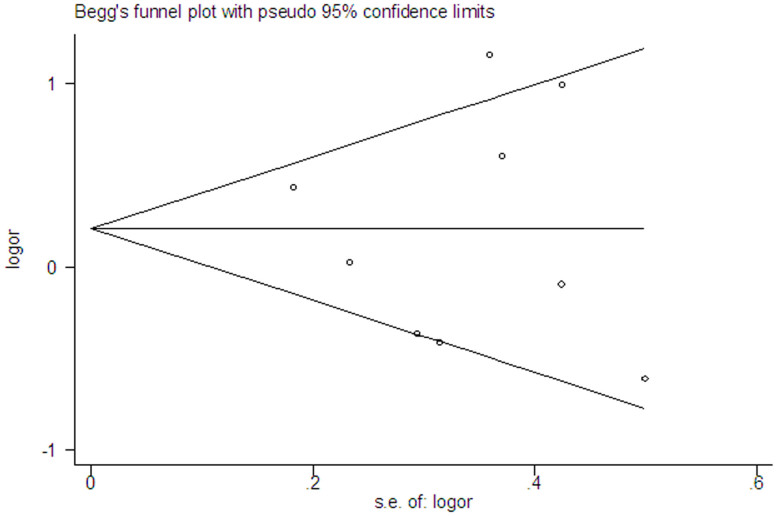
Publication bias
Since publication bias was hard to detect when the number of studies were small, we selected the IL1B (−511) polymorphism to assess publication bias (nine studies included). The shape of the funnel plot seemed symmetrical (Fig. 5). Both Begg's test and Egger's test suggested no evidence of publication bias (P = 1.000 and P = 0.865, respectively).
Figure 5. Begg's funnel plot for the IL1B (-511) polymorphism and COPD risk.
Discussion
The IL1B and IL1RN polymorphisms have been implicated in the pathogenesis of COPD and have been investigated in numerous case-control association studies. However, the results are controversial, possibly because single studies may have been underpowered. Given the amount of accumulated data and the still equivocal role of the IL-1 gene polymorphisms in the etiology of COPD, we performed the present meta-analysis of 12 case-control studies for a total of 1530 COPD patients and 1524 controls to investigate the association between the four most studied IL1B and IL1RN polymorphisms and COPD. The main findings of the present meta-analysis are: (1) the T allele carriers of the IL1B (−511) polymorphism are associated with an increased risk for developing COPD in East Asians; (2) the C allele carriers of the IL1B (−31) polymorphism are associated with an increased risk for COPD in East Asians; (3) compared with the LL homozygotes, the 22 homozygotes of the IL1RN (VNTR) polymorphism are associated with an increased risk for developing COPD in East Asians; and (4) there is no significant association between the IL1B (+3954) polymorphism and COPD risk.
IL-1B is a pro-inflammatory cytokine which is synthesized by a variety of cell types, including blood monocytes and tissue macrophages. IL-1B is thought to be an important mediator in cigarette smoke-induced inflammation and COPD. In BALB/c mice, IL-1B production was significantly up-regulated in lung homogenates after smoke exposure22. In human subjects, elevated IL-1B levels were observed among smokers23 and in primary explant cultures of bronchial epithelial cells derived from COPD patients24. IL-1B expression in COPD neutrophils was correlated with disease severity as measured by forced expiratory volume in 1 second (FEV1)25. In addition, sputum IL-1B was showed to be a potential biomarker for bacteria-associated exacerbations of COPD26. IL-1B plays a central role in the regulation of immune responses and inflammatory processes, including promotion of the movement of inflammatory cells from the blood to inflamed tissues, regulation of the extracellular matrix, induction of the expression of a variety of inflammatory mediators such as IL-6, IL-8 and tumor necrosis factor-α(TNF-α), and promotion of the differentiation of inflammatory cells5. Since IL-1B regulates a variety of inflammatory processes, any alteration in its blood or tissue concentration may significantly affect these processes. The IL1B (−511) T and (−31) C alleles are associated with higher levels of IL-1B and with severe inflammation, in comparison to (−511) C and (−31) T alleles, which are associated with lower levels of IL-1B27,28. Genetic variation in the level of gene expression within the IL1B gene may alter susceptibility to COPD. In the present meta-analysis, we found that the T allele carriers of the IL1B (−511) SNP and the C allele carriers of the IL1B (−31) polymorphism had an increased risk for COPD in East Asians, suggesting an effect of the IL1B gene on COPD in this ethnic group. Due to limited availability of published data, subgroup analysis was only conducted in East Asians. To test whether this association is population dependent, future studies should be performed in other ethnic groups besides East Asians.
