Abstract
A whole genome association (WGA) study was carried out to find quantitative trait loci (QTL) for sensory evaluation traits in Hanwoo. Carcass samples of 250 Hanwoo steers were collected from National Agricultural Cooperative Livestock Research Institute, Ansung, Gyeonggi province, Korea, between 2011 and 2012 and genotyped with the Affymetrix Bovine Axiom Array 640K single nucleotide polymorphism (SNP) chip. Among the SNPs in the chip, a total of 322,160 SNPs were chosen after quality control tests. After adjusting for the effects of age, slaughter-year-season, and polygenic effects using genome relationship matrix, the corrected phenotypes for the sensory evaluation measurements were regressed on each SNP using a simple linear regression additive based model. A total of 1,631 SNPs were detected for color, aroma, tenderness, juiciness and palatability at 0.1% comparison-wise level. Among the significant SNPs, the best set of 52 SNP markers were chosen using a forward regression procedure at 0.05 level, among which the sets of 8, 14, 11, 10, and 9 SNPs were determined for the respectively sensory evaluation traits. The sets of significant SNPs explained 18% to 31% of phenotypic variance. Three SNPs were pleiotropic, i.e. AX-26703353 and AX-26742891 that were located at 101 and 110 Mb of BTA6, respectively, influencing tenderness, juiciness and palatability, while AX-18624743 at 3 Mb of BTA10 affected tenderness and palatability. Our results suggest that some QTL for sensory measures are segregating in a Hanwoo steer population. Additional WGA studies on fatty acid and nutritional components as well as the sensory panels are in process to characterize genetic architecture of meat quality and palatability in Hanwoo.
Keywords: Hanwoo, Single Nucleotide Polymorphism, Meat Quality, Palatability, Whole Genome Association
INTRODUCTION
In the last several decades, quantitative trail loci (QTL) or genes with causal polymorphism for economic traits such as milk production, carcass traits, and meat qualities in farm animals have been extensively studied (Seaton et al., 2002). Many of the economic traits are polygenic i.e. affected by multiple genes, each of which contributes a small effect on phenotype expression, which did not allow for efficient identification of causal mutations in the trait of interest.
However, as sequencing technologies such as next generation sequencing have evolved, whole genome sequencing enabled of the generation of many genetic markers, e.g. single nucleotide polymorphism (SNP) using high-throughput genotyping platforms (Hartwell, 2000). Also, the whole genome of the Hereford breed was sequenced by the Bovine Genome Sequencing and Analysis Consortium led by NIH and USDA, and several kinds of high density SNP chips or arrays developed by Illumina or Affymetrix are commercially available (Rincon et al., 2011), enabling the implementation of whole genome association (WGA) studies (Goddard and Hayes, 2009).
Korean beef consumers prefer Hanwoo (a Korean native cattle) beef to the imported breeds from US or Australia, mainly due to high intramuscular fat content, as well as thin muscle fibers, minimal connective tissues, and good flavor (Jo et al., 2012). To meet the consumers’ demands, the selection of the Hanwoo breed has been focused on high marbling and thus high meat quality (Choy et al., 2012). There were some reports about WGA studies on marbling or meat quality in Hanwoo (Lee et al., 2010; Kim et al., 2011; Lee et al., 2013). However, no WGA study about sensory measurements has been reported in Hanwoo.
Herein, we first report WGA mapping results to detect QTL for sensory traits in Hanwoo using high density SNP chips.
MATERIALS AND METHODS
Animals and phenotypes
Samples of 250 Hanwoo steers were randomly collected at the farm of Livestock Research Center, National Agricultural Cooperative Federation, Ansung, Gyeonggi province, Korea. The steers were transported after weaning (about 6 to 7 months of age) from Hanwoo producers across South Korea, raised under similar management practices, and fed with the same diet ad libitum. The steers were treated according to the recommendations described in “The Guide for the Care and Use of Laboratory Animals” published by the institutional Animal Care and Use Committee (IACUC) of NIAS (2012-C-037) in Korea. The steers were slaughtered at about 31 months of age at a local municipal slaughterhouse between November, 2011 and February, 2012. The carcasses were cut into two parts without electric stimulation, washed, and immediately cooled at 0°C for 24 h in a chilling room without packaging. The average weight (standard deviation) of the carcasses was 409 (54) kg, ranging from 213 to 484 kg. In accordance with the current Animal Product Grading System of Korea, the carcasses were dissected at the last rib and the first lumber vertebra. Carcass yield and quality grades were measured, i.e. as A (carcass weight 372 kg), B (404 kg), C (440 g) for 71, 102, and 77 individuals, and 1++, 1+, 1, 2 (marbling) for 35, 53, 120, and 42 individuals, respectively. Samples were cut into 10mm thickness and deeply heated up to 72°C temperature on a both side grill before checking for color, aroma, tenderness, juiciness and overall palatability (scaling from 1, lowest to 9, greatest) by eight trained sensory panels. Average values of nine point likert scale method were measured for the sensory tests. Summary statistics of the five sensory traits were described in Table 1.
Table 1.
