Abstract
Background:
Recently, a prospective study reported improved clinical outcomes for recurrent ovarian cancer patients treated with chemotherapies indicated to be sensitive by a chemoresponse assay, compared with those patients treated with non-sensitive therapies, thereby demonstrating the assay's prognostic properties. Due to cross-drug response over different treatments and possible association of in vitro chemosensitivity of a tumour with its inherent biology, further analysis is required to ascertain whether the assay performs as a predictive marker as well.
Methods:
Women with persistent or recurrent epithelial ovarian cancer (n=262) were empirically treated with one of 15 therapies, blinded to assay results. Each patient's tumour was assayed for responsiveness to the 15 therapies. The assay's ability to predict progression-free survival (PFS) was assessed by comparing the association when the assayed therapy matches the administered therapy (match) with the association when the assayed therapy is randomly selected, not necessarily matching the administered therapy (mismatch).
Results:
Patients treated with assay-sensitive therapies had improved PFS vs patients treated with non-sensitive therapies, with the assay result for match significantly associated with PFS (hazard ratio (HR)=0.67, 95% confidence interval (CI)=0.50–0.91, P=0.009). On the basis of 3000 simulations, the mean HR for mismatch was 0.81 (95% range=0.66–0.99), with 3.4% of HRs less than 0.67, indicating that HR for match is lower than for mismatch. While 47% of tumours were non-sensitive to all assayed therapies and 9% were sensitive to all, 44% displayed heterogeneity in assay results. Improved outcome was associated with the administration of an assay-sensitive therapy, regardless of homogeneous or heterogeneous assay responses across all of the assayed therapies.
Conclusions:
These analyses provide supportive evidence that this chemoresponse assay is a predictive marker, demonstrating its ability to discern specific therapies that are likely to be more effective among multiple alternatives.
Keywords: chemoresponse assay, chemosensitivity assay, ovarian cancer, predictive marker, personalised medicine
Clinical management in the recurrent epithelial ovarian cancer (EOC) setting is not standardized. Although numerous chemotherapeutic regimens are currently recommended (National Comprehensive Cancer Network, 2013a), there is insufficient evidence from clinical trials to demonstrate that any single treatment is superior to any other, particularly in terms of overall survival (OS; Coleman et al, 2013), and, in practice, the choice of treatment remains empiric. Chemoresponse assays are one approach that has the potential to improve the selection of a clinically effective therapy among the many options. Although there have been multiple studies providing encouraging data, the use of chemoresponse assays in the treatment of recurrent EOC continues to be debated (Schrag et al, 2004; Burstein et al, 2011).
Most of the previous chemoresponse assay studies in EOC were retrospective analyses evaluating patients who had tissues assayed over many years, making it difficult to ensure the quality of the assay data and its consistency with the clinical data (Holloway et al, 2002; Matsuo et al, 2010; Pant et al, 2010). Recently, a large, prospective study evaluating the clinical relevance of a chemoresponse assay (ChemoFx, Precision Therapeutics, Inc., Pittsburgh, PA, USA) in recurrent or persistent EOC was reported (Rutherford et al, 2013). The study showed a significant improvement in the clinical outcome for patients treated with therapies to which their tumours were assayed as sensitive compared with those treated with therapies assayed as non-sensitive, supporting the clinical benefit of the assay. However, questions remain regarding this assay's prognostic and/or predictive utility given that (1) cross-drug response is common in ovarian cancer (i.e., if a tumour is sensitive to treatment A, then the same tumour may also be sensitive to treatments B, C, D, etc.), making it challenging to select a specific therapy among the multiple available treatments; and (2) the assay may simply reflect the intrinsic biology of a tumour (i.e., patients with assay-sensitive treatment results may simply have better prognoses than patients with assay-resistant results, regardless of the treatment clinically administered) and, thus, this assay may have only a prognostic role. As such, it is important to further evaluate the ability of this assay to function as a predictive marker, that is, select specific effective therapies for personalised treatment.
A prognostic marker is a clinical or biological characteristic that ‘identifies patients with differing risks of a specific outcome, such as progression or death' (Sargent et al, 2005), regardless of treatment administered. Prognostic markers are helpful for identifying patients that are at high risk of relapse and therefore are potential candidates for alternative management strategies. In contrast, predictive markers ‘are associated with response (benefit) or lack of response to a particular therapy relative to other available therapy' (McShane, 2012) and can be used to identify patients most likely to benefit from a specific therapy. In a clinical setting where a number of treatment options with similar impact on patient outcome are available, the utility of a predictive marker for individualised therapy would generally be greater than the utility of a prognostic marker.
The current study sought to address whether this chemoresponse assay could function as a predictive marker, with the capacity to discern specific therapies that are likely to be more effective and those that are not. The study results reported herein are consistent with REMARK guidelines (McShane et al, 2005).