IL-RA binds to the IL-1 receptor and acts as a competitive inhibitor of IL-1B. Allele 2 (two repeats) of the IL1RN (VNTR) polymorphism is associated with increased IL-1ra levels29. In addition, allele 2 strongly enhances in vitro production of IL-1B and plays a pivotal role in determining the balance between secreted IL-1B and IL-1RA30. The IL1-RA/IL-1B ratio is critical in determining the severity of inflammatory responses. The IL1RN allele 2 has been showed to be associated with a low IL1-RA/IL-1B ratio, thereby leading to a more prolonged and more severe pro-inflammatory immune response31.The frequency of this allele was found to be increased in diseases of autoimmune or inflammatory nature, including ankylosing spondylitis32, diabetic nephropathy33, and ulcerative colitis34. Our meta-analysis showed that the IL1RN allele 2 homozygotes had an increased risk for COPD, suggesting a significant pathogenetic effect of the IL1RN allele 2 in COPD.
There are several limitations to this meta-analysis. First, between-study heterogeneity was found in some pooled analyses. We were unable to further identify the exact sources of heterogeneity by meta-regression analysis in that there were limited relevant data provided by the included studies. Some potentially relevant factors, such as ethnicity, genotyping method, gender, phenotype of the disease may account for heterogeneity. We further performed subgroup analysis according to ethnicity, which greatly reduced heterogeneity. Second, we only included publications written in English or Chinese in the present meta-analysis, which may have missed potentially eligible studies published in other languages. Third, few included studies reported the percentage of smokers in COPD patients and controls. Because of limited availability of published data, we were unable to evaluate the effect of smoking on the association between the IL1 gene polymorphisms and COPD. Fourth, there has been considerable strength of linkage disequilibrium (LD) in the IL-1 gene cluster. Feakes et al. found by family studies significant LD between the IL1B (−511) and IL1RN (VNTR) polymorphisms35. Joos et al. found that the IL1B (−511) T allele in combination of the IL1RN allele 2 was protective against a rapid decline in lung function in smokers36. These results suggest that haplotype analysis, as well as genotype analysis, may be necessary in evaluating genetic effects of the IL-1 gene cluster on COPD. We did not address this issue in our meta-analysis since few included studies performed haplotype analysis and there were discrepancies in constructing haplotypes among them12,16,18. Association studies may provide more insights for the role of the IL-1 gene in COPD if researchers can investigate haplotype association.
In summary, our meta-analysis showed that the IL1B (−511), (−31) and IL1RN (VNTR) polymorphisms are associated with COPD risk in East Asians. There is no significant association between the IL1B (+3954) polymorphism and COPD risk. Further studies with large sample size are needed to establish a more definitive conclusion. In addition, more studies should be performed in other ethnic groups besides East Asians.
Author Contributions
J.H. and Z.F.X. designed the study. Z.K.X., Q.P.H., J.H. and Z.F.X. collected data, performed the statistical analyses and drafted the manuscript. All authors read and approved the final manuscript.