Summary statistics for the five sensory related traits in the 250 Hanwoo steers
| Traits | n | Mean | SDa | Min | Max | CVb |
|---|---|---|---|---|---|---|
| Aroma | 249 | 5.09 | 2.34 | 1.00 | 7.67 | 46.0 |
| Color | 250 | 5.20 | 0.88 | 1.67 | 7.33 | 17.0 |
| Juiciness | 250 | 5.11 | 1.33 | 1.67 | 8.67 | 26.1 |
| Tenderness | 250 | 4.99 | 1.39 | 1.00 | 8.67 | 27.9 |
| Overall acceptability | 250 | 4.95 | 1.34 | 1.00 | 8.33 | 27.0 |
SD, standard deviation; Min, minimum; Max, maximum; CV, coefficient of variation (%).
Molecular data
The Affymetrix Bovine Axiom Array 640k SNP chips were used for genotyping the Hanwoo steers. Quality controls were tested to screen available SNPs with PLINK software version 1.07 (Purcell et al., 2007), and those SNPs were removed with the following four criteria; i) the frequency of the least genotype was less than ten, ii) the call rate was smaller than 95%, iii) p-value of Hardy-Weinberg equilibrium test was less than 0.001, iv) minor allele frequency was less than 5%.
Statistical analysis
Genome-wide association analyses were performed according to Aulchenko et al. (2007). At the first step, slaughter-year-season and age of month were fitted as a fixed effect and a covariate, using SAS general linear model procedure of SAS v9.1. Then, the fixed or covariate with statistical significance at 0.1 level was fitted into a mixed model with a genome-relationship (G) matrix (van Raden et al., 2008), because the pedigree information of the 250 steers was limited. The G-matrix was constructed using the R subroutine (version 2.15.0), and the residuals of each phenotype were obtained from the mixed model equation using ASREML program (version 3.0). At the second step, the residuals were regressed on each SNP using a simple linear regression model by PLINK version 1.07 software. For each individual, the SNP genotype values for BB, BA, and AA were assigned as −1, 0, and 1, such that allele substitution effect replacing B with A allele was estimated. To set threshold values of statistical significance, 0.1% point-wise p value from the F distributions was applied for each SNP test.
Among the significant SNPs for each trait, the best set of SNP markers were selected by applying a forward regression procedure, because some of the significant SNPs would yield redundant information due to the relationships between closely linked SNPs, i.e. linkage disequilibrium (LD), a non-random association between alleles of different SNPs. Inclusion of each SNP into the model was determined at 0.05 level.
The variation explained by each SNP (S2SNP) was calculated as , where i denotes each genotype, αi is allele substitution effect (= −â, 0, and + â for BB, AB, and AA, respectively, in which â was estimated from the linear regression analysis for the SNP), fi is the frequency of ith genotype, μ is the population mean that can be expressed as (fAA-fBB)â (Falconer and Mackay, 1996). Proportion of phenotypic variance due to the SNP was then estimated as SSNP/S2P, in which S2P, phenotypic variance, was obtained from residual values of the trait after adjusting the fixed or covariate effect. Therefore, the estimate of the proportion of phenotype variance due to all of the significant SNPs was ∑S2SNPi/S2P.
RESULTS AND DISCUSSION
The summary statistics for the five sensory measures are shown in Table 1. The coefficients of variations of the sensory traits ranged between 17% and 46% with the greatest for Aroma (Table 1). This result showed that, in general, the sensory traits had greater variation than body conformations or carcass quality traits in Hanwoo (Lee et al., 2010; Alam et al., 2011).
After quality control processes, a total of 322,160 SNPs were selected from the 647,866 SNPs in the Affymetrix Bovine Axiom Array 640k chip. The number of available SNPs per chromosome and average interval sizes between flanking SNPs are shown in Table 2. The physical map of the available SNPs spanned 2,654 Mbp and the average distance between adjacent SNPs was 8.24±14.38 kb. More than 10,000 SNPs were available in autosomes of 1 through 15 except 14. However, the available SNPs were very low, i.e. 49.7% (Table 2), relative to the number of SNPs that were embedded in the Affymetrix Bovine Axiom array 640k chip. One of the reasons may be that the contents of the SNPs in the array were based on twenty multi-breeds, such that the availability of the SNPs on the array was limited in Hanwoo breed, which has quite different genetic characteristics compared to European or Continental commercial breeds (Decker et al., 2009).
Table 2.
