Abstract
Members of serpin superfamily are involved in wide array of cellular processes to control proteolytic activities of eukaryotic organisms. Vertebrate serpins are extensively studied and reported to be classified into six groups (V1-V6) based on gene structures. However, there is no study conducted for serpins in urochordates (the closest living invertebrates related to vertebrates) to date. To unravel further the phylogenetic history of serpin genes, we characterized serpin genes from two urochordates (Ciona intestinalis and Ciona savignyi). There are 11 and 5 serpins in the C. intestinalis and C. savignyi, respectively. The exon/intron structures and genomic locus comparisons together with sequence phylogenetic analysis, suggested that urochordate serpins are classified into six groups (U1-U6), different from six groups (V1-V6) of vertebrate serpins. Human α1-antitrypsin shared lower sequence identities and similarities with urochordates serpins ranged from 14-29% and 30-49%, respectively. Based on protein sequences, genes and genomic architectures, we conclude that these two urochordates do not contain a single copy of genuine ortholog of the vertebrate serpins.
Keywords: Bayesian phylogeny, Gene structure, Microsynteny, Serpins, Sequence analysis, Synteny, Urochordates
1. Introduction
The regulations of proteolytic activity of serine proteases are critical steps to maintain balanced homeostasis and the serine protease inhibitors (serpins) possess capabilities to regulate the proteolytic activities of these serine proteases by inhibitory mechanism 1. The members of serpin superfamily have instrumental roles in a variety of physiological and cellular functions and are associated with the vertebrate blood coagulation cascade, complement activation, inflammation, programmed cell death, cell development, and fibrinolysis 1-3. Serpins are single domain proteins with an average size: 350-400 amino acids and molecular weights of 40-60 kDa 2-4. These proteins are classified into two functional categories - inhibitory (majority) such as antithrombin III 5, while some of them are non-inhibitory, which adopt other function than inhibitory roles such as angiotensinogen, which lost inhibition during vertebrate evolution 6. Serpins are thought to have evolved through gene duplication and divergence events, giving rise to a large number of serpin genes within an organism, each encoding a protein with a unique reactive center region and physiological function(s) 7. They also tend to form a gene cluster after tandem duplication events and these cluster expand in a species-specific manner. This broad family of proteins was initially identified through similarities between the primary structure of 3 human serpins; antithrombin III, α1-protease inhibitor and chicken egg white albumin 7. Apart from 36 human serpins, there are more than 3000 serpins, which have now been described from viruses, bacteria, archaea, and other eukaryotes. These serpin members represent the largest superfamily and most diverse family of protease inhibitors 3. Many additional serpins are likely to be identified as more sequenced genomes become available in the era of rapid and desktop based next-generation DNA sequencing methods. Based on sequence analyses, all serpins so far described have been classified into one of 16 clades, designated A to P, plus 10 unclassified 'orphan' sequences and this classification system is called as “clade-based classification” system 8. These genes are characterized by specific pattern of gene structures dividing theses into six groups (V1-V6) in vertebrates and this system is called as “group-based classification” system 9.
Ciona species live in flat water areas of the oceans and go through two phases of the life cycle - an adult stage, which metamorphoses from free-swimming tadpole stage. The tadpole is built of approximately 2500 cells, whose development can be observed easily under the microscope on the basis of the transparency of the larva 10. Additionally, this organism has the relatively short life cycle of approximately three months, making it a good system for developmental research. Urochordate is a non-vertebrate chordate that diverged very early from the other chordates, namely cephalochordates and vertebrates, approximately 550 million years ago. Therefore, it is considered highly important for the understanding of evolution of the vertebrates. There is no study of serpins exists in this evolutionary and developmentally important lineage of invertebrates, close to vertebrates. Hence, there is an urgent requirement to study serpins in urochordates. Herein, we described the properties of serpins from two urochordates. By combining genes and genomic organizational comparisons coupled with sequence and phylogenetic analyses, we corroborated that urochordate serpins are classified into six groups (U1-U6), which are distinguishable from six vertebrate serpin groups (V1-V6).
2. Results
2.1. Catalogue of serpins from urochordates
There are eleven and five serpins in the C. intestinalis and C. savignyi as summarized in Tables 1-2, respectively. These two urochordates serpins are further characterized in next sections.
Table 1.
List of serpins from Ciona intestinalis genome.
| Gene name | JGI protein accession id | Protein length | RCL |
|---|---|---|---|
| P1-P1' | |||
| Ci-Spn-1 | ci0100132788 | 449 | R-S |
| Ci-Spn-2 | ci0100132818 | 412 | R-S |
| Ci-Spn-3 | ci0100134682 | 402 | R-S |
| Ci-Spn-4 | ci0100141118 | 441 | R-S |
| Ci-Spn-5 | ci0100143209 | 413 | S-V |
| Ci-Spn-6 | ci0100146394$ | 377 | R-S |
| Ci-Spn-7 | ci0100146394$ | 380 | S-M |
| Ci-Spn-8 | ci0100146394$ | 379 | R-S |
| Ci-Spn-9 | ci0100148346 | 409 | D-S |
| Ci-Spn-10A | ci0100154072# | 408 | R-S |
| Ci-Spn-10B | ci0100154072# | 407 | P-L |
$ci0100146394 is accession id for Ci-Spn-6, Ci-Spn-7 and Ci-Spn-8 in database for Ciona intestinalis genome.
