Abstract
Bipolar disorder (BD) is a highly heritable psychiatric disease characterized by recurrent episodes of mania and depression. To identify new BD genes and pathways, the present study employed a three-step approach. First, gene-expression profiles of BD patients were assessed during both a manic and an euthymic phase. These profiles were compared intra-individually and with the gene-expression profiles of controls. Second, those differentially expressed genes that were considered potential trait markers of BD were validated using data from the Psychiatric Genomics Consortiums' genome-wide association study (GWAS) of BD. Third, the implicated molecular mechanisms were investigated using pathway analytical methods. In the present patients, this novel approach identified: (i) sets of differentially expressed genes specific to mania and euthymia; and (ii) a set of differentially expressed genes that were common to both mood states. In the GWAS data integration analysis, one gene (STAB1) remained significant (P=1.9 × 10−4) after adjustment for multiple testing. STAB1 is located in close proximity to PBMR1 and the NEK4-ITIH1-ITIH3-ITIH4 region, which are the top findings from GWAS meta-analyses of mood disorder, and a combined BD and schizophrenia data set. Pathway analyses in the mania versus control comparison revealed three distinct clusters of pathways tagging molecular mechanisms implicated in BD, for example, energy metabolism, inflammation and the ubiquitin proteasome system. The present findings suggest that STAB1 is a new and highly promising candidate gene in this region. The combining of gene expression and GWAS data may provide valuable insights into the biological mechanisms of BD.
Introduction
Bipolar disorder (BD) is a complex and highly heritable psychiatric disorder with a lifetime prevalence of 0.5–1.5%, and ranks among the top 10 leading causes of years lost because of disability worldwide.1 BD is characterized by the occurrence of recurrent episodes of mania and depression. Mania is a state of elevated mood, which is accompanied by a variety of distinctive symptoms such as increased activity, reduced need for sleep, exaggerated estimation of ability, rapid thoughts and speech, and irritability.2 In contrast, the depressive phases of BD are characterized by sadness, anhedonia and fatigue. In between these episodes, the mood state is described as euthymic, that is, diagnostic criteria for mania and depression are not met. However, research suggests that subclinical mood symptoms may persist during euthymic periods.3
At the time of writing, the etiology and underlying biological mechanisms of BD remain largely unknown, and this lack of knowledge limits the development of more effective treatments, objective diagnostic tools and biological markers of disease susceptibility. However, genome-wide association study (GWAS) analyses have identified novel genetic risk variants for BD that had not been previously implicated in the disorder, and thus provided promising new leads into the biology of BD.4,5 Polygenic score analyses of these data have shown that many more contributing variants remain undetected, as most do not pass the threshold of genome-wide significance because of multiple testing.6 Furthermore, gene-expression studies and state-of-the-art analytical strategies, such as pathway- and gene-network analyses, have generated new insights into the etiology of BD through the identification of novel biological mechanisms.7 To obtain additional information, various authors have advocated a convergent approach, involving the combining of data from different sources.8, 9, 10
The aim of the present feasibility study was to apply a novel three-step convergent approach to identify new candidate genes and pathways for BD. First, the gene-expression profiles of a sample of BD patients were analyzed during both a manic phase (BD manic) and an euthymic phase (BD euthymic). These profiles were first compared on an intrapatient basis, and then with the gene-expression profiles of controls. Second, those differentially expressed genes from step one that were considered potential trait markers of BD were integrated with existing GWAS data. Third, the implicated molecular mechanisms were investigated using pathway analytical methods.
Materials and methods
Subjects
All patients and controls were of German descent. To enhance sample comparability, all blood samples were drawn in the morning during routine clinical phlebotomy. The study was approved by the respective local ethics committees. Written informed consent was obtained from all subjects following a detailed explanation of the study. For patients recruited in a manic phase who lacked the capacity to provide informed consent, preliminary consent was obtained before blood sampling by the treating psychiatrist. The patient was then interviewed after recovery by a member of the study team, and was provided with a detailed explanation of the study and asked to sign the full informed consent documentation. Blood samples were only processed after full informed consent had been obtained. In cases where this consent was refused, the respective blood samples were destroyed. All subjects included in the present study had participated in a previous GWAS of BD (for details see Cichon et al.4).
BD patients
Blood from both a manic and an euthymic phase was collected for 95 BD in-patients. The diagnosis was assigned using the Structured Clinical Interview for the DSM-IV axis I disorders (SCID-I),11 and the Operational Criteria Checklist for Psychotic Illness (OPCRIT v3.32),12 and through the evaluation of medical records and family history. Mania was evaluated using the Young Mania Rating Scale (YMRS), a diagnostic questionnaire used to evaluate manic symptoms in patients.13 Details of the medications prescribed at each blood sampling time point are indicated in Table 1. No statistically significant differences in terms of the type of medication prescribed were observed between mood states.
Table 1. Demographics of patients and controls, and clinical characteristics and prescribed medication, at the time of blood sampling for patients.
| Euthymia | Mania | Controls | Group differences | |
|---|---|---|---|---|
| Demographics | ||||
| N | 11 | 10 | ||
| Age, years | 48.3±12.0 | 47.6±10.3 | n.s. | |
| Clinical characteristics | ||||
| YMRS Score | 4.3±4.7 | 25.2±10.2 | 0.00 (0.01) | |
| Medication n (%) | ||||
| Antipsychotics all | 11 (100) | 11 (100) | n.s. | |
| Typical antipsychotics | 2 (18) | 5 (45) | n.s. (0.021) | |
| Atypical antipsychotics | 10 (91) | 10 (91) | n.s. | |
| Mood stabilizers | 4 (36) | 4 (36) | n.s. | |
| Anticonvulsants | 9 (82) | 10 (91) | n.s. | |
| Antidepressants | 1 (9) | 0 (0) | n.s. (0.038) | |
| Benzodiazepines | 5 (45) | 5 (45) | n.s. | |
Abbreviations: n.s., not significant; YMRS, young mania rating scale.