The number of available SNPs in the 250 Hanwoo steers and average interval distance between adjacent SNPs that are embedded in the bovine Affymetrix 640k chip
| BTA | Number of SNPsa | %b | Average interval size (kb) | Standard deviation (kb) | Total distance (kb)c |
|---|---|---|---|---|---|
| 1 | 19,829 | 50.3 | 8.0 | 10.2 | 158,300 |
| 2 | 17,110 | 50.8 | 8.0 | 10.9 | 136,520 |
| 3 | 14,912 | 51.8 | 8.1 | 11.4 | 121,351 |
| 4 | 16,534 | 52.0 | 7.3 | 10.1 | 120,629 |
| 5 | 13,765 | 51.6 | 8.8 | 13.5 | 121,164 |
| 6 | 15,347 | 54.0 | 7.8 | 14.6 | 119,272 |
| 7 | 13,508 | 50.8 | 8.3 | 17.1 | 112,623 |
| 8 | 14,206 | 50.5 | 8.0 | 11.2 | 113,334 |
| 9 | 14,001 | 51.7 | 7.5 | 10.1 | 105,584 |
| 10 | 14,433 | 50.8 | 7.2 | 11.2 | 104,236 |
| 11 | 13,015 | 48.9 | 8.2 | 15.3 | 107,190 |
| 12 | 11,916 | 52.4 | 7.6 | 20.2 | 91,116 |
| 13 | 10,239 | 50.7 | 8.2 | 12.9 | 84,132 |
| 14 | 9,221 | 50.0 | 9.1 | 14.0 | 83,465 |
| 15 | 11,154 | 51.3 | 7.6 | 10.9 | 85,173 |
| 16 | 9,753 | 49.1 | 8.3 | 14.7 | 81,403 |
| 17 | 8,500 | 48.2 | 8.8 | 16.0 | 74,634 |
| 18 | 7,821 | 49.7 | 8.4 | 16.0 | 65,898 |
| 19 | 5,987 | 44.5 | 10.7 | 17.6 | 63,915 |
| 20 | 9,553 | 52.2 | 7.5 | 10.4 | 71,762 |
| 21 | 9,255 | 50.4 | 7.7 | 13.4 | 71,461 |
| 22 | 7,917 | 52.1 | 7.7 | 12.8 | 61,277 |
| 23 | 9,746 | 55.3 | 5.4 | 9.7 | 52,491 |
| 24 | 8,796 | 51.4 | 7.1 | 10.2 | 62,411 |
| 25 | 6,082 | 53.2 | 7.0 | 16.9 | 42,764 |
| 26 | 7,628 | 50.1 | 6.7 | 10.3 | 50,830 |
| 27 | 7,629 | 51.8 | 6.0 | 11.0 | 45,400 |
| 28 | 7,161 | 52.4 | 6.5 | 9.5 | 46,206 |
| 29 | 7,095 | 50.6 | 7.2 | 14.3 | 51,072 |
| X | 0 | 0 | - | - | - |
| Pseudo | 47 | 17.1 | 3,227.5 | 11,526.1 | 148,466 |
| Total: 322,160 | Mean: 49.7 | Mean: 8.2 | Mean: 14.4 | Total: 2,654,080 |
SNP, single nucleotide polymorphism; BTA, bovine chromosome number.
Among the 647,866 SNPs in the Affymetrix 640 SNP chip, the number of SNPs were chosen after quality control tests.
Proportion of available SNPs in each BTA.
Distance between the first and the last SNPs that was located on each BTA.
A total of 1,631 SNPs were detected, and the numbers of significant SNPs were 325, 301, 309, 335, and 361 SNPs for color, aroma, tenderness, juiciness and palatability, respectively (results not shown). For the significant SNPs, a forward selection procedure was applied to determine independent sets of SNPs for each trait. Fifty two significant SNPs were chosen as the best marker set, for which 8, 14, 11, 10, and 9 SNPs were determined for the respective traits (Table 3). The significant SNPs explained about 18% to 31% of phenotypic variance, with the maximum for aroma (31%) and the minimum for color (18%). Figure 1 displays profiles of the test statistics after WGA analyses for the five sensory traits.
Table 3.
Identities, positions, and effect of the SNPs associated with sensory test traits in a Hanwoo steer population
| Trait/SNP maker | SNPa | BTA | Position (bp) | Estimateb | SE | −log10Pc | % σ2pd |
|---|---|---|---|---|---|---|---|
| Aroma | 30.7 | ||||||
| AX-22028107 | [A/G] | 2 | 6,300,508 | −0.49 | 0.18 | 1.74 | |
| AX-22310754 | [A/G] | 2 | 80,382,416 | −0.36 | 0.18 | 1.30 | |
| AX-22504510 | [A/G] | 2 | 128,694,058 | 0.60 | 0.17 | 2.34 | |
| AX-25044518 | [T/C] | 3 | 39,776,491 | 0.34 | 0.12 | 2.12 | |
| AX-25252417 | [T/C] | 3 | 94,943,252 | 0.31 | 0.11 | 1.34 | |
| AX-25283035 | [T/C] | 3 | 102,534,659 | 0.22 | 0.12 | 1.30 | |
| AX-26832968 | [A/G] | 7 | 14,142,752 | 0.30 | 0.10 | 2.24 | |
| AX-26932949 | [T/C] | 7 | 38,591,573 | 0.29 | 0.12 | 1.35 | |