Materials and Methods
Study Population
The current analyses were conducted using the evaluable cohort of 262 women with persistent or recurrent EOC from a recent prospective, multisite, noninterventional clinical study (Trial registration ID: NCT00288275; Rutherford et al, 2013). All patients were enrolled under an IRB approved protocol and provided written informed consent consistent with all federal, state and local regulations before participating in the study. The study design, patient eligibility, chemoresponse assay methodology and patient outcome assessment have been described elsewhere (Rutherford et al, 2013). Briefly, patients were treated with one of 15 prospectively defined, common chemotherapy options available for recurrent EOC (National Comprehensive Cancer Network, 2013a), based on the medical judgment of the oncologist who was blinded to the assay results. Fresh tissue tumour samples were collected at the time of recurrence and then assayed for chemoresponse against this panel of therapies. Each therapy for each patient tumour sample was classified by the assay as sensitive (S), intermediate (I) or resistant (R). A CONSORT diagram describing the study population is presented as Supplementary Figure 1.
Statistical analysis
The primary endpoint of this study was progression-free survival (PFS), defined as the length of time from the start of therapy until the date of first-documented disease progression or death. Disease progression was measured by radiologic examination, physical examination and CA-125 measurements using RECIST or GCIC criteria. Assessment was performed every other cycle during the treatment, every 3 months for the first 2 years, every 6 months for the next 3 years and annually thereafter. The association of chemoresponse assay results with PFS was assessed using the Cox proportional hazards model (Cox, 1972); the proportional hazards assumption was tested by examining the relationship between scaled Schoenfeld residuals and time (Grambsch and Therneau, 1994). The Kaplan–Meier method was used to estimate the probability and duration of PFS (Kaplan and Meier, 1958), and the log-rank test was used to test for the differences between PFS curves by assay category (Peto and Peto, 1972).
Several novel approaches were used to evaluate the predictive value of the assay (Supplementary Figure 2). First, association of the assay with PFS was compared between match and mismatch analyses. The match analysis was performed using the assay result for the administered therapy (assayed therapy=administered chemotherapy) for each patient; the mismatch analysis was performed using the assay results for a treatment randomly selected from all assayed treatments, not necessarily matching the administered therapy, for each patient (assayed therapy≠administered chemotherapy). Both univariate and multivariate analyses controlling for other clinical factors were conducted. Using this approach, if the assay is neither prognostic nor predictive, the HR of PFS for assay-sensitive vs assay-non-sensitive for both match and mismatch would be 1.0; if the assay is only prognostic, then both HRs for match and mismatch would be <1.0 and have identical values. However, if the assay has both prognostic and predictive value, then both HRs for PFS for match and mismatch would be <1.0, and the HR for match would be lower than HR for mismatch. In short, if the assay is a predictive marker, then the association of PFS with match assays is expected to be stronger than with mismatch assays. To obtain the HR for mismatch, the assay result for one therapy for each patient was randomly selected with equal probability from the (up to) 15 study-designated therapies, and the association with PFS was calculated. This procedure was repeated 3000 times and the mean HR for mismatch was assessed.
Second, the influence of cross-drug response (either sensitivity or resistance to all assayed therapies) was evaluated by calculating a multiple drug response index (MDRI). This index represents the percentage of all assayed therapies to which a patient scored as sensitive  . Patients were classified into four groups based on their MDRI and administered clinical treatment: sensitive to all assayed therapies (MDRI=100%) and therefore treated with a sensitive therapy (SA), sensitive to some therapies (0<MDRI<100%) and treated with a sensitive therapy (SP), non-sensitive to all therapies (MDRI=0) and therefore treated with a non-sensitive therapy (RA), and non-sensitive to some therapies (0<MDRI<100%) and treated with a non-sensitive therapy (RP). This analysis investigates if the observed association between assay result and clinical outcome is driven by the inherent chemosensitivity of the tumour (i.e., homogeneous response to all therapies) or is due to the ability of the assay to specifically identify sensitive therapies. In this approach, the chemoresponse assay is suggested to be a predictive marker when the PFS for sensitive categories (SA, SP) differs from the PFS for non-sensitive categories (RA, RP) and no substantial difference is observed within each category (SA vs SP and RA vs RP). In other words, PFS should be associated with the chemosensitivity of the tumour for the administered therapy, independent of MDRI (i.e., regardless of homogeneous or heterogeneous assay responses across all of the assayed therapies).