Selection of BD patients for the gene-expression analyses
An extreme group design using male patients was applied. This approach was selected as the detection of differential gene expression is optimized by comparisons between two groups that show extreme differences in terms of the phenotype of interest, but are otherwise homogeneous. Thus, patients with the highest differences in YMRS score between manic and euthymic states (mean difference: 20.09±7.3) were selected (n=11; mean age=48.27±10.29 years, Table 1). Patients had a mean YMRS score of 25.2 (±10.2) when manic, and 4.3 (±4.7) when euthymic (Table 1).
Controls for the gene-expression analyses
Male, age-matched healthy controls (mean age=47.6±11.98 years) were recruited for the gene-expression analyses (Table 1).
Blood sampling, RNA preparation and microarray procedure
Blood samples were collected in RNA-stabilizing PAXgene tubes (PreAnalytix, BD, Heidelberg, Germany), and stored at −80 °C until analysis. RNA was isolated with the PAXgene Blood RNA Kit 50 (PreAnalytix, Qiagen, Hilden, Germany) according to standard protocols. Total RNA yield was determined using the Quant-iT RiboGreen RNA Reagent and Kit (Life Technologies, Darmstadt, Germany), and a Wallac Victor 2 1420 Multilabel Counter (Perkin Elmer, Rodgau, Germany). Total RNA purity was evaluated via OD measurements (260 nm/280 nm) in a NanoDrop (peqLab, Erlangen, Germany). RNA integrity was determined by RNA integrity number measurement using RNA 6000 Nano Assay RNA chips run in an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). The inclusion criteria were as follows: a ratio of 1.9–2.2 (OD 260/280); RNA integrity number>8.0, and the absence of a peak of genomic DNA contamination in electropherograms. Samples were transcribed to cDNA and hybridized to an Affymetrix Gene Chip Human Exon 1.0 ST Array (Affymetrix, Santa Clara, CA, USA) using the Whole Transcript Sense Target Labeling Assay protocol and 100 ng of total RNA from each sample.
Microarray data analyses
Gene-expression profiling was performed using Affymetrix Human Exon 1.0 ST arrays. Biotinylated antisense cRNA was prepared according to the Affymetrix standard labeling protocol. One control sample performed poorly and was excluded from the analyses. Hybridization on the chip was performed in a GeneChip Hybridization oven 640. Chips were dyed in a GeneChip Fluidics Station 450 and scanned with a GeneChip Scanner 3000. All equipments were manufactured by Affymetrix (Affymetrix, UK). Data importing was performed using the Affymetrix Expression Console Software package and was restricted to core probe sets that correspond to genes. The core set on the raw fluorescence intensity values was normalized by applying robust multi-array average. Differential gene expression was analyzed using analysis of variance and the commercial software package SAS JMP7 Genomics, V4 (SAS Institute, Cary, NC, USA). A false-positive rate of a=0.05 with false discovery rate correction was used as the level of significance (threshold: P<2.59 × 10−3). All raw and normalized data have been deposited in the Gene-expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/; accession no: GSE46416).
The following gene-expression profile comparisons were performed: (1) BD euthymic versus controls; (2) BD manic versus controls; and (3) BD euthymic versus BD manic.
Validation using qRT-PCR
cDNA was generated per Life Technologies High-Capacity cDNA Reverse Transcription Kit. Triple qRT-PCR reactions per sample were performed using Taqman gene expression probes (Life Technologies) for stabilin1/STAB1 (Hs01109068_m1/Exon 25–26) by the 7900HT QRT-PCR system (Life Technologies). Fast program: 50 °C, 2:00 min; 95 °C, 0:20 min, and 40 repeats of 95 °C, 0:01 min and 60 °C, 0:20 min. β-Actin/ACTB (Hs9999903_m1) normalized the relative expression of the STAB1 gene. Fold change differences were calculated by the Δ Ct method.
Integration of GWAS data
To explore whether the genes that were differentially expressed in the present BD patients and controls were significantly enriched for association in patients and controls from a GWAS of BD, gene-based association tests were performed on the Psychiatric Genomics Consortium (PGC)-BD primary data set.5 The gene-based tests were calculated using VEGAS, which accounts for gene size, single-nucleotide polymorphism (SNP) density and linkage disequilibrium between SNPs.14 To capture regulatory regions and SNPs in linkage disequilibrium, the default setting in VEGAS was used.
Pathway analysis
To detect whether a global expression pattern of a set of genes was significantly related to BD, pathway analysis was performed using Globaltest.15 Gene set information was retrieved from the KEGG database (215 pathways; KEGG.db, version 2.5.0). The probe IDs were mapped to ENTREZ gene IDs using the Bioconductor annotation library Homo sapiens hugene10stv1 V12.0.016 for Molecular and Behavioral Neuroscience Institute Custom Chip Definition Files (MBNI CDF).
Pathway scores were corrected for multiple testing using the Benjamini–Hochberg method. To account for the size of the gene set, a probe label permutation test was performed.17 The empirical P-value was calculated as the number of permutations obtaining a test statistics greater than, or equal to, the one observed in the original data, divided by the overall number of permutations applied.
Detecting the associated genes in replicated pathways
Any probe with a single probe P-value of⩽0.05 (as calculated by the Global Test) was considered to contribute to the significance of a pathway. The resulting list of probe IDs was mapped to ENTREZ gene IDs. To visualize potential crosstalk between significant pathways, a hierarchical clustering algorithm was used (average linkage method with the Jaccard coefficient as the distance measure) to group pathways with similar contributory gene content. An implicated SNP was defined as an SNP with a single SNP P-value of⩽0.05.
Results
Gene-expression differences in BD patients during euthymic and manic phases and in comparison with controls
After correction for multiple testing, (i) 262 genes were differentially regulated in the comparison between euthymic patients and controls (177 downregulated (68%) and 85 upregulated (32%) in BD euthymic); (ii) 216 genes were differentially regulated in the comparison between manic patients and controls (44 downregulated (20%) and 172 upregulated (80%)); and (iii) 22 genes were differentially regulated in the intrapatient comparison of euthymia and mania (all upregulated in BD manic; threshold: P<2.59 × 10−3).