| AX-27236015 | [A/G] | 8 | 2,138,386 | −0.25 | 0.13 | 1.63 | |
| AX-18755438 | [T/C] | 10 | 35,548,753 | 0.19 | 0.10 | 1.32 | |
| AX-20705361 | [C/G] | 15 | 48,461,742 | −0.56 | 0.21 | 1.43 | |
| AX-21567050 | [A/C] | 18 | 22,783,742 | 0.50 | 0.18 | 1.70 | |
| AX-23995926 | [A/G] | 25 | 25,630,340 | −0.24 | 0.09 | 1.74 | |
| AX-24413580 | [T/C] | 27 | 30,873,451 | 0.37 | 0.10 | 1.52 | |
| Color | 18.4 | ||||||
| AX-18487566 | [A/G] | 1 | 129,176,475 | 0.13 | 0.08 | 1.38 | |
| AX-27133183 | [G/A] | 7 | 88,729,387 | 0.18 | 0.07 | 1.71 | |
| AX-27889364 | [A/G] | 9 | 58,658,454 | 0.23 | 0.09 | 2.01 | |
| AX-19559065 | [A/C] | 12 | 20,239,702 | −0.20 | 0.07 | 2.30 | |
| AX-19613598 | [T/C] | 12 | 33,269,516 | −0.14 | 0.07 | 1.41 | |
| AX-21113987 | [G/A] | 16 | 63,613,670 | −0.13 | 0.06 | 1.39 | |
| AX-21117675 | [G/A] | 16 | 64,372,169 | 0.16 | 0.06 | 1.92 | |
| AX-21733351 | [T/C] | 18 | 61,989,245 | 0.18 | 0.08 | 1.64 | |
| Juiciness | 25.6 | ||||||
| AX-18342275 | [C/T] | 1 | 90,088,898 | −0.55 | 0.23 | 1.31 | |
| AX-22292276 | [A/G] | 2 | 75,388,033 | 0.36 | 0.18 | 1.32 | |
| AX-26352284 | [T/C] | 6 | 7,549,442 | 0.66 | 0.23 | 1.75 | |
| AX-26703353 | [T/C] | 6 | 101,662,167 | 0.42 | 0.11 | 2.61 | |
| AX-26742891 | [C/T] | 6 | 110,948,656 | 0.32 | 0.11 | 1.83 | |
| AX-27728564 | [A/G] | 9 | 15,441,609 | 0.35 | 0.19 | 1.33 | |
| AX-19104043 | [C/A] | 11 | 15,600,024 | 0.32 | 0.15 | 1.52 | |
| AX-22572250 | [G/A] | 20 | 6,155,235 | 0.31 | 0.11 | 2.25 | |
| AX-23535517 | [C/G] | 23 | 33,495,661 | 0.40 | 0.19 | 1.84 | |
| AX-24016864 | [T/G] | 25 | 30,386,628 | 0.30 | 0.14 | 1.74 | |
| Palatability | 23.5 | ||||||
| AX-26703353 | [T/C] | 6 | 101,662,167 | 0.30 | 0.08 | 4.30 | |
| AX-26742891 | [C/T] | 6 | 110,948,656 | 0.18 | 0.08 | 2.01 | |
| AX-27056668 | [A/G] | 7 | 69,048,276 | 0.18 | 0.08 | 1.53 | |
| AX-18624743 | [A/G] | 10 | 3,581,326 | −0.40 | 0.19 | 1.64 | |
| AX-20558433 | [C/T] | 15 | 13,361,044 | −0.23 | 0.09 | 1.65 | |
| AX-21807591 | [T/C] | 19 | 16,225,261 | −0.89 | 0.38 | 1.81 | |
| AX-21807600 | [A/G] | 19 | 16,226,761 | −1.10 | 0.39 | 1.67 | |
| AX-23847051 | [T/C] | 24 | 54,680,656 | −0.33 | 0.12 | 1.56 | |
| AX-23856093 | [C/T] | 24 | 56,668,495 | 0.25 | 0.12 | 1.39 | |
| Tenderness | 28.1 | ||||||
| AX-22345506 | [T/C] | 2 | 90,197,319 | 0.20 | 0.11 | 1.70 | |
| AX-26703353 | [T/C] | 6 | 101,662,167 | 0.23 | 0.11 | 2.63 | |
| AX-26742891 | [C/T] | 6 | 110,948,656 | 0.30 | 0.10 | 1.92 | |
| AX-27646347 | [C/T] | 8 | 107,014,241 | −0.20 | 0.10 | 1.33 | |
| AX-27809598 | [T/C] | 9 | 36,731,857 | 0.18 | 0.11 | 1.49 | |
| AX-18624743 | [A/G] | 10 | 3,581,326 | −0.51 | 0.23 | 1.77 | |
| AX-19181219 | [C/T] | 11 | 35,096,159 | −0.21 | 0.10 | 1.43 | |
| AX-19331523 | [G/A] | 11 | 73,007,955 | −0.21 | 0.10 | 1.52 | |
| AX-22865134 | [A/G] | 21 | 8,651,267 | 0.27 | 0.11 | 2.36 | |
| AX-24194530 | [C/T] | 26 | 30,461,302 | 0.21 | 0.10 | 1.38 | |
| AX-24734214 | [A/C] | 29 | 15,421,430 | −0.24 | 0.11 | 1.96 |
SNP, single nucleotide polymorphism; BTA, bovine chromosome number; SE, standard error.
Nucleotides of substitution.
Estimate is for allele substitution effect replacing the latter with the former allele (nucleotide) in the SNP column.
Negative logarithm of the comparison-wise p-value of the test statistic against the null hypothesis of no SNP effect at the SNP position.
Proportion of phenotypic variance explained by the SNP. The values on the rows of trait name are the sum of the % σ2p values of all SNPs.
Figure 1.