#Ci-Spn-10 shows two variations in the RCL exon, named A and B, respectively
Table 2.
List of serpins from Ciona savignyi genome from Ensembl 74 (December 2013).
| Given Name | Gene Accession Id | Localization | Protein Length | RCL P1-P1' |
|
|---|---|---|---|---|---|
| Cs-Spn-1 | ENSCSAVG00000004408 | reftig_72:471765-473162 | 388 | R-S | |
| Cs-Spn-2 | ENSCSAVG00000006721 | reftig_65:1008461-1013265 | 409 | P-G | |
| Cs-Spn-3 | ENSCSAVG00000005029 | reftig_194:201970-204904 | 412 | R-S | |
| Cs-Spn-4 | ENSCSAVG00000005022 | reftig_194:194913-200294 | 300 | R-S | |
| Cs-Spn-5 | ENSCSAVG00000004177 | reftig_99:92392-98988 | 429 | R-S |
2.2. Urochordate serpins are classified into six groups U1-U6 based on intron-exon structures
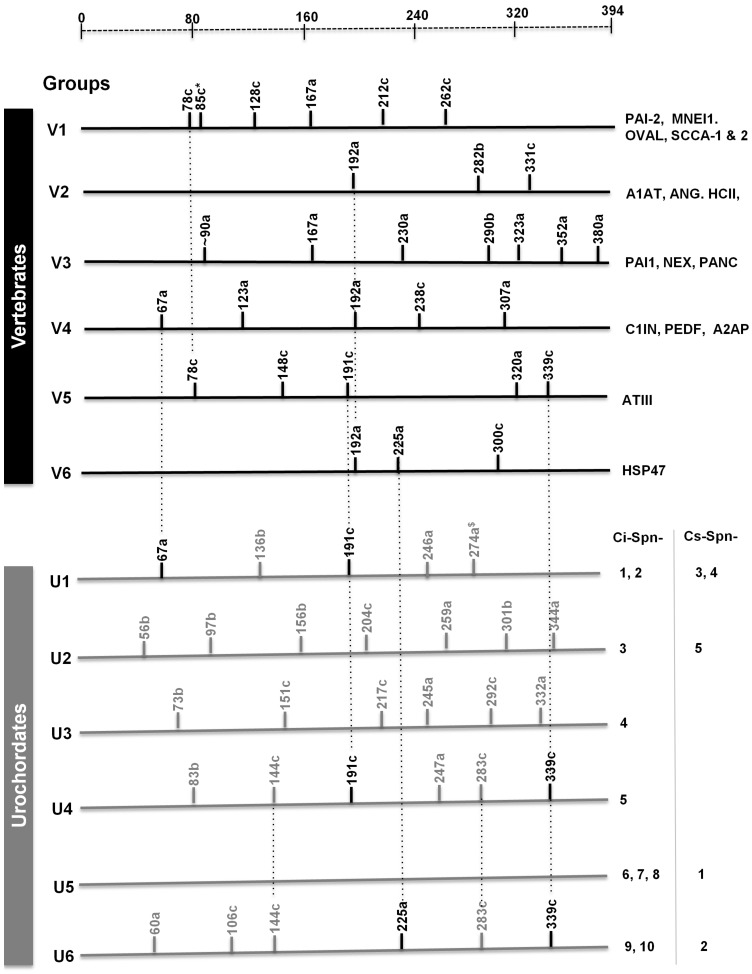
Serpins from urochordates have unique gene structures as compared to vertebrate serpins (Fig. 1). Serpins from two urochordates are classified in six distinct groups and are named as group U1-U6. Group U1 serpins have five introns at positions 67a, 136b, 191c, 246a and 274a (missing in Ci-Spn-1). These intron positions are based on amino acid numbering of mature (without signal peptide) human A1AT, followed by intron phasing with suffixes a-c as reported in previous studies 5, 9. Two of these introns (at the positions 67c and 191c) are shared with vertebrate groups V4 and V5, respectively. Group U2 has a single serpin in two Ciona species with seven introns at the positions 56b, 97b, 156b, 204c, 259a, 301b and 344a. Group U3 has a single serpin in Ciona intestinalis only with six introns at the positions 73b, 151c, 217c, 254a, 292c and 322a. Similarly, group U4 has a single serpin in Ciona intestinalis only and it has six introns at the positions 83b, 144c, 191c, 247a, 283c and 339a, of which two positions are shared with vertebrate group V5. Group U5 genes are intron-less. Whereas group U6 have serpins with six introns at the positions 60a, 106c, 144c, 225a, 283c and 339a, with fourth and sixth introns are shared with vertebrate groups V6 and V5, respectively. Groups U4 and U6 share three introns at the positions 144c, 283c and 339a. By and large, Ciona serpins have a different set of intron positions as compared to vertebrate serpins 9 and only very few positions are shared with vertebrate serpins. Orthologs of vertebrate and urochordate serpins are listed in Fig 1; these orthologs are maintained in respective groups.
Fig 1.
Intron-based classification system for serpin genes from vertebrates and two urochordates illustrates six distinct groups namely V1-V6 and U1-U6, respectively. Black line and grey lines indicates vertebrate-specific and urochordate-specific intron positions, respectively. *indicates that the intron at the position 85c can differentiate between groups V1a and V1b. $ indicates that intron at position 274a is not found in Ci-Spn-2. Introns shared in more than one groups are marked. Only introns mapping to the conserved serpin fold (amino acids 33 to 394 of human α1-antitrypsin) are taken into consideration. Group wise distributions of major serpins are indicated.