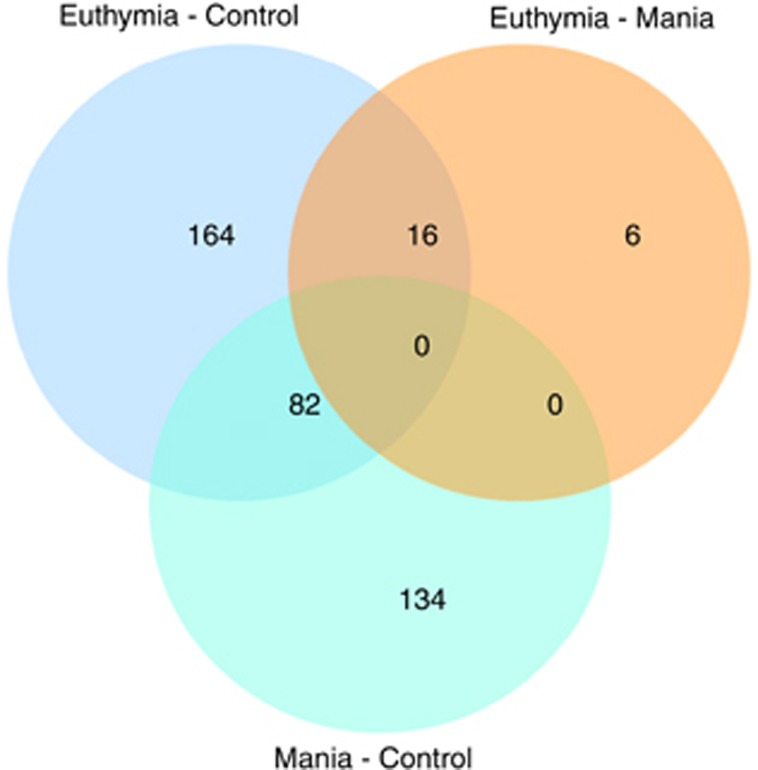
A set of 82 genes was dysregulated (40 upregulated, 42 downregulated) in both BD euthymic versus controls and BD manic versus controls. A set of 16 genes was dysregulated in both the comparison of BD euthymic versus controls and BD euthymic versus BD manic (all upregulated; Figure 1).
Figure 1.
Venn diagram. Number of differentially expressed genes across considered comparisons.
qRT-PCR
qRT-PCR results for STAB1 show a trend towards the direction of mRNA changes observed in the microarray experiments (Supplementary Figure 1). In view of the small sample size, these do not surpass the significance level obtained through the performance of a t-test.
Integration of gene-expression data with GWAS data
As with genetic variation, genes that are differentially expressed in both the manic and the euthymic phases versus controls are not representative of a specific BD state but rather can be considered trait markers of the disease. Instead, these genes are more likely to be implicated in the etiology of BD per se. Therefore, the gene-based analyses focused on the 82 genes that were dysregulated in both the BD euthymic versus controls and the BD manic versus control comparisons. Data for 59 of these genes were available in the PGC-BD primary data set. Significantly, more genes were associated with BD than would be expected by chance (P=0.04). A list of the top 10 associated genes, that is, those with a nominally significant gene-based P-value in the analysis of the full set of SNPs, is provided in Table 2. One gene—Stabilin1 (STAB1)—had an empirical P-value of 1.9 × 10−4, which exceeds a Bonferroni-corrected threshold of P<6.1 × 10−4 (that is, 0.05/82).
Table 2. List of the top 10 associated genes from GWAS of BD patients and controls.
| Gene | Chr | nSNPs | P | minP |
|---|---|---|---|---|
| STAB1 | 3 | 71 | 1.9 × 10−4 | 5.5 × 10−7 |
| RBM14 | 11 | 23 | 1.0 × 10−2 | 1.0 × 10−4 |
| YWHAG | 7 | 70 | 1.2 × 10−2 | 8.3 × 10−4 |
| SLPI | 20 | 108 | 1.3 × 10−2 | 8.2 × 10−4 |
| HIST1H3H | 6 | 60 | 3.2 × 10−2 | 2.8 × 10−3 |
| HNRNPC | 14 | 107 | 3.9 × 10−2 | 1.7 × 10−4 |
| PCID2 | 13 | 90 | 4.0 × 10−2 | 1.5 × 10−2 |
| CDKN2D | 19 | 47 | 4.0 × 10−2 | 9.8 × 10−4 |
| ANKRD36 | 2 | 18 | 4.6 × 10−2 | 8.3 × 10−3 |
| CHST14 | 15 | 52 | 5.0 × 10−2 | 2.0 × 10−2 |
Abbreviations: BD, bipolar disorder; Chr, Chromosome; GWAS, genome-wide association study; minP, the original association P-value for the best SNP within the gene; P, the gene-based P-value considering the full set of SNPs; SNP, single-nucleotide polymorphisms.
Pathway analysis
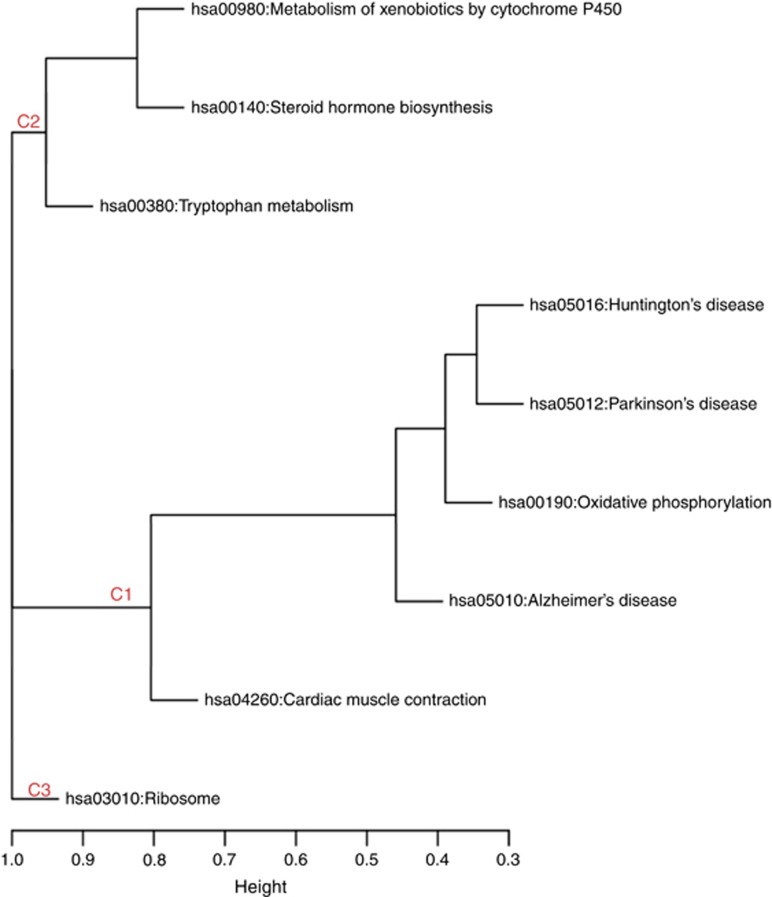
Analysis of KEGG pathways was performed for both the BD euthymic versus control and the BD manic versus control comparison groups. After correction for multiple testing, no pathway had a significant P-value in the BD euthymic versus control analysis. In contrast, the BD manic versus control analysis yielded nine significant pathways (Table 3). Clustering of significant pathways on the basis of gene-content similarity generated three pathway clusters (Figure 2). Cluster 1 consisted of pathways classified as being involved in human diseases, cluster 2 consisted of metabolism pathways and cluster 3 contained the ribosome pathway from the class of KEGG pathways with involvement in genetic information processing. Owing to the small number of dysregulated genes, pathway analyses were not possible for the BD manic versus euthymic comparison.