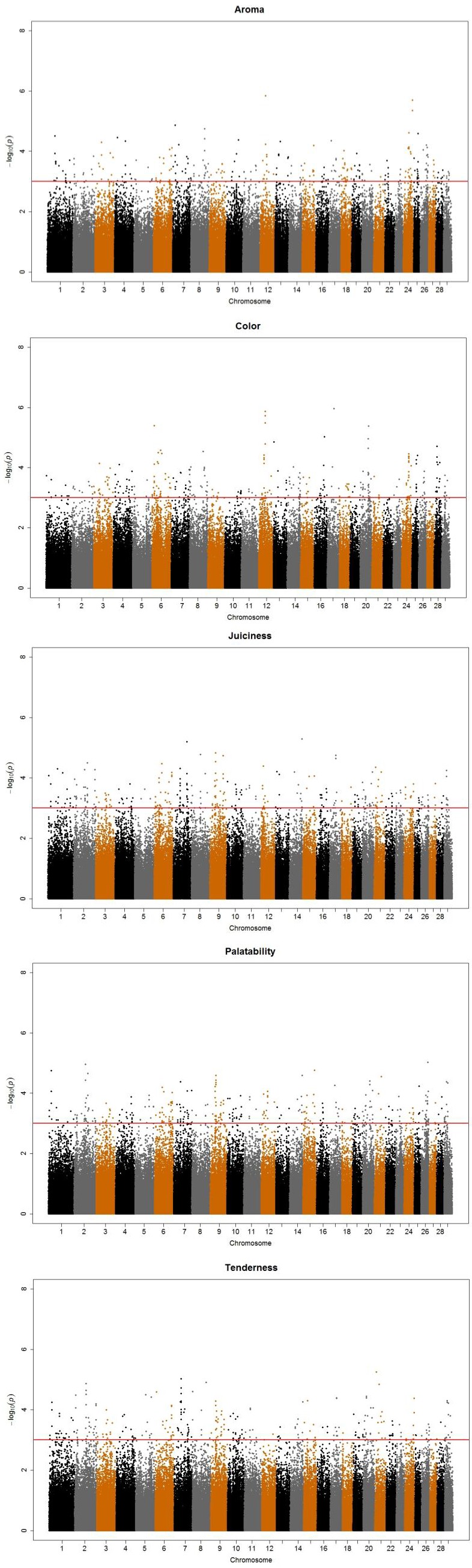
i) The Manhattan plots for the five sensory evaluation results after genome-wise association analyses. X-axis indicates chromosome number and Y-axis p values (-log10P) from the F test statistics for each single nucleotide polymorphism. ii) The Manhattan plots for the five sensory evaluation GWAS results after genome-wise association analyses. X-axis indicates chromosome number and Y-axis p values (-log10P) from the F test statistics for each single nucleotide polymorphism.
In general, the detected SNPs for each trait were distributed across autosomal chromosomes, and the SNPs that were detected on the same chromosomes were located at good distances, e.g. more than 9 Mbp, except the two SNPs on BTA16 for color and the four SNPs on BTA19 and BTA24 for palatability, respectively (Table 3). This may be partly due to the statistical analysis process, i.e. the forward regression procedure eliminated the significant SNPs that were closely located, and had great LD with the significant markers that were chosen (Table 3).
The number of significant markers for the sensory traits was smaller than the QTL mapping reports in other beef cattle (Gill et al., 2010; Allais et al., 2011). One of the main reasons may be the small sample size (n = 250) used in this study, which is a crucial factor in detecting significant QTL (Goddard and Hayes, 2009). Therefore, further collection of genotypes and phenotypes for the sensory traits is in process using Hanwoo steers.
Another reason may be low LD between markers in Hanwoo population. While intensive selection programs for western commercial breeds such as Angus or Holstein have been implemented for the last 100 years, the breeding program of Hanwoo for growth and carcass quality started less than 40 years ago (Jo et al., 2012). This would cause LDs between closely linked markers to be low in Hanwoo cattle compared to the western cattle breeds, which would not allow for the detection of trait-related SNPs, unless the SNPs are very close and tightly linked to the causal mutations for the traits.
To evaluate the extent of LD in Hanwoo and to determine the number of markers that is sufficient to have enough power for QTL detection, LDs between syntenic SNPs in the bovine Affymetrix axiom array were measured using Gold program (1.1.0 version) (McKay et al., 2007). A LD measure, r2 value, was used for all pairs of syntenic SNPs in the autosomal chromosomes (Devlin and Risch, 1995). The r2 values were binned according to the physical distance separating the loci (Table 4). The average r2 values (bin size) were 0.32 (1 kb), 0.24 (10 kb), 0.18 (20 kb), 0.14 (30 kb), 0.17 (40 kb), 0.18 (50 kb), 0.16 (60 kb), 0.08 (70 kb), 0.18 (80 kb), 0.07 (90 kb), 0.16 (100 kb), 0.17 (200 kb), 0.16 (300 kb), and less than 0.03 (>400 kb), respectively, showing that the LDs in Hanoo were lower between close distance loci, compared to other Bos taurus commercial breeds (Goddard and Hayes, 2009). McKay et al. (2007) also reported that the average r2 value was greater than 0.4 when two DNA markers were 1 kb apart in six breeds, i.e. Charolaise, Brahman, Holstein, Japanese Black, and Limousine.
Table 4.