2.3. Micro-synteny analysis supports gene structure-based six groups of urochordate serpins
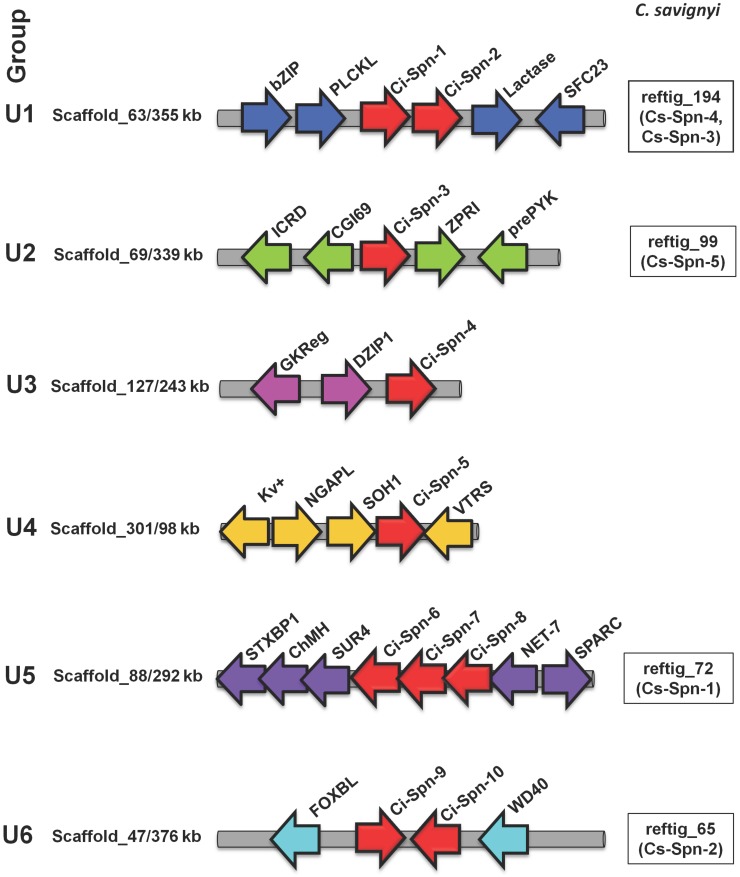
To support the grouping of serpins based on gene structures in two urochordates and to compare it with vertebrate serpins, we carried out micro-syntenic analyses. Ci-Spn-1 and Ci-Spn-2 were found to be adjacent to each other and having the same orientation in C. intestinalis. These genes are located in scaffold_63, flanked by a dyad of genes namely a basic-leucine zipper (bZIP) transcription factor and a Pleckstrin-like (PLCKL) gene on the one side, while on the other side, there is a dyad of genes - the lactase and vesicle coat complex COPII, subunit SEC23 (SEC23) (Fig. 2). In similar fashion, the reftig_194 of C. savignyi genome possess Cs-Spn-4 and Cs-Spn-3 with the same flanking. These members maintained on this syntenic organization form group U1. Ci-Spn-3 was located on scaffold_69, flanked by a dyad of genes - secreted frizzled-related protein (fCRD) and mitochondrial carrier protein CGI69 (CGI69) on the one side, while a dyad of genes - ZPR1 type Zn-finger (ZFR1) and RYK receptor-like tyrosine kinase precursor (preRYK) is located on the other side (Fig. 2). This genomic architecture is maintained in the reftig_99 with Cs-Spn-5 for C. savignyi. These serpins form group U2. The Ci-Spn-4 was located on scaffold_127 as a single serpin gene, flanked by a dyad of glucokinase regulatory protein (GKReg) and Zn-finger, C2H2 type (DZIP1) (Fig. 2), forming group U3. Group U4 has one serpin as Ci-Spn-5, which is located on scaffold_301 flanked by a triad of potassium voltage gated Kv channel (Kv+), NGAP like protein (NGAPL) and transcriptional regulator SOH1 (SOH1) on the one side, while the singe gene valyl-tRNA synthetase (VTRS) is situated on the other side (Fig. 2). The Ci-Spn-6, Ci-Spn-7, and Ci-Spn-8 are found on scaffold_88 adjacent to each other in the same orientation (Fig. 2) in C. intestinalis. These three serpins are flanked by a triad of genes - syntaxin 5 (STXBP1), chondromodulin-1 precursor (ChMH) and surfeit locus protein 4 (SUR4) on the one side and the other side has a dyad of genes as tetraspanin 15 (NET7) and secreted protein acidic and rich in cysteine (SPARC). This genomic structure is found in the reftig_72 with only Cs-Spn-1 for C. savignyi. These five serpins belongs to group U5.
Fig 2.
Genomic organizations of Ciona serpins. Ciona serpins (red arrows) were identified on different scaffolds surrounded by marker genes (arrows in different colors as they are non-homologous in different loci). White boxes indicate the corresponding locus on specific reftig with corresponding serpins in C. savignyi.
C. intestinalis serpins Ci-Spn-9 and Ci-Spn-10 are found in opposite orientations on scaffold_47 flanked by a Fbox-like gene (FOXBL) and a G-protein beta WD40 protein (WD40) (Fig. 2), while C savignyi possess this fragment on the reftig_65 with a single serpin, Cs-Spn-2. These serpins constitute group U6.