Table 3. Significantly dysregulated KEGG pathways in the comparison group mania vs controls.
| Pathway ID | KEGG pathway | P | No. of probes |
|---|---|---|---|
| hsa05010 | Alzheimer's disease | 2.5 × 10−3 | 157 |
| hsa05012 | Parkinson's disease | 2.8 × 10−3 | 113 |
| hsa05016 | Huntington's disease | 4.4 × 10−3 | 171 |
| hsa04260 | Cardiac muscle contraction | 1.9 × 10−3 | 70 |
| hsa00380 | Tryptophan metabolism | 1.9 × 10−3 | 41 |
| hsa00190 | Oxidative phosphorylation | 2.6 × 10−3 | 116 |
| hsa00140 | Steroid hormone biosynthesis | 3.9 × 10−4 | 50 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | 8.7 × 10−4 | 64 |
| hsa03010 | Ribosome | 5.2 × 10−3 | 75 |
Figure 2.
Mania associated KEGG pathways clustered on the basis of implicated genes. C1, Cluster 1; C2, Cluster 2; C3, Cluster 3.
Discussion
To identify new BD genes and pathways, the present study applied a novel, three-step convergent approach. First, candidate genes were identified through intrapatient comparisons of gene-expression profiles during euthymic and manic states, and case–control comparisons. Second, differentially expressed genes were tested for association with the disease in existing BD GWAS data. Proof of a genetic association strengthens the hypothesis that a gene is implicated in the etiology of the disorder, and that its differential expression is not merely a downstream epiphenomenon. Third, the implicated molecular mechanisms were investigated using pathway analytical methods. In total, 262 genes were dysregulated in BD euthymic versus controls (85 upregulated, 177 downregulated), and 216 genes were differentially expressed in BD manic versus controls (172 upregulated and 44 downregulated. Gene-expression profiling identified 82 genes that were dysregulated in both the BD euthymic and BD manic versus control comparisons (40 upregulated, 42 downregulated). Intrapatient comparisons of the two mood states revealed 22 dysregulated genes (all upregulated in euthymia). Of these, 16 were downregulated in BD euthymic patients compared with controls (Figure 1).
Whether euthymic BD patients can be viewed as ‘healthy' is a matter of ongoing debate.3 In the present sample, euthymic patients differed from controls in terms of gene expression, and a large number of genes were dysregulated in patients irrespective of mood phase. Thus, our data underline differences between euthymic patients and controls, and suggest that similar, as well as specific, genes are dysregulated during euthymic and manic phases.
In the second step, differentially regulated genes were tested for association in GWAS data using a gene-based approach. This analysis focused on genes that were dysregulated in both mood states versus controls. Of 10 nominally significant genes, STAB1 remained significant after correction for multiple testing (Table 2). Interestingly, STAB1 is located in close proximity to genes on chromosome 3p21.1 that have shown genome-wide significance in previous studies, that is, PBMR1 (~20 kb 3′ of STAB1) in a GWAS of mood disorder (P=3.6 × 10−8),18 and NEK4-ITIH1-ITIH3-ITIH4 (~190 kb 3′ of STAB1) in a meta-analysis combining BD and schizophrenia GWAS data (Praw=8.4 × 10−9; Supplementary Figure 1).19 To date, no specific gene has been conclusively shown to account for these association findings. The present results, however, suggest that STAB1 is a new and highly promising candidate gene in this region. A recent pathway analysis of a schizophrenia GWAS, which identified STAB1 as one of 15 candidate genes, provides further support for this hypothesis.20 Possible approaches to the further investigation of this hypothesis are genetic fine mapping of the associated regions, and studies of effects of the associated SNPs on STAB1 expression.
STAB1 encodes a multifunctional scavenger receptor, and its expression is induced during chronic inflammation and tumor progression.21 Post-mortem gene-expression studies in the dorsolateral prefrontal cortex have revealed differential expression of STAB1 in BD and schizophrenia patients compared with controls (see www.stanleygenomics.org), although the direction of these changes were inconsistent across studies. Enigma, which is the largest consortium conducting meta-analyses of neuroimaging and genetic data worldwide, performed meta-analyses of structural neuroimaging data, and reported one significant association in the above mentioned 3p21.1 region, that is, between average caudate volume and an SNP in STAB1 (P=7 × 10−3, Supplementary Figure 2).22 The caudate nucleus is part of the ventral striatum, a brain region implicated in BD.23 STAB1 binds and internalizes SPARC (secreted protein acidic and rich in cysteine), a protein which is also involved in inflammation.24 SPARC has been implicated in the regulation of depression and affect anxiety-related behavior in mice.25 As STAB1 was upregulated in both the BD euthymic- and the BD manic versus control comparisons (fold change=1.5; P=1.3 × 10−4 and P=2.5 × 10−4, respectively), this gene could be a marker for BD per se, irrespective of mood phase. Further studies are warranted to elucidate the role of STAB1 and its interplay with SPARC in BD. Mouse models with STAB1 deficiency are available26 and represent a potential tool for further analyses.