Weighted mean and standard deviation of r2, a measure of linkage disequilibrium, between pairs of SNPs
| Bina | Interval size between SNP pairs | Whole genome level | |
|---|---|---|---|
|
| |||
| Average r2 | SD | ||
| 1 | 0–1 kbp | 0.320 | 0.327 |
| 2 | 1–10 kbp | 0.244 | 0.284 |
| 3 | 10–20 kbp | 0.178 | 0.232 |
| 4 | 20–30 kbp | 0.142 | 0.198 |
| 5 | 30–40 kbp | 0.169 | 0.187 |
| 6 | 40–50 kbp | 0.176 | 0.194 |
| 7 | 50–60 kbp | 0.157 | 0.178 |
| 8 | 60–70 kbp | 0.082 | 0.132 |
| 9 | 70–80 kbp | 0.177 | 0.194 |
| 10 | 80–90 kbp | 0.067 | 0.113 |
| 11 | 90–100 kbp | 0.160 | 0.182 |
| 12 | 100–200 kbp | 0.174 | 0.193 |
| 13 | ~300 kbp | 0.164 | 0.183 |
| 14 | ~400 kbp | 0.023 | 0.041 |
| 15 | ~500 kbp | 0.020 | 0.035 |
| 16 | ~600 kbp | 0.019 | 0.031 |
| 17 | ~700 kbp | 0.018 | 0.029 |
| 18 | ~800 kbp | 0.017 | 0.028 |
| 19 | ~900 kbp | 0.017 | 0.027 |
| 20 | ~1,000 k(1Mb) | 0.016 | 0.026 |
| 21 | ~2 Mb | 0.015 | 0.023 |
| 22 | ~3 Mb | 0.014 | 0.021 |
| 23 | ~4 Mb | 0.013 | 0.020 |
| 24 | ~5 Mb | 0.013 | 0.019 |
| 25 | ~6 Mb | 0.012 | 0.018 |
| 26 | ~7 Mb | 0.012 | 0.017 |
| 27 | ~8 Mb | 0.011 | 0.017 |
| 28 | ~9 Mb | 0.011 | 0.016 |
| 29 | ~10 Mb | 0.010 | 0.015 |
SNP, single nucleotide polymorphism; SD, standard deviation.
The r2 values for pairs of SNPs were binned according to the physical distance separating the two SNPs.
Since the r2 values were greater than 0.2 for the pair of SNPs with the distance less than 10 kb, which was a sufficient distance to capture enough LD information in cattle (Goddard and Hayes, 2009), it was expected that the marker map density is not a determining factor to influence the power to detect QTL in this WGA study.
Chromosome- or genome-wise threshold values to take multiple tests into account that were obtained by permutation tests or false discovery rate methods were not considered (Churchill and Doerge, 1994; Benjamini and Hochberg, 1995), because it would reach too stringent threshold values causing very limited number of significant SNPs to be detected in this study.
Three SNPs affected more than two traits, i.e. AX-26703353 (BTA6, 101 Mb) and AX-26742891 (BTA6, 110 Mb) for tenderness, juiciness, and palatability. Another SNP, AX-18624743 that was positioned at 3.5 Mb of BTA10 was associated with tenderness and palatability (Table 3), indicating pleiotropic effect, i.e. one gene influencing multiple phenotypes.
Some QTL regions in this study were located in the regions where previous studies reported QTL for the same or related traits. To compare our results with the cattle QTL website (http://www.animalgenome.org/cgi-bin/QTLdb/index), in which more than 8,305 QTLs have been reported, the Btau_4.6 version was used, which enabled conversion of genetic distance to physical distance on the website.
Alexander et al. (2007) and Gutiérrez-Gil et al. (2008) found aroma QTL on BTA2 in an F2 population from Wagyu×Limousine and BTA7 in an F2 population from Charolaise×Holstein cross, respectively. The two QTL were positioned between 5.8 to 6.5 Mb (p = 0.11) and 72.5 to 96.5 Mb (p = 0.0238). Our study found five SNPs on the same BTAs, one of which was AX-22028107 at 6.3 Mb of BTA2, the exact position of the QTL in Alexander et al. (2007).
Cacas et al. (1998;2000;2003) detected tenderness QTL on BTA2 (5.1 to 5.3 Mb) in a Piedmontese × Angus cross, on BTA29 (36.4 to 46.5 Mb) in Belgian Blue and Piedmontese, and on BTA29 (27.4 to 48.2 Mb) in an F2 population from Brahman×Hereford cross, respectively. Allais et al. (2010;2011) found tenderness QTL at 5.9 to 6.1 Mb of BTA2 (p<0.01) and at 43.7 to 43.9 Mb of BTA 29 (p = 0.002) in Charolaise, Limousine and Blonde d’Aquitaine young bulls. Gutiérrez-Gil et al. (2008) found two tenderness QTL at 56.2 to 77.4 Mb of BTA10 (p = 0.01) and at 34.2 to 51.6 Mb of BTA 29 (p = 0.0079) in F2s from bull calves of a Charolaise×Holstein cross. Gill et al. (2009; 2010) reported two tenderness QTL at 43.7 to 43.9 Mb of BTA29 (p<0.05) and at 82.1 to 82.2 Mb of BTA11 in a commercial Angus-cross cattle. In the chromosomal region where the tenderness QTLs were detected by Gill et al. (2010) and Casas et al. (2003), AX-19331523 for tenderness was detected at 73 Mb of BTA11 (Table 3).
We detected a SNP for color, AX-27133183 at 88.7 Mb of BTA7. In the similar region, Reardon et al. (2010) detected a QTL in crossbred Bos taurus cattle, i.e. at 96.8 to 97 Mb (p = 0.0002) for the color.
One of the factors that mainly contribute to meat taste is fatty acid. For example, fat tissue containing abundant mono-unsaturated fatty acid contributes to beef flavor (Dryden and Marchello, 1970; Melton et al., 1982; Sturdivant et al., 1992; Jeremiah et al., 1996). In particular, oleic acid (C18:1) is positively correlated with the overall palatability of beef (Waldman et al., 1968; Westerling and Hedrick, 1979). Therefore, characterization of the relationship between sensory traits and fatty acid components in meat would facilitate palatability improvement in Hanwoo meat. Furthermore, fatty acid components that are related to health and nutrition, such as stearic acid (C18:0) and oleic acid (C18:1) have effects on low density lipoprotein cholesterol (Bonanome and Grundy, 1988; Berner, 1993), for which Hanwoo meat could be genetically improved through genome selection.