By comparing these syntenies with previous studies on syntenies analyses of vertebrate serpin groups (namely V1 11, 12, V2 6, 13, 14, V3 9, 14, V4 14, 15, V5 14, 5 and V6 14, 16), it is clear that there is no synteny relationship is shared among serpins of vertebrates and urochordates. Hence, this chromosomal mapping analysis supports that gene structure based six (U1-U6) groups as six groups maintained on six distinct chromosomal localizations and conserved in two urochordate genomes.
2.4. Bayesian phylogenetic analysis also supports classification of six groups
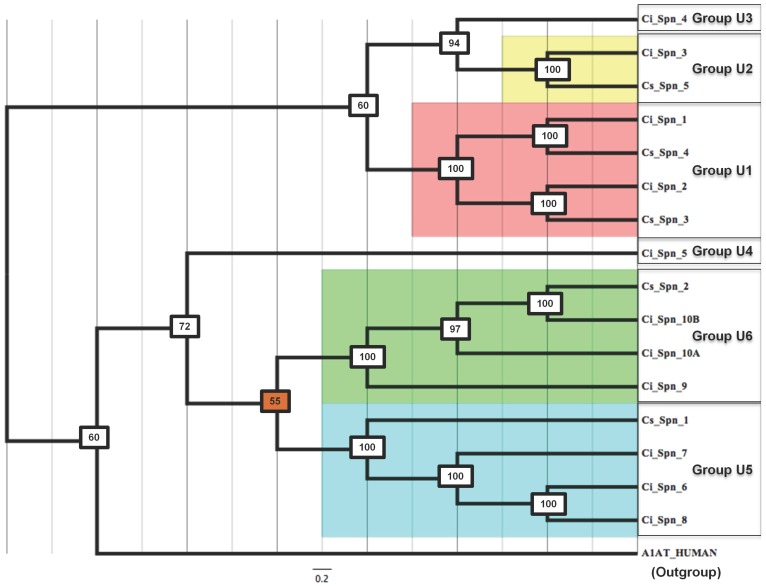
Furthermore, a Bayesian phylogenetic tree of urochordate serpins (Fig. 3) was created, which corroborates with clustering into six groups U1-U6 as supported by gene structures and chromosomal mapping. This tree has lowest percentage posterior probabilities of 55. Human α1-antitrypsin was used as outgroup. Hence, this tree also supports urochordate-specific classification system of serpins.
Fig 3.
Bayesian phylogenetic history of serpins from C. intestinalis and C. savignyi. Human α1-antitrypsin was used as the outgroup. The color boxes represent Ciona serpins in different scaffolds, which cluster in this phylogenetic tree together.
2.5. Sequence and structural analysis of these serpins
Supplementary Material: Fig. S1 illustrates protein sequence alignment of serpins from urochordates, demonstrating amino acid conservations are marked by black, brown and yellow colors, corresponding for 70% or more, 50-69%, and 30-49% identities, respectively.
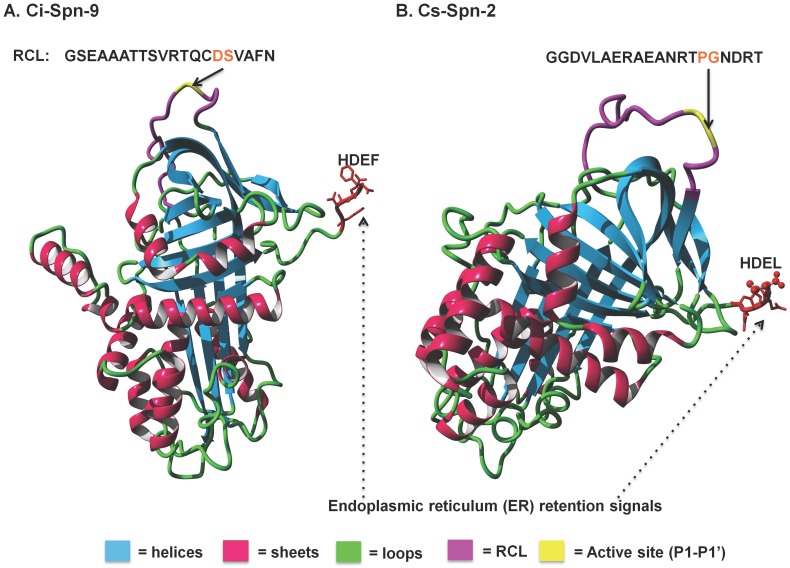
These serpins have conserved three serpin motifs and reactive center loop (RCL) with P1-P2 (red color). Four urochordate serpins (Ci-Spn-9, Ci-Spn-10A, Ci-Spn-10B and Cs-Spn-1) have C-terminal ER-retention signals (blue color), respectively.
Table 3 illustrates sequence identities and similarities amongst serpins from urochordates and human A1AT. Human A1AT has protein sequence identities and similarities with urochordates serpins ranged from 14-29% and 30-49%, respectively. Urochordates serpins are present on the same genomic fragments share higher sequence identities and similarities with each other. Ci-Spn-1 and Cs-Spn-4 shares 56%/73% identities/similarities. Similarly, Ci-Spn-2 and Cs-Spn-3 shares 54%/72% identities/similarities and these serpins are localized on same chromosomal locus (Fig. 2). Ci-Spn-6, Ci-Spn-7, Ci-Spn-8 and Cs-Spn-1 localized on same genomic fragment (Fig. 2) and these serpins share sequence identities and similarities ranged between 50-90% and 67-96%, respectively. In similar fashion, four serpins Ci-Spn-9, Ci-Spn-10A, Ci-Spn-10B and Cs-Spn-2 localized on same genomic fragment (Fig. 2) and these serpins share sequence identities and similarities ranged between 69-88% and 74-91%, respectively. Interestingly, the ER-retention signals are features of the group U6 (Supplementary Material: Fig. S1)
Table 3.