Pathway analyses of genes from the BD manic versus control comparison revealed nine pathways with statistically significant differential expression (Table 3). Clustering of these pathways on the basis of gene-content similarity generated three clusters (Figure 2). These clusters were mainly driven by dysregulated genes whose biological functions have been implicated in BD by previous research.
In cluster 1, enrichment of genes involved in energy metabolism and the mitochondrial complex (Table 4) was observed. Transcriptional studies of multiple brain regions have implicated these genes in BD (for review see Konradi et al.27). Mitochondrial dysfunction is an important component of the pathophysiology of BD,28 and mitochondrial modulators are considered potential therapeutic targets.28 The cluster 1 pathways were also enriched in ubiquitin proteasome genes (Table 4). Dysregulation of these genes has been reported in brain- and blood-based gene-expression studies of patients with psychosis.29 Cluster 1 pathways also contained a number of genes involved in inflammation (Table 4). Cytokines have been implicated in BD and other psychiatric disorders, although these data are limited (for example, Guloksuz et al.30). Finally, genes from the pathways in cluster 1 have been implicated in protein trafficking (for example, clathrin), and protein trafficking abnormalities have been linked to the pathophysiology of BD (for a review, Schubert et al.31).
Table 4. Genes in the pathways of interest.
|
Cluster 1 |
Human disease |
Gene |
| hsa04260 | Cardiac muscle contraction | ATP1A1, ATP1B3, ATP2A2, CACNA2D4, COX4I1, COX5A, COX5B, COX6B1, COX6C, COX7A2, COX7A2L, COX7B, COX7C, COX8A, CYC1, SLC9A6, TPM4, UQCR10, UQCR11, UQCRC1, UQCRC2 |
| hsa05010 | Alzheimer's disease | ADAM10, ADAM17, APH1A, APP, ATP2A2, ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5E, ATP5F1, ATP5G2, ATP5G3, ATP5H, ATP5J, ATP5O, BID, CALM1, CALM2, CALM3, CAPN1, CAPN2, CASP3, CDK5, CHP, COX4I1, COX5A, COX5B, COX6B1, COX6C, COX7A2, COX7A2L, COX7B, COX7C, COX8A, CYC1, FADD, FAS, GNAQ, HSD17B10, IL1B, LPL, LRP1, MAPK3, NAE1, NCSTN, NDUFA1, NDUFA2, NDUFA3, NDUFA4, NDUFA5, NDUFA6, NDUFA7, NDUFA8, NDUFA9, NDUFB10, NDUFB11, NDUFB2, NDUFB3, NDUFB4, NDUFB5, NDUFB7, NDUFB8, NDUFC1, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS5, NDUFS7, NDUFS8, PLCB3, PPP3CB, PPP3R1, PSEN1, PSENEN, SDHB, TNF, TNFRSF1A, UQCR10, UQCR11, UQCRC1, UQCRC2 |
| hsa00190 | Oxidative phosphorylation | ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5E, ATP5F1, ATP5G2, ATP5G3, ATP5H, ATP5I, ATP5J, ATP5J2, ATP5L, ATP5O, ATP6AP1, ATP6V0B, ATP6V0D1, ATP6V0E1, ATP6V1A, ATP6V1B2, ATP6V1C1, ATP6V1D, ATP6V1E1, ATP6V1G1, COX17, COX4I1, COX5A, COX5B, COX6B1, COX6C, COX7A2, COX7A2L, COX7B, COX7C, COX8A, CYC1, NDUFA1, NDUFA2, NDUFA3, NDUFA4, NDUFA5, NDUFA6, NDUFA7, NDUFA8, NDUFA9, NDUFB10, NDUFB11, NDUFB2, NDUFB3, NDUFB4, NDUFB5, NDUFB7, NDUFB8, NDUFC1, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS5, NDUFS7, NDUFS8, PPA1, SDHB, TCIRG1, UQCR10, UQCR11, UQCRC1, UQCRC2 |
| hsa05012 | Parkinson's disease | SEPT5, ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5E, ATP5F1, ATP5G2, ATP5G3, ATP5H, ATP5J, ATP5O, CASP3, COX4I1, COX5A, COX5B, COX6B1, COX6C, COX7A2, COX7A2L, COX7B, COX7C, COX8A, CYC1, HTRA2, NDUFA1, NDUFA2, NDUFA3, NDUFA4, NDUFA5, NDUFA6, NDUFA7, NDUFA8, NDUFA9, NDUFB10, NDUFB11, NDUFB2, NDUFB3, NDUFB4, NDUFB5, NDUFB7, NDUFB8, NDUFC1, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS5, NDUFS7, NDUFS8, PARK7, SDHB, SLC18A2, SLC25A5, UBA7, UBE2G1, UBE2J1, UBE2J2, UBE2L6, UQCR10, UQCR11, UQCRC1, UQCRC2, VDAC3 |
| hsa05016 | Huntington's disease | AP2S1, ATP5A1, ATP5B, ATP5C1, ATP5D, ATP5E, ATP5F1, ATP5G2, ATP5G3, ATP5H, ATP5J, ATP5O, CASP3, CLTB, CLTC, COX4I1, COX5A, COX5B, COX6B1, COX6C, COX7A2, COX7A2L, COX7B, COX7C, COX8A, CYC1, DCTN1, DCTN2, DCTN4, DLG4, DNAL4, GNAQ, HDAC2, NDUFA1, NDUFA2, NDUFA3, NDUFA4, NDUFA5, NDUFA6, NDUFA7, NDUFA8, NDUFA9, NDUFB10, NDUFB11, NDUFB2, NDUFB3, NDUFB4, NDUFB5, NDUFB7, NDUFB8, NDUFC1, NDUFS1, NDUFS2, NDUFS3, NDUFS4, NDUFS5, NDUFS7, NDUFS8, PLCB3, POLR2B, POLR2E, POLR2H, POLR2K, POLR2L, PPARG, REST, SDHB, SLC25A5, SOD1, UCP1, UQCR10, UQCR11, UQCRC1, UQCRC2, VDAC3 |
|
Cluster 2 |
Metabolism |
Gene |
| hsa00140 | Steroid hormone biosynthesis | COMT, CYP1B1, HSD17B12, HSD17B7, SRD5A1, SRD5A3, UGT2A1, UGT2B15 |
| hsa00980 | Metabolism of xenobiotics by cytochrome P450 | ADH5, LDH3B1, CYP1B1, CYP2E1, GSTK1, GSTM5, GSTP1, MGST1, MGST2, MGST3, UGT2A1, UGT2B15 |
| hsa00380 | Tryptophan metabolism | ACAT1, ACAT2, AFMID, CYP1B1, ECHS1, GCDH, HADHA, MAOA, MAOB, OGDH, TPH1, WARS |
|
Cluster 3 |
Genetic information processing |
Gene |
| hsa03010 | Ribosome | FAU, RPL10, RPL10A, RPL11, RPL12, RPL15, RPL17, RPL18, RPL19, RPL23, RPL24, RPL26, RPL27, RPL28, RPL29, RPL3, RPL30, RPL31, RPL34, RPL35, RPL35A, RPL36AL, RPL37A, RPL38, RPL4, RPL41, RPL5, RPL7A, RPL8, RPLP0, RPLP1, RPLP2, RPS11, RPS13, RPS15, RPS15A, RPS16, RPS20, RPS23, RPS24, RPS25, RPS27L, RPS4X, RPS4Y1, RPS5, RPS6, RPS7, RPS9, RSL24D1, UBA52 |
In the cluster 2 pathways, differential expression was mainly driven by cytochrome P450 proteins, which catalyze many of the reactions involved in drug metabolism and the synthesis of lipids.32 Cluster 2 also contained proteins involved in the biosynthesis or degradation of neurotransmitters that have been implicated in BD previously (Table 4).33,34