Currently, phenotype and genotypic data for fatty acids as well as additional sensory measurements from Hanwoo steers are being collected for further study regarding the relationship between fatty acids and sensory traits, as well as for GWA analyses.
CONCLUSION
Using the Affymetrix bovine 640k SNP chip, a whole genome scan results for sensory traits are reported in a Hanwoo population. Even though the number of significantly detected SNPs was limited, partly due to a small sample size, our results provide preliminary genetic basis for characterizing genetic architecture of the sensory traits in Hanwoo. Additional study and validation tests are required, in order to apply the SNPs for sensory traits to commercial Hanwoo populations through marker-assisted breeding programs.
ACKNOWLEDGMENTS
This research was supported by the Technology Development Program for Agriculture and Forestry (Project No. 311016-3), Ministry of Agriculture, Forestry, and Fisheries, Republic of Korea, 2013.
REFERENCES
- Alam M, Lee YM, Park BL, Kim JH, Lee SS, Shin HD, Kim KS, Kim NS, Kim JJ. A whole genome association study to detect single nucleotide polymorphisms for body conformation traits in a Hanwoo population. Asian Australas J Anim Sci. 2011;24:322–329. [Google Scholar]
- Alexander LJ, Macneil MD, Geary TW, Snelling WM, Rule DC, Scanga JA. Quantitative trait loci with additive effects on palatability and fatty acid composition of meat in a Wagyu-Limousin F2population. Anim Genet. 2007;38:506–513. doi: 10.1111/j.1365-2052.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- Allais S, Levéziel H, Payet-Duprat N, Hocquette JF, Lepetit J, Rousset S, Denoyelle C, Bernard-Capel C, Journaux L, Bonnot A, Renand G. The two mutations, Q204X and nt821, of the myostatin gene affect carcass and meat quality in young heterozygous bulls of French beef breeds. J Anim Sci. 2010;88:446–454. doi: 10.2527/jas.2009-2385. [DOI] [PubMed] [Google Scholar]
- Allais S, Journaux L, Levéziel H, Payet-Duprat N, Raynaud P, Hocquette JF, Lepetit J, Rousset S, Denoyelle C, Bernard-Capel C, Renand G. Effects of polymorphisms in the calpastatin and μ-calpain genes on meat tenderness in 3 French beef breeds. J Anim Sci. 2011;89:1–11. doi: 10.2527/jas.2010-3063. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, de Koning DJ, Haley C. Genomewide rapid association using mixed model and regression: a fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177:577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rat: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Berner LA. Defining the role of milk fat in balanced diets. Adv Food Nutr Res. 1993;37:131–257. doi: 10.1016/s1043-4526(08)60117-5. [DOI] [PubMed] [Google Scholar]
- Bonanome A, Grundy SM. Effect of dietary stearic acid on plasma cholesterol and lipoprotein levels. N Engl J Med. 1988;318:1244–1248. doi: 10.1056/NEJM198805123181905. [DOI] [PubMed] [Google Scholar]
- Casas E, Keele JW, Shackelford SD, Koohmaraie M, Sonstegard TS, Smith TP, Kappes SM, Stone RT. Association of the muscle hypertrophy locus with carcass traits in beef cattle. J Anim Sci. 1998;76:468–473. doi: 10.2527/1998.762468x. [DOI] [PubMed] [Google Scholar]
- Casas E, Shackelford SD, Keele JW, Koohmaraie M, Smith TPL, Stone RT. Detection of quantitative trait loci for growth and carcass composition in cattle. J Anim Sci. 2003;81:2976–2983. doi: 10.2527/2003.81122976x. [DOI] [PubMed] [Google Scholar]
- Casas E, Shackelford SD, Keele JW, Stone RT, Kappes SM, Koohmaraie M. Quantitative trait loci affecting growth and carcass composition of cattle segregating alternate forms of myostatin. J Anim Sci. 2000;78:560–569. doi: 10.2527/2000.783560x. [DOI] [PubMed] [Google Scholar]
- Choy YH, Park BH, Choi TJ, Choi JG, Cho KH, Lee SS, Choi YL, Koh KC, Kim HS. Estimation of relative economic weights of Hanwoo carcass traits based on carcass market price. Asian Australras J Anim Sci. 2012;25:1667–1673. doi: 10.5713/ajas.2012.12397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. Empirical threshold values for quantitative trait mapping. Genetics. 1994;138:963–971. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JE, Pires JC, Conant GC, McKay SD, Heaton MP, Chen K, Cooper A, Vilkki J, Seabury CM, Caetano AR, Johnson GS, Brenneman RA, Hanotte O, Eggert LS, Wiener P, Kim J-J, Kim KS, Sonstegard TS, van Tassell C, Neibergs HL, McEwan JC, Brauning R, Coutinho LL, Babar ME, Wilson GA, McClure MC, Rolf MM, Kim J, Schnabel RD, Taylor JF. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc. Natl. Acad. Sci USA. 2009;106:18644–18649. doi: 10.1073/pnas.0904691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- Dryden FD, Maechello JA. Influence of total lipid and fatty acid composition upon the palatability of three bovine muscles. J Anim Sci. 1970;31:36–41. [Google Scholar]