References
- Decramer M., Janssens W. & Miravitlles M. Chronic obstructive pulmonary disease. Lancet. 379, 1341–1351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molfino N. A. Current thinking on genetics of chronic obstructive pulmonary disease. Curr Opin Pulm Med. 13, 107–113 (2007). [DOI] [PubMed] [Google Scholar]
- Bossé Y. Updates on the COPD gene list. Int J Chron Obstruct Pulmon Dis. 7, 607–631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. et al. Genetic polymorphism of matrix metalloproteinase family and chronic obstructive pulmonary disease susceptibility: a meta-analysis. Sci Rep. 3, 2818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinarello C. A. Interleukin-1. Cytokine Growth Factor Rev. 8, 253–265 (1997). [DOI] [PubMed] [Google Scholar]
- Nicklin M. J., Weith A. & Duff G. W. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, andinterleukin-1 receptor antagonist genes. Genomics. 19, 382–384 (1994). [DOI] [PubMed] [Google Scholar]
- Luomala M. et al. A study of interleukin-1 cluster genes in susceptibility to and severity of multiple sclerosis. J Neurol Sci. 185, 123–127 (2001). [DOI] [PubMed] [Google Scholar]
- Shete A. R., Joseph R., Vijayan N. N., Srinivas L. & Banerjee M. Association of single nucleotide gene polymorphism at interleukin-1beta +3954, -511, and -31 in chronic periodontitis and aggressive periodontitis in Dravidian ethnicity. J Periodontol. 81, 62–69 (2010). [DOI] [PubMed] [Google Scholar]
- MANTEL N. & HAENSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 22, 719–748 (1959). [PubMed] [Google Scholar]
- DerSimonian R. & Laird N. Meta-analysis in clinical trials. Control Clin Trials. 7, 177–188 (1986). [DOI] [PubMed] [Google Scholar]
- Ishii T. et al. Neither IL-1beta, IL-1 receptor antagonist, nor TNF-alpha polymorphisms are associated with susceptibility to COPD. Respir Med. 94, 847–851 (2000). [DOI] [PubMed] [Google Scholar]
- Hegab A. E. et al. Polymorphisms of TNFalpha, IL1beta, and IL1RN genes in chronic obstructive pulmonary disease. Biochem Biophys Res Commun. 329, 1246–1252 (2005). [DOI] [PubMed] [Google Scholar]
- Broekhuizen R. et al. Pulmonary cachexia, systemic inflammatory profile, and the interleukin 1beta -511 single nucleotide polymorphism. Am J Clin Nutr. 82, 1059–1064 (2005). [DOI] [PubMed] [Google Scholar]
- Shi Y. Z., Liu B., Huo J. M. & Zhang W. Relationship between interleukin-1 cytokine gene polymorphisms and genetic susceptibility to chronic obstructive pulmonary diseases. Zhongguo Lin Chuang Kang Fu. 10, 67–69 (2006). [Google Scholar]
- Hsieh M. H. et al. Lack of associations between several polymorphisms in cytokine genes and the risk ofchronic obstructive pulmonary diseases in Taiwan. Kaohsiung J Med Sci. 24, 126–137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M. et al. Polymorphisms in interleukin-1B and its receptor antagonist genes and the risk of chronic obstructive pulmonary disease in a Korean population: a case-control study. Respir Med. 102, 1311–1320 (2008). [DOI] [PubMed] [Google Scholar]
- Liu Y. Q., Liu X. Y., Li H. F. & Wang L. H. Relationship between polymorphism of IL-1β gene and chronic obstructive pulmonary disease in Han population. Zhongguo Man Xing Bing Yu Fang Yu Kong Zhi. 20, 128–131 (2012). [Google Scholar]
- Shukla R. K., Kant S., Bhattacharya S. & Mittal B. Association of cytokine gene polymorphisms in patients with chronic obstructive pulmonary disease. Oman Med J. 27, 285–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Q., Wang L. H., Liu X. Y. & Li H. F. Association study between polymorphism of IL1β-31T/C,-511C/T and chronic obstructive pulmonary disease. Zhongguo Zong He Lin Chuang. 29, 1169–1173 (2013). [Google Scholar]