The cluster 3 pathway consisted of ribosomal genes. The function of the ribosome is protein synthesis, and research suggests that disturbances in protein synthesis may be related to psychiatric disorders. For example, the control of protein synthesis via eIF2-alpha kinases is activated by memory processes, as mediated by glutamate receptors, and many of the genes in this network have been associated with BD.35,36 In this context, it is of note that during the mania phase, 80% of the differentially regulated genes were upregulated, whereas during the euthymia phase, 68% were downregulated in the present patients. BD patients may become more physically active during a manic phase, and comparable changes in gene expression have been found in studies of exercise and training.37,38 Even if dysregulation of these ‘exercise genes' is only a marker for dysregulated processes during mania and does not represent a causal mechanism, it is nonetheless of potential value in a clinical context, as early awareness of such dysregulation, together with prompt and effective counter strategies, could prevent the development of a fully-fledged manic episode.39, 40 Combining the assessment of clinical symptoms and biological biomarkers could enhance the specificity and selectivity of mood-change prediction.
Although differential pathway expression was found in the BD manic versus control group, no pathway showed association in the BD euthymic versus control group. One reason for this may be that euthymia is a more heterogeneous state than mania. Thus, during mania, when patients exhibit very pronounced and severe symptoms, specific dysregulated genes are pronounced. As manic and euthymic gene expressions were compared on an intrapatient basis, the results are not attributable to inter-individual differences.
A major limitation of the present study was a lack of knowledge concerning the influence of medication-related factors. Thus, the possibility that the differences observed between patients and controls were because medication cannot be excluded. In particular, some of the alterations discussed above are influenced by lithium,41, 42, 43 which was taken by 50% of the present patients in both the manic and the euthymic phases. As the direction of gene dysregulation substantially differs between the two states, it is unlikely that these differences were entirely caused by medication. Although no significant differences were observed in terms of the classes of medication prescribed during the manic and euthymic phases, the dosages differed significantly between the two mood phases. Therefore, the possibility that differential gene expression was partly because differences in medication dosages cannot be excluded. Nevertheless, the findings of the parallel GWAS approach, which is trait-specific and robust against such medication effects, support our results.
Further limitations were the small sample size and the fact that the patients were not stringently assessed with respect to time points, medications, and alcohol and tobacco use. In addition, patients did not have the same scores in their manic and euthymic phases but were selected in a top down manner with respect to the highest difference in YMRS scores. All of these factors increased the heterogeneity of the sample and—given the stringent threshold for significance—decreased the chances of identifying significant gene-expression differences.
Another limitation is that gene expression was analyzed in peripheral tissue, which does not entirely reflect gene-expression changes in the brain.44 However, many of the genes that were differentially expressed are also found in the brain.45 Furthermore, previous authors have demonstrated the value of analyzing peripheral tissue to explore molecular mechanisms and detect new genes and pathways for BD.7,46 Finally, intrapatient comparisons of mania and euthymia were only possible using peripheral tissue, and all potential biomarkers derived from transcriptional profiling must be measurable in easily accessible tissue if they are to be developed for routine clinical use.
In summary, the present study applied a novel three-step convergent approach that involved intrapatient and case–control comparisons of euthymic and manic gene-expression profiles. To our knowledge, this is the first study to compare gene-expression profiles and dysregulated pathways in this manner. The findings suggest that besides differing from controls during their manic phases, BD patients differ from controls during euthymic phases. The results provide support for previously implicated molecular mechanisms, and suggest new BD genes and mechanisms. We hypothesize that genes and pathways implicated in the manic phase relate to the mania state rather than to disease-specific BD traits. Differentiation between trait- and state-specific gene expression through intrapatient comparisons may be of value in identifying novel vulnerability genes and medication targets, and the delineation of biomarkers for the early detection of mania onset. Our data suggest STAB1 as a new and highly promising candidate gene for BD. As STAB1 was upregulated during mania and euthymia, further investigation is warranted to determine whether this gene is a potential medication target or biomarker for BD.
Acknowledgments
This work was supported by the German Federal Ministry of Education and Research, within the context of the MooDS-Net of the National Genome Research Network Plus (grant 01GS08144 to Sven Cichon and Markus Nöthen; grant 01GS08147 to Marcella Rietschel); the German Research Foundation (FOR926, BI 1227/3-1, SFB636-Z4 and SFB779-TPA08). Markus Nöthen received support from the Alfried Krupp von Bohlen und Halbach-Stiftung. Marcella Rietschel was supported by the Seventh Framework Programme of the European Union (ADAMS project, HEALTH-F4-2 009-242257). Jana Strohmaier was supported by the German Research Foundation (GRK 793). JensTreutlein was supported by the German Research Foundation (TR 920/2-1). We thank Christine Hohmeyer and Slavica Radosavljvic-Bjelic for their assistance with the experiments, and Christine Schmäl for critically reviewing the manuscript.