. Patients were classified into four groups based on their MDRI and administered clinical treatment: sensitive to all assayed therapies (MDRI=100%) and therefore treated with a sensitive therapy (SA), sensitive to some therapies (0<MDRI<100%) and treated with a sensitive therapy (SP), non-sensitive to all therapies (MDRI=0) and therefore treated with a non-sensitive therapy (RA), and non-sensitive to some therapies (0<MDRI<100%) and treated with a non-sensitive therapy (RP). This analysis investigates if the observed association between assay result and clinical outcome is driven by the inherent chemosensitivity of the tumour (i.e., homogeneous response to all therapies) or is due to the ability of the assay to specifically identify sensitive therapies. In this approach, the chemoresponse assay is suggested to be a predictive marker when the PFS for sensitive categories (SA, SP) differs from the PFS for non-sensitive categories (RA, RP) and no substantial difference is observed within each category (SA vs SP and RA vs RP). In other words, PFS should be associated with the chemosensitivity of the tumour for the administered therapy, independent of MDRI (i.e., regardless of homogeneous or heterogeneous assay responses across all of the assayed therapies).
In addition, a two-dimensional clustering analysis was conducted using the chemoresponse assay scores for seven single-agent therapies (carboplatin, cisplatin, gemcitabine, pegylated liposomal doxorubicin (PLD), paclitaxel, docetaxel and topotecan). These seven agents, alone or in combination, comprise the 15 treatments included in this study. This analysis examines the level of correlation between assay results for different therapies and also explores whether patients could be clustered into distinct groups, based on their assay results. If the assay simply reflects the intrinsic biology of a tumour, these clusters may have different prognostic profiles. The clustering analysis was performed using a hierarchical clustering algorithm based on the complete linkage method (Kaufman and Rousseeuw, 1990).
All analyses were performed using SAS version 9.3 (SAS Institute, Cary, NC, USA), except for clustering analysis which was implemented using R version 3.01 (cran.r-project.org). All reported P-values are two-sided with P<0.05 considered statistically significant.
Results
A total of 335 patients were clinically eligible for inclusion in the study, with 262 of them evaluable for analysis. Patient characteristics for the 262 patient cohort have been previously reported (Rutherford et al, 2013) (Table 1) and are comparable to the superset of clinically eligible patients (Supplementary Table 1). Of the 15 therapies prospectively defined in the protocol, 12 were administered clinically, including both single agent and combination therapies (Rutherford et al, 2013).
Table 1. Patient characteristics.
| Number of patients (%) N=262 | |
|---|---|
|
Age (years) | |
| Median (range) |
61 (24–93) |
|
Race | |
| Caucasian | 234 (89.3) |
| African American | 14 (5.3) |
| Others |
14 (5.3) |
|
ECOG performance status | |
| 0 | 183 (69.6) |
| 1 | 66 (25.2) |
| 2 |
13 (5.0) |
|
Histology | |
| Serous | 178 (67.9) |
| Endometrioid | 19 (7.3) |
| Clear cell | 19 (7.3) |
| Mucinous | 3 (1.3) |
| Adenocarcinoma, NOS | 30 (11.5) |
| Others |
13 (5.0) |
|
Tumour grade | |
| 1 | 12 (4.6) |
| 2 | 33 (12.6) |
| 3 | 168 (64.1) |
| Unknown |
49 (18.7) |
|
Platinum sensitivity status | |
| Platinum-sensitive | 145 (55.3) |
| Platinum-resistant |
117 (44.7) |
|
Type of chemotherapy | |
| Carboplatin/Paclitaxel | 79 (30.2) |
| Liposomal Doxorubicin | 55 (21.0) |
| Carboplatin/Gemcitabine | 25 (9.5) |
| Topotecan | 22 (8.4) |
| Carboplatin | 15 (5.7) |
| Carboplatin/Docetaxel | 15 (5.7) |
| Cisplatin/Gemcitabine | 15 (5.3) |
| Cisplatin/Paclitaxel | 13 (5.0) |
| Gemcitabine | 9 (3.4) |
| Paclitaxel | 5 (1.9) |
| Carboplatin/Topotecan | 5 (1.9) |
| Cisplatin |
4 (1.5) |
|
Clinical prognoses | |
| Alive without progression | 33 (12.6) |
| Alive with progression | 67 (25.6) |
| Dead | 162 (61.8) |
Abbreviations: ECOG=Eastern Cooperative Oncology Group; NOS=not otherwise specified.
Reproduced by kind permission of Elsevier from Rutherford et al (2013).