Comparison of serpins from two urochordates illustrating sequence identity and sequence similarity. i and s indicate Ci-Spn- and Cs-Spn-, respectively. A1AT - human α1-antitripsin.
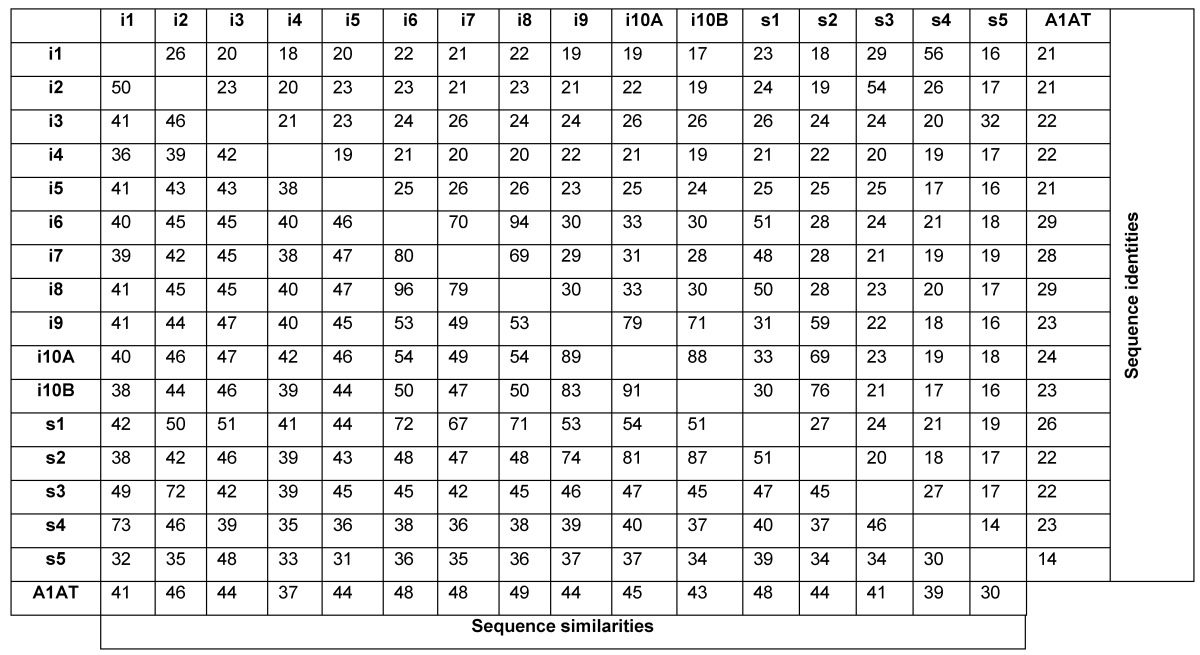
Furthermore, Ci-Spn-3 and Cs-Spn-5 shares same syntenies and have sequence identity and similarity 32% and 48%, respectively. Although these values are lower than other examples above, these values are highest in comparison to other serpins in table 3. This reveals that serpins on same syntenic organizations have originated by tandem duplication events and hence share higher sequence identities and similarities.
To further examine the relationships of vertebrate and urochordate groups of serpins, we carried out sequence and phylogenetic analyses by combining six representative member of group V1 to V6 from human, namely monocyte/neutrophil elastase inhibitor (MNEI), α1-antitrypsin (A1AT), plasminogen activator inhibitor 1 (PAI1), pigment epithelium-derived factor (PEDF), antithrombin III (ATIII) and heat shock protein 47 (HSP47). The six-serpin groups of vertebrates and two urochordates are maintained as evident in the Bayesian phylogenetic tree (Fig. 4). The group U5 is close to vertebrate group V1 and it is evident from sequence identity scores in Table 4, with members of the group U5 shared 37.1%-41.4% sequence identities with human MNEI. Similarly, the group U6 is closely related to the vertebrate group V5, also evident on sequence identity scores in table 4. These identities are slightly higher and it is common that several invertebrate serpins have slightly higher sequence identities with vertebrate groups V1 and V5, as evident for Hv-Spn-1 (from Hydra vulgaris) shares 35.5% and 33.2% identities, respectively. Only sequence identities of 25-40% cant not assign orthologous status for serpins in such a large superfamily. Other traits of serpins (gene structure pattern and genomic localizations) are not matching between vertebrates and two urochordates. Hence, it is not possible to assign orthology across vertebrates and urochordate species for serpins.
Fig 4.
Bayesian evolutionary history of serpins from vertebrates (V1-V6) and urochordates (U1-U6) illustrates separate groups are maintained. Six representatives of vertebrate serpin groups were used for this analysis. We used Hv- Spn-1 (Genbank ID: XP_002156931.1) from Hydra vulgaris as an outgroup.
Table 4.