- Falconer DS, Mackay TFC. Introduction To Quantitative Genetics. 4th ed. Pearson/Prentice Hall; London, UK: 1996. [Google Scholar]
- Gill JL, Bishop SC, McCorquodale C, Williams JL, Wiener P. Association of selected SNP with carcass and taste panel assessed meat quality traits in a commercial population of Aberdeen Angus-sired beef cattle. Genet Sel Evol. 2009;41:36. doi: 10.1186/1297-9686-41-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill JL, Bishop SC, McCorquodale C, Williams JL, Wiener P. Associations between single nucleotide polymorphisms in multiple candidate genes and carcass and meat quality traits in a commercial Angus-cross population. Meat Sci. 2010;86:985–993. doi: 10.1016/j.meatsci.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Goddard ME, Hayes BJ. Mapping genes for complex traits in domestic animals and their use in breeding programs. Nat Rev Genet. 2009;10:381–391. doi: 10.1038/nrg2575. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Gil B, Wiener P, Nute GR, Burton D, Gill JL, Wood JD, Williams JL. Detection of quantitative trait loci for meat quality traits in cattle. Anim Genet. 2008;39:51–61. doi: 10.1111/j.1365-2052.2007.01682.x. [DOI] [PubMed] [Google Scholar]
- Hartwell L, Hood L, Goldberg ML, Silver LM, Reynolds A. Genetics: from Genes to Genomes. 2nd ed. McGraw-Hill College; Boston, MA, USA: 2000. pp. 302–307. [Google Scholar]
- Jeremiah LE. The influence of subcutaneous fat thickness and marbling on beef: Palatability and consumer acceptability. Food Res Int. 1996;29:513–520. [Google Scholar]
- Jo C, Cho SH, Chang J, Nam KC. Keys to production and processing of Hanwoo beef: A perspective of tradition and science. Anim Front. 2012;2:32–38. [Google Scholar]
- Kim Y, Ryu J, Woo J, Kim JB, Kim CY, Lee C. Genome-wide association study reveals five nucleotide sequence variants for carcass traits in beef cattle. Anim Genet. 2011;42:361–365. doi: 10.1111/j.1365-2052.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- Lee YM, Han CM, Li Y, Lee JJ, Kim LH, Kim JH, Kim DI, Lee SS, Park BL, Shin HD, Kim KS, Kim NS, Kim JJ. A whole genome association study to detect single nucleotide polymorphism for carcass traits in Hanwoo populations. Asian Australas J Anim Sci. 2010;23:417–424. [Google Scholar]
- Lee SH, Choi BH, Lim D, Gondro C, Cho YM, Dang CG, Sharma A, Jang GW, Lee KT, Yoon D, Lee HK, Yeon SH, Yang BS, Kang HS, Hong SK. Genome-wide association study identifies major loci for carcass weight on BTA14 in Hanwoo (Korean cattle) PLoS ONE. 2013;8(10):e74677. doi: 10.1371/journal.pone.0074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay SD, Schnabel RD, Murdoch BM, Matukumalli LK, Aerts J, Coppieters W, Crews D, Dias Neto E, Gill CA, Gao C, Mannen H, Stothard P, Wang Z, Van Tassell CP, Williams JL, Taylor JF, Moore SS. Whole genome linkage disequilibrium maps in cattle. BMC Genet. 2007;8:74. doi: 10.1186/1471-2156-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton SL, Amiri M, Davis GW, Backus WR. Flavor and chemical characteristics of ground beef from grass-, forage-grain- and grain-finished steers. J Anim Sci. 1982;55:77–87. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon W, Mullen AM, Sweeney T, Hamill RM. Association of polymorphisms in candidate genes with colour, water-holding capacity, and composition traits in bovine M. longissimus and M. semimembranosus. Meat Sci. 2010;86:270–275. doi: 10.1016/j.meatsci.2010.04.013. [DOI] [PubMed] [Google Scholar]
- Rincon G, Weber KL, Van Eenennaam AL, Golden BL, Medrano JF. Hot topic: Performance of bovine high-density genotyping platforms in Holsteins and Jerseys. J Dairy Sci. 2011;94:6116–6121. doi: 10.3168/jds.2011-4764. [DOI] [PubMed] [Google Scholar]
- Seaton G, Haley CS, Knott SA, Kearsey M, Visscher PM. QTL express: Mapping quantitative trait loci in simple and complex pedigree. Bioinformatics. 2002;18:339–340. doi: 10.1093/bioinformatics/18.2.339. [DOI] [PubMed] [Google Scholar]
- Sturdivant CA, Lunt DK, Smith GC, Smith SB. Fatty acid composition of subcutaneous and intramuscular adipose tissues and M. longissimusdorsi of Wagyu cattle. Meat Sci. 1992;32:449–458. doi: 10.1016/0309-1740(92)90086-J. [DOI] [PubMed] [Google Scholar]
- van Raden PM. Efficient methods to compute genomic predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Waldman RC, Suess GG, Brungardt VH. Fatty acids of certain bovine tissue and their association with growth, carcass and palatability traits. J Anim Sci. 1968;27:632–635. [Google Scholar]
- Westerling DB, Hedrick HB. Fatty acid composition of bovine lipids as influenced by diet, sex and anatomical location and relationship to sensory characteristics. J Anim Sci. 1979;48:1343–1348. [Google Scholar]