Michael Deuschle and his research group have received speaker and consulting fees from BristolMyers Squibb, Otsuka Pharma, and Servier. Michael Deuschle is a national coordinator and principal investigator of phase II and III trials for Lilly Pharma and Roche. The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- WHO The World Health Report. 2008.
- Anderson IM, Haddad PM, Scott J. Bipolar disorder. Bmj. 2012;345:e8508. doi: 10.1136/bmj.e8508. [DOI] [PubMed] [Google Scholar]
- Linke J, Sonnekes C, Wessa M. Sensitivity to positive and negative feedback in euthymic patients with bipolar I disorder: the last episode makes the difference. Bipolar Disord. 2011;13:638–650. doi: 10.1111/j.1399-5618.2011.00956.x. [DOI] [PubMed] [Google Scholar]
- Cichon S, Muhleisen TW, Degenhardt FA, Mattheisen M, Miro X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. 2011;88:372–381. doi: 10.1016/j.ajhg.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Ripke S, Scott LJ, Andreassen OA, Cichon S, Craddock N, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Harold D, Nyholt DR, Consortium AN, International Endogene C, Genetic, Environmental Risk for Alzheimer's disease C et al. Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer's disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22:832–841. doi: 10.1093/hmg/dds491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedroso I, Lourdusamy A, Rietschel M, Nothen MM, Cichon S, McGuffin P, et al. Common genetic variants and gene-expression changes associated with bipolar disorder are over-represented in brain signaling pathway genes. Biological psychiatry. 2012;72:311–317. doi: 10.1016/j.biopsych.2011.12.031. [DOI] [PubMed] [Google Scholar]
- Miro X, Meier S, Dreisow ML, Frank J, Strohmaier J, Breuer R, et al. Studies in humans and mice implicate neurocan in the etiology of mania. Am J Psychiatry. 2012;169:982–990. doi: 10.1176/appi.ajp.2012.11101585. [DOI] [PubMed] [Google Scholar]
- Niculescu AB, 3rd, Kelsoe JR. Convergent functional genomics: application to bipolar disorder. Ann Med. 2001;33:263–271. doi: 10.3109/07853890108998755. [DOI] [PubMed] [Google Scholar]
- Patel SD, Le-Niculescu H, Koller DL, Green SD, Lahiri DK, McMahon FJ, et al. Coming to grips with complex disorders: genetic risk prediction in bipolar disorder using panels of genes identified through convergent functional genomics. Am J Med Genet. 2010;153B:850–877. doi: 10.1002/ajmg.b.31087. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Robert L, Gibbon M, Williams JB. Structured Clinical Interview for DSM-IV-TR Axis I Disorders, Research Version, Patient Edition. Biometrics Research, New York State Psychiatric Institute: New York, NY, USA; 2002. [Google Scholar]
- McGuffin P, Farmer A, Harvey I. A polydiagnostic application of operational criteria in studies of psychotic illness. Development and reliability of the OPCRIT system. Arch Gen Psychiatry. 1991;48:764–770. doi: 10.1001/archpsyc.1991.01810320088015. [DOI] [PubMed] [Google Scholar]
- Young RC, Biggs JT, Ziegler VE, Meyer DA. A rating scale for mania: reliability, validity and sensitivity. Br J Psychiatry. 1978;133:429–435. doi: 10.1192/bjp.133.5.429. [DOI] [PubMed] [Google Scholar]
- Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, et al. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, van de Geer SA, de Kort F, van Houwelingen H. A global test for groups of genes: testing association with a clinical outcome. Bioinformatics. 2004;20:93–99. doi: 10.1093/bioinformatics/btg382. [DOI] [PubMed] [Google Scholar]
- De Groot PJ, Reiff C, Mayer C, Muller M. NuGO contributions to GenePattern. Genes Nutr. 2008;3:143–146. doi: 10.1007/s12263-008-0093-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Smyth GK.Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn Stat Appl Genet Mol Biol 20109, Article 39. [DOI] [PubMed] [Google Scholar]
- McMahon FJ, Akula N, Schulze TG, Muglia P, Tozzi F, Detera-Wadleigh SD, Steele CJ, Breuer R, Strohmaier J, Wendland JR, Mattheisen M, Muhleisen TW, Maier W, Nothen MM, Cichon S, Farmer A, Vincent JB, Holsboer F, Preisig M, Rietschel M. Meta-analysis of genome-wide association data identifies a risk locus for major mood disorders on 3p21.1. Nat Genet. 2010;42:128–131. doi: 10.1038/ng.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GCBDWG Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YH, Kim JH, Song GG. Pathway analysis of a genome-wide association study in schizophrenia. Gene. 2013;525:107–115. doi: 10.1016/j.gene.2013.04.014. [DOI] [PubMed] [Google Scholar]
- Kzhyshkowska J. Multifunctional receptor stabilin-1 in homeostasis and disease. TheScientificWorldJournal. 2010;10:2039–2053. doi: 10.1100/tsw.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak NM, Stein JL, Medland SE, Hibar DP, Thompson PM, Toga AW. EnigmaVis: online interactive visualization of genome-wide association studies of the Enhancing NeuroImaging Genetics through Meta-Analysis (ENIGMA) consortium. Twin Res Hum Genet. 2012;15:414–418. doi: 10.1017/thg.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand WR, Yurgelun-Todd D. Striatal structure and function in mood disorders: a comprehensive review. Bipolar Disord. 2010;12:764–785. doi: 10.1111/j.1399-5618.2010.00874.x. [DOI] [PubMed] [Google Scholar]
- Workman G, Sage EH. Identification of a sequence in the matricellular protein SPARC that interacts with the scavenger receptor stabilin-1. J Cell Biochem. 2011;112:1003–1008. doi: 10.1002/jcb.23015. [DOI] [PubMed] [Google Scholar]
- Campolongo M, Benedetti L, Podhajcer OL, Pitossi F, Depino AM. Hippocampal SPARC regulates depression-related behavior. Genes Brain Behav. 2012;11:966–976. doi: 10.1111/j.1601-183X.2012.00848.x. [DOI] [PubMed] [Google Scholar]