Sequence identities between serpins from vertebrate and two urochordates.
| 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. | 9. | 10. | 11. | 12. | 13. | 14. | 15. | 16. | 17. | 18. | 19. | 20. | 21. | 22. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.Hs-MNE1_(V1) | ||||||||||||||||||||||
| 2. Hs-A1AT_(V2) | 29.7 | |||||||||||||||||||||
| 3. Hs-PAI1_(V3) | 29.1 | 26.9 | ||||||||||||||||||||
| 4. Hs-PEDF_(V4) | 24.3 | 24.1 | 20.3 | |||||||||||||||||||
| 5. Hs-ATIII_(V5) | 38.8 | 25.8 | 23.5 | 22.1 | ||||||||||||||||||
| 6. Hs-HSP47_(V6) | 28.5 | 25.6 | 21.9 | 24.4 | 19.2 | |||||||||||||||||
| 7. Ci-Spn-1_(U1) | 25.7 | 22.4 | 22.5 | 20.3 | 24.0 | 20.4 | ||||||||||||||||
| 8. Ci-Spn-2_(U1) | 27.3 | 22.5 | 24.3 | 18.8 | 24.3 | 18.9 | 25.8 | |||||||||||||||
| 9. Ci-Spn-3_(U2) | 25.5 | 24.7 | 23.9 | 18.4 | 21.1 | 18.6 | 20.4 | 24.1 | ||||||||||||||
| 10. Ci-Spn-4_(U3) | 24.4 | 22.7 | 23.0 | 17.9 | 19.9 | 18.7 | 18.2 | 20.0 | 20.4 | |||||||||||||
| 11. Ci-Spn-5_(U4) | 28.5 | 22.7 | 23.5 | 21.0 | 25.6 | 21.6 | 19.0 | 23.1 | 25.0 | 20.4 | ||||||||||||
| 12. Ci-Spn-6_(U5) | 39.0 | 29.5 | 27.0 | 22.9 | 30.9 | 24.2 | 23.0 | 22.8 | 24.8 | 20.9 | 25.4 | |||||||||||
| 13. Ci-Spn-7_(U5) | 38.7 | 28.4 | 28.5 | 23.0 | 30.3 | 24.5 | 22.3 | 22.1 | 27.0 | 20.3 | 25.2 | 69.3 | ||||||||||
| 14. Ci-Spn-8_(U5) | 38.7 | 29.6 | 27.3 | 22.1 | 31.0 | 23.1 | 22.9 | 22.4 | 24.6 | 20.3 | 25.8 | 95.0 | 68.1 | |||||||||
| 15. Ci-Spn-9_(U6) | 37.1 | 25.6 | 26.5 | 20.8 | 31.3 | 24.4 | 21.2 | 22.9 | 24.2 | 22.3 | 23.4 | 31.7 | 30.2 | 31.1 | ||||||||
| 16. Ci-Spn-10A_(U6) | 38.0 | 26.9 | 28.0 | 21.1 | 32.8 | 27.3 | 21.1 | 23.0 | 25.5 | 21.5 | 25.4 | 34.5 | 32.4 | 34.3 | 80.0 | |||||||
| 17. Ci-Spn-10B_(U6) | 34.6 | 25.7 | 26.4 | 20.9 | 29.3 | 25.5 | 19.1 | 20.9 | 25.7 | 19.5 | 23.7 | 31.0 | 29.9 | 30.8 | 71.4 | 89.2 | ||||||
| 18. Cs-Spn-1_(U5) | 41.4 | 28.5 | 29.7 | 22.9 | 32.1 | 23.4 | 24.6 | 24.3 | 26.6 | 21.2 | 25.3 | 52.1 | 49.4 | 50.5 | 32.8 | 34.6 | 31.3 | |||||
| 19. Cs-Spn-2_(U6) | 34.6 | 23.7 | 24.9 | 20.9 | 29.1 | 26.1 | 18.8 | 20.1 | 24.3 | 21.7 | 23.3 | 30.2 | 29.7 | 30.1 | 60.1 | 70.3 | 76.4 | 29.8 | ||||
| 20. Cs-Spn-3_(U1) | 24.6 | 22.7 | 24.0 | 20.7 | 23.5 | 20.0 | 29.1 | 54.9 | 24.2 | 19.6 | 24.8 | 24.0 | 21.1 | 23.4 | 24.3 | 24.3 | 22.2 | 25.3 | 21.4 | |||
| 21. Cs-Spn-4_(U1) | 25.3 | 25.1 | 22.1 | 17.6 | 23.8 | 20.5 | 55.1 | 25.5 | 20.1 | 19.1 | 17.7 | 22.7 | 20.9 | 21.9 | 20.0 | 20.1 | 18.8 | 22.7 | 18.7 | 27.3 | ||
| 22. Cs-Spn-5_(U2) | 22.5 | 16.6 | 22.5 | 12.8 | 20.0 | 14.4 | 18.1 | 20.0 | 36.4 | 18.4 | 18.8 | 20.3 | 20.5 | 19.9 | 18.7 | 20.1 | 18.8 | 22.1 | 19.1 | 19.5 | 17.1 | |
| 23. Hv-Spn-1 | 35.5 | 24.4 | 26.1 | 19.7 | 33.2 | 22.2 | 23.2 | 25.4 | 28.5 | 26.5 | 24.0 | 34.1 | 32.9 | 34.1 | 30.2 | 31.9 | 28.5 | 32.8 | 28.5 | 25.1 | 20.5 | 22.0 |
Additionally, homology models of Ci-Spn-9 and Cs-Spn-2 illustrate the conserved serpin core with endoplasmic reticulum retention signals as HDEF and HDEL (Fig. 5), respectively. These two serpins have different RCL sequences.