- Sun L. R. et al. Relationship between the genetic polymorphism of IL-1, IL-6 and pulmonary hypertension in patient with COPD. Yi Xue Yan Jiu Za Zhi. 42, 80–84 (2013). [Google Scholar]
- Issac M. S., Ashur W. & Mousa H. Genetic Polymorphisms of Surfactant Protein D rs2243639, Interleukin (IL)-1β rs16944 and IL-1RN rs2234663 in Chronic Obstructive Pulmonary Disease, Healthy Smokers, and Non-Smokers. Mol Diagn Ther, 10.1007/s40291-014-0084-5 (2014). [DOI] [PubMed] [Google Scholar]
- Botelho F. M. et al. Innate immune processes are sufficient for driving cigarette smoke-induced inflammation in mice. Am J Respir Cell Mol Biol. 42, 394–403 (2010). [DOI] [PubMed] [Google Scholar]
- Kuschner W. G., D'Alessandro A., Wong H. & Blanc P. D. Dose-dependent cigarette smoking-related inflammatory responses in healthy adults. Eur Respir J. 9, 1989–1994 (1996). [DOI] [PubMed] [Google Scholar]
- Rusznak C. et al. Effect of cigarette smoke on the permeability and IL-1beta and sICAM-1 release from cultured human bronchial epithelial cells of never-smokers, smokers, and patients with chronic obstructive pulmonary disease. Am J Respir Cell Mol Biol. 23, 530–536 (2000). [DOI] [PubMed] [Google Scholar]
- Sapey E. et al. Imbalances between interleukin-1 and tumor necrosis factor agonists and antagonists in stable COPD. J Clin Immunol. 29, 508–516 (2009). [DOI] [PubMed] [Google Scholar]
- Bafadhel M. et al. Acute exacerbations of chronic obstructive pulmonary disease: identification of biologic clusters and their biomarkers. Am J Respir Crit Care Med. 184, 662–671 (2011). [DOI] [PubMed] [Google Scholar]
- El-Omar E. M. et al. The role of interleukin-1 polymorphisms in the pathogenesis of gastric cancer. Nature. 412, 99 (2001). [DOI] [PubMed] [Google Scholar]
- Kim S. H., Mok J. W., Kim H. S. & Joo C. K. Association of -31T>C and -511 C>T polymorphisms in the interleukin 1 beta (IL1B) promoter in Korean keratoconus patients. Mol Vis. 14, 2109–2116. (2008). [PMC free article] [PubMed] [Google Scholar]
- Danis V. A., Millington M., Hyland V. J. & Grennan D. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol. 99, 303–310 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santtila S., Savinainen K. & Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 47, 195–198 (1998). [DOI] [PubMed] [Google Scholar]
- Witkin S. S., Gerber S. & Ledger W. J. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin Infect Dis. 34, 204–209 (2002). [DOI] [PubMed] [Google Scholar]
- van der Paardt M. et al. Interleukin-1beta and interleukin-1 receptor antagonist gene polymorphisms in ankylosing spondylitis. Rheumatology (Oxford). 41, 1419–1423 (2002). [DOI] [PubMed] [Google Scholar]
- Lee S. H. et al. Polymorphisms in interleukin-1 beta and Interleukin-1 receptor antagonist genes are associated with kidney failure in Korean patients with type 2 diabetes mellitus. Am J Nephrol. 24, 410–414 (2004). [DOI] [PubMed] [Google Scholar]
- Tountas N. A. et al. Functional and ethnic association of allele 2 of the interleukin-1 receptor antagonist gene in ulcerative colitis. Gastroenterology. 117, 806–813 (1999). [DOI] [PubMed] [Google Scholar]
- Feakes R. et al. Interleukin 1 receptor antagonist (IL-1ra) in multiple sclerosis. J Neuroimmunol. 105, 96–101 (2000). [DOI] [PubMed] [Google Scholar]
- Joos L. et al. Association of IL-1beta and IL-1 receptor antagonist haplotypes with rate of decline in lung function in smokers. Thorax. 56, 863–866 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]