- Arnold SA, Rivera LB, Carbon JG, Toombs JE, Chang CL, Bradshaw AD, et al. Losartan slows pancreatic tumor progression and extends survival of SPARC-null mice by abrogating aberrant TGFbeta activation. PLoS ONE. 2012;7:e31384. doi: 10.1371/journal.pone.0031384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konradi C, Sillivan SE, Clay HB. Mitochondria, oligodendrocytes and inflammation in bipolar disorder: evidence from transcriptome studies points to intriguing parallels with multiple sclerosis. Neurobiol Dis. 2012;45:37–47. doi: 10.1016/j.nbd.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierenberg AA, Kansky C, Brennan BP, Shelton RC, Perlis R, Iosifescu DV. Mitochondrial modulators for bipolar disorder: A pathophysiologically informed paradigm for new drug development. Aust N Z J Psychiatry. 2012;47:26–42. doi: 10.1177/0004867412449303. [DOI] [PubMed] [Google Scholar]
- Bousman CA, Chana G, Glatt SJ, Chandler SD, May T, Lohr J, et al. Positive symptoms of psychosis correlate with expression of ubiquitin proteasome genes in peripheral blood. Am J Med Genet. 2010;153B:1336–1341. doi: 10.1002/ajmg.b.31106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guloksuz S, Cetin EA, Cetin T, Deniz G, Oral ET, Nutt DJ. Cytokine levels in euthymic bipolar patients. J Affect Disord. 2010;126:458–462. doi: 10.1016/j.jad.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Schubert KO, Focking M, Prehn JH, Cotter DR. Hypothesis review: are clathrin-mediated endocytosis and clathrin-dependent membrane and protein trafficking core pathophysiological processes in schizophrenia and bipolar disorder. Mol Psychiatry. 2012;17:669–681. doi: 10.1038/mp.2011.123. [DOI] [PubMed] [Google Scholar]
- Miksys S, Tyndale RF. Cytochrome P450-mediated drug metabolism in the brain. J Psychiatry Neurosci. 2012;37:120133. doi: 10.1503/jpn.120133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EP, Nurnberger JI., Jr Molecular genetics of bipolar disorder. Genes Brain Behavior. 2006;5:85–95. doi: 10.1111/j.1601-183X.2005.00138.x. [DOI] [PubMed] [Google Scholar]
- Preisig M, Ferrero F, Malafosse A. Monoamine oxidase a and tryptophan hydroxylase gene polymorphisms: are they associated with bipolar disorder. Am J Pharmacogenomics. 2005;5:45–52. doi: 10.2165/00129785-200505010-00004. [DOI] [PubMed] [Google Scholar]
- Carter CJ. eIF2B and oligodendrocyte survival: where nature and nurture meet in bipolar disorder and schizophrenia. Schizophr Bull. 2007;33:1343–1353. doi: 10.1093/schbul/sbm007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter CJ. Multiple genes and factors associated with bipolar disorder converge on growth factor and stress activated kinase pathways controlling translation initiation: implications for oligodendrocyte viability. Neurochem Int. 2007;50:461–490. doi: 10.1016/j.neuint.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Moon MK, Cho BJ, Lee YJ, Choi SH, Lim S, Park KS, et al. The effects of chronic exercise on the inflammatory cytokines interleukin-6 and tumor necrosis factor-alpha are different with age. Appl Physiol Nutr Metab. 2012;37:631–636. doi: 10.1139/h2012-039. [DOI] [PubMed] [Google Scholar]
- Sakharov DA, Maltseva DV, Riabenko EA, Shkurnikov MU, Northoff H, Tonevitsky AG, et al. Passing the anaerobic threshold is associated with substantial changes in the gene expression profile in white blood cells. Eur J Appl Physiol. 2012;112:963–972. doi: 10.1007/s00421-011-2048-3. [DOI] [PubMed] [Google Scholar]
- Lobban F, Solis-Trapala I, Symes W, Morriss R, Erp Group UoL Early warning signs checklists for relapse in bipolar depression and mania: utility, reliability and validity. J Affect Disord. 2011;133:413–422. doi: 10.1016/j.jad.2011.04.026. [DOI] [PubMed] [Google Scholar]
- Morriss RK, Faizal MA, Jones AP, Williamson PR, Bolton C, McCarthy JP. Interventions for helping people recognise early signs of recurrence in bipolar disorder. Cochrane Database Syst Rev. 2007. p. CD004854. [DOI] [PMC free article] [PubMed]
- Beaulieu JM, Marion S, Rodriguiz RM, Medvedev IO, Sotnikova TD, Ghisi V, et al. A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell. 2008;132:125–136. doi: 10.1016/j.cell.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Bosetti F, Seemann R, Rapoport SI. Chronic lithium chloride administration to rats decreases brain protein level of epsilon (epsilon) subunit of eukaryotic initiation factor-2B. Neurosci Lett. 2002;327:71–73. doi: 10.1016/s0304-3940(02)00354-3. [DOI] [PubMed] [Google Scholar]
- Chiu CT, Wang Z, Hunsberger JG, Chuang DM. Therapeutic potential of mood stabilizers lithium and valproic acid: beyond bipolar disorder. Pharmacol Rev. 2013;65:105–142. doi: 10.1124/pr.111.005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins B, Martin MV, Morgan L, Vawter MP. Analysis of whole genome biomarker expression in blood and brain. Am J Med Genet. 2010;153B:919–936. doi: 10.1002/ajmg.b.31062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt SH, Sommer WH, Hanssons AC, Sticht C, Rietschel M, Witt CC. Comparison of gene expression profiles in the blood, hippocampus and prefronal cortex of rats. In Silico Pharmacol. 2013;1:1–6. doi: 10.1186/2193-9616-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajer K, Andrus BM, Gardner W, Lourie A, Strange B, Campo J, et al. Discovery of blood transcriptomic markers for depression in animal models and pilot validation in subjects with early-onset major depression. Transl Psychiatry. 2012;2:e101. doi: 10.1038/tp.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.