Fig 5.
Protein structural models of two Ciona serpins. These two protein models are based on the crystal structure of human α1-antitrypsin (PDB Id: 1OPH, chain:A) with RMSD of 0.67 Å and 0.66 Å, respectively.
3. Discussion
Serpins from urochordates are classified into six groups (U1-U6) on the basis of exon-intron structures, chromosomal localization and Bayesian phylogenetic analysis. Vertebrate serpins possesses four features: (a) Serpin genes exhibit a variety of distinct exon-intron patterns, which divides vertebrate serpins into six groups (V1-V6) 9. (b) Extensive gene clustering is found such as in A1AT-like serpins 13 and in ov-serpins 17. (c) Serpins exhibit dynamic structural properties and they exist in three forms such as active, cleaved, and latent forms. (d) Considerable functional radiations exist in serpins. Serpins from urochordates possess these four features in similar manner. These serpins encodes for 30 different introns positions (Fig. 1), which lead into six distinct exon-intron patterns, dividing into six groups (U1-U6). This classification is similar to vertebrate classification of groups V1-V6, but these are distinct groups. Lancelets also have three distinct serpin groups (L1 and L3) 16, which are different from both groups of vertebrate serpins and urochordate serpins. Lancelet group L2 harbors two introns at the positions 283c and 339a 16, shared with urochordate groups U2 and U6. Intron at the position 339c appears to be frequently occurring intron position, reported in various serpins from different eukaryotic lineages 16. However, currently known metazoan genomes are insufficient to corroborate the phylogenetic origin of this intron 5.
Group U2 to U4 are single member groups and possesses single member on genomic loci (Fig. 2). Similarly, groups V5 5 and V6 are also single member serpins possessing single members on the genomic loci. In the vertebrate's group V2 - two serpins namely angiotensinogen 6 and heparin cofactor 18, group V3 -- all five members 14, C1 inhibitor in the group V4 20, single group V5 5 and V6 16 have single serpin gene on one genomic loci. Group U4 member, Ci-Spn-5 is sharing two intron positions with vertebrate group V5 (Fig. 1). To understand whether this is incidental or due to common ancestry, but they do not possess any other characteristics of vertebrate ATIII 5. Urochordate group U6 and vertebrate group V6 shared an intron at position 225a (Fig. 1) and endoplasmic reticulum retention signal in the respective proteins.
The urochordate group U5 and U6 have slightly higher sequence identities with vertebrate groups V1 and V5 (table 4) also evident in the phylogenetic classification (Fig. 4). However, they do not share any other features, hence orthologous status were not confirmed. There is difference of numbers of serpins between two urochordate genomes. Best explanation for this variation is the differences of extend of gene duplications within these two species, such as in case of tandem serpin duplications are lesser in C. savignyi for groups U5 and U6 with ratio among two species is 3:1 and 2:1 for C. intestinalis:C. savignyi. In contrast, groups U3-U4 have no members in C. savignyi, suggested lesser extent of segmental duplications. The genomes of C. intestinalis and C. savignyi have 16,658 and 11,616 coding genes (Ensembl 74 [December 2013]), respectively. This corroborates that about 5000 genes are lesser in C. savignyi in comparison C. intestinalis. Hence, tandem and segmental duplications are accountable of these differences.
These six group members of urochordates serpins (U1-U6) are localized into six different genomic fragments (Fig. 2) and separating branching of these six groups in Bayesian phylogenetic tree (Fig. 3), also supported exon-encoded classification of urochordate serpins. Serpins on same syntenic fragments share higher sequence identities and similarities, as these serpins have originated by tandem duplication events (Table 3). Although Ci-Spn-9 and Cs-Spn-2 shares the conserved serpin core with endoplasmic reticulum retention signals, however, these two serpins have different RCL sequences such as GSEAAATTSVRTQCD-SVAFN and GGDVLAERAEANRTP-GNDRT with different P1-P1' active sites (Fig. 5), respectively. These two serpins hints that these serpins have different physiological roles.
Collectively the data presented here revealed that serpins from these two urochordates are forming six groups U1-U6, which are different from vertebrate six groups V1-V6. These two urochordates harbor serpins that are highly diverged and that have no orthologs in vertebrates based on analysis of synteny, gene structures, and protein sequences.
4. Material and methods
4.1. Serpins catalogue from two Ciona species
The JGI's C. intestinalis genome database and the C. savignyi genome at the Ensembl database release 74 (December 2013) were scanned for serpins with BLAST suite 20, 21 using human α1-antitrypsin (A1AT).
4.2. Gene structure prediction
To ensure accuracy, gene structure prediction within the JGI database and the Ensembl 22 was taken and combined with predictions of AUGUSTUS gene prediction tool 23. Mature human A1AT was used as standard sequence for intron position mapping and numbering of intron positions, followed by suffixes a-c for their location as reported previously 5, 9.
4.3. Synteny analysis
Syntenies were constructed by illustrating flanking genes and their orientations using the JGI genome browser and the Ensembl browser for C. intestinalis and C. savignyi, respectively.
4.4. Construction of Bayesian phylogenetic tree
MUSCLE program 24, 25 was used to align the serpin protein sequences from Ciona for the purpose of phylogenetic analyses. Two phylogenetic trees was constructed using the Bayesian approach (5 runs, until average standard deviation of split frequencies was lower than 0.0098, 25% burn-in-period, WAG+G+I matrix-based model 26) in the MrBayes 3.2.1 program 27.