Association of clinical outcome with chemoresponse assay results: match/mismatch analysis
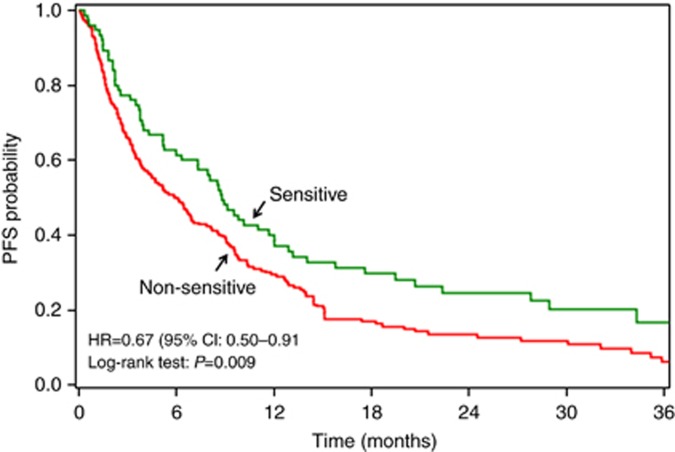
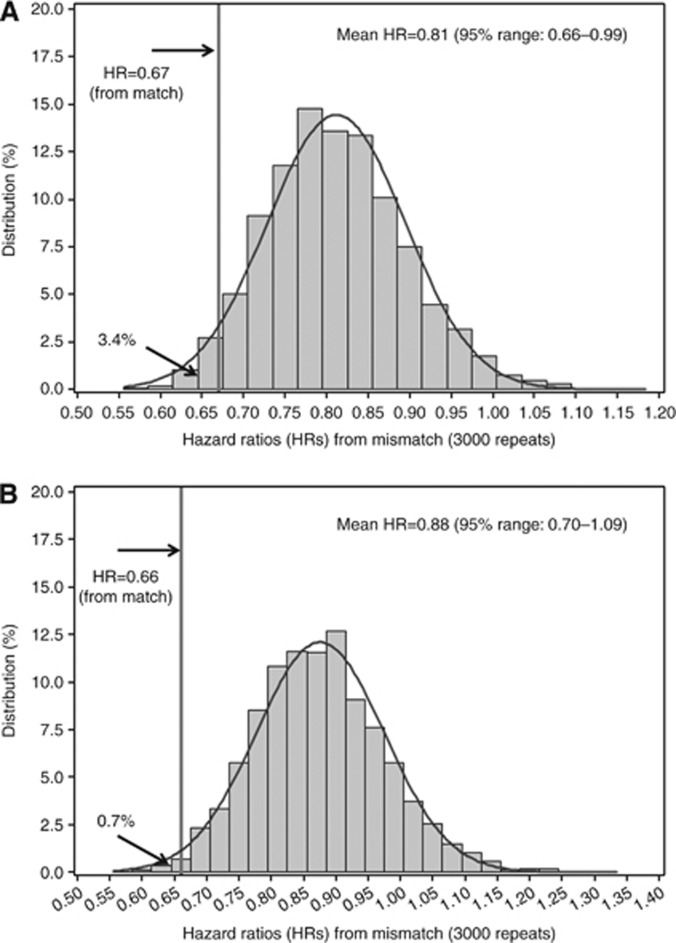
As previously reported, the assay result for match was significantly associated with PFS (HR=0.67, 95% CI=0.50–0.91, P=0.009), with patients treated with an assay-sensitive therapy showing an improvement in PFS as compared with assay-non-sensitive (I+R) patients (Figure 1). This association was consistent after controlling for clinical covariates (HR=0.66, 95% CI=0.47–0.94, P=0.020; Rutherford et al, 2013). The average prognostic value of assay results for multiple different therapies was examined using the assay results for mismatch, in which the assay result for one treatment was randomly selected from the (up to) 15 designated therapies with equal probability for each patient, and the association with PFS was estimated. Based on 3000 repeated re-samplings, the mean HR for mismatch was 0.81 (95% range=0.66–0.99) (Figure 2A). Based on the distribution of HRs for mismatch, only 3.4% of HRs were <0.67 (Figure 2A). When multivariate analysis was performed, the HRs were 0.66 and 0.88, based on match and mismatch, respectively, with only 0.7% of HRs from mismatch <0.66 (Figure 2B). The results for OS were similar. In univariate analysis, the HRs for death were 0.61 and 0.76, based on match and mismatch, respectively, with 5.3% of HRs from mismatch <0.61; in multivariate analysis, the HRs were 0.59 and 0.79, based on match and mismatch, respectively, with 3.6% of HRs from mismatch <0.59. The HRs obtained from both match and mismatch analyses demonstrate the prognostic value of the assay (i.e., HR for both match and mismatch are significantly less than 1.0). More importantly, these results also indicate that the HR for match is materially lower than that for mismatch which supports the predictive nature of the assay.
Figure 1.
Association of clinical treatment assay result (match) with PFS. Patients treated with assay-sensitive treatments experienced a median PFS of 8.8 months, whereas those treated with assay-non-sensitive treatments experienced a median PFS of 5.9 months. Reproduced by kind permission of Elsevier from Rutherford et al (2013).
Figure 2.
Association of randomly selected assay result (mismatch) with PFS. The mean HR for mismatch was calculated from repeated (3000) simulations in univariate (A) and multivariate (B) analyses.
Cross-drug response and impact on clinical outcome