4.5. Construction of homology models and visualization
Homology models of Ci-Spn-9 and Cs-Spn-2 were generated using the I-TASSER 28 and visualized the resulting model using YASARA 29.
Supplementary Material
Figure S1.
References
- 1.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–6. doi: 10.1038/35038119. [DOI] [PubMed] [Google Scholar]
- 2.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–804. doi: 10.1021/cr010170+. [DOI] [PubMed] [Google Scholar]
- 3.Silverman G, Bird P, Carrell R, Church F, Coughlin P, Gettins P. et al. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–6. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- 4.van Gent D, Sharp P, Morgan K, Kalsheker N. Serpins: structure, function and molecular evolution. The international journal of biochemistry & cell biology. 2003;35:1536–47. doi: 10.1016/s1357-2725(03)00134-1. [DOI] [PubMed] [Google Scholar]
- 5.Kumar A, Bhandari A, Sarde SJ, Goswami C. Sequence, phylogenetic and variant analyses of antithrombin III. Biochemical and biophysical research communications. 2013;440:714–24. doi: 10.1016/j.bbrc.2013.09.134. doi:10.1016/j.bbrc.2013.09.134. [DOI] [PubMed] [Google Scholar]
- 6.Kumar A, Sarde SJ, Bhandari A. Revising angiotensinogen from phylogenetic and genetic variants perspectives. Biochemical and biophysical research communications. 2014;446:504–18. doi: 10.1016/j.bbrc.2014.02.139. doi:10.1016/j.bbrc.2014.02.139. [DOI] [PubMed] [Google Scholar]
- 7.Hunt LT, Dayhoff MO. A surprising new protein superfamily containing ovalbumin, antithrombin-III, and alpha 1-proteinase inhibitor. Biochem Biophys Res Commun. 1980;95:864–71. doi: 10.1016/0006-291x(80)90867-0. [DOI] [PubMed] [Google Scholar]
- 8.Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–64. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- 9.Kumar A, Ragg H. Ancestry and evolution of a secretory pathway serpin. BMC Evol Biol. 2008;8:250.. doi: 10.1186/1471-2148-8-250. doi:1471-2148-8-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development (Cambridge, England) 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 11.Benarafa C, Remold-O'Donnell E. The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11367–72. doi: 10.1073/pnas.0502934102. doi:10.1073/pnas.0502934102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiserman D, Bird PI. Analysis of vertebrate genomes suggests a new model for clade B serpin evolution. BMC genomics. 2005;6:167. doi: 10.1186/1471-2164-6-167. doi:10.1186/1471-2164-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forsyth S, Horvath A, Coughlin P. A review and comparison of the murine α1-antitrypsin and α1-antichymotrypsin multigene clusters with the human clade A serpins. Genomics. 2003;81:336–45. doi: 10.1016/s0888-7543(02)00041-1. doi:10.1016/S0888-7543(02)00041-1. [DOI] [PubMed] [Google Scholar]
- 14.Kumar A. Phylogenomics of vertebrate serpins. 2010; Ph.D. thesis, University of Bielefeld, Bielefeld, Germany. urn:nbn:de:hbz:361-17480.
- 15.Xu X, Zhang SS-M, Barnstable CJ, Tombran-Tink J. Molecular phylogeny of the antiangiogenic and neurotrophic serpin, pigment epithelium derived factor in vertebrates. BMC genomics. 2006;7:248. doi: 10.1186/1471-2164-7-248. doi:10.1186/1471-2164-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ragg H, Kumar A, Koster K, Bentele C, Wang Y, Frese MA. et al. Multiple gains of spliceosomal introns in a superfamily of vertebrate protease inhibitor genes. BMC Evol Biol. 2009;9:208.. doi: 10.1186/1471-2148-9-208. doi:10.1186/1471-2148-9-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benarafa C, Remold-O'Donnell E. The ovalbumin serpins revisited: perspective from the chicken genome of clade B serpin evolution in vertebrates. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11367–72. doi: 10.1073/pnas.0502934102. doi:10.1073/pnas.0502934102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar A, Bhandari A, Sarde SJ, Goswami C. Genetic variants and evolutionary analyses of heparin cofactor II. Immunobiology. 2014 doi: 10.1016/j.imbio.2014.05.003. doi:10.1016/j.imbio.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 19.Kumar A, Bhandari A, Sarde SJ, Goswami C. Molecular phylogeny of C1 inhibitor depicts two immunoglobulin-like domains fusion in fishes and ray-finned fishes specific intron insertion after separation from zebrafish. Biochemical and biophysical research communications. 2014 doi: 10.1016/j.bbrc.2014.05.097. doi:10.1016/j.bbrc.2014.05.097. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Lipman DJ. Protein database searches for multiple alignments. Proc Natl Acad Sci U S A. 1990;87:5509–13. doi: 10.1073/pnas.87.14.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S. et al. Ensembl 2013. Nucl Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. doi:Doi 10.1093/Nar/Gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanke M, Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Research. 2005;33:W465–W7. doi: 10.1093/nar/gki458. doi:10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113.. doi: 10.1186/1471-2105-5-113. doi:10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. doi:10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–9. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 27.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 28.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–38. doi: 10.1038/nprot.2010.5. doi:10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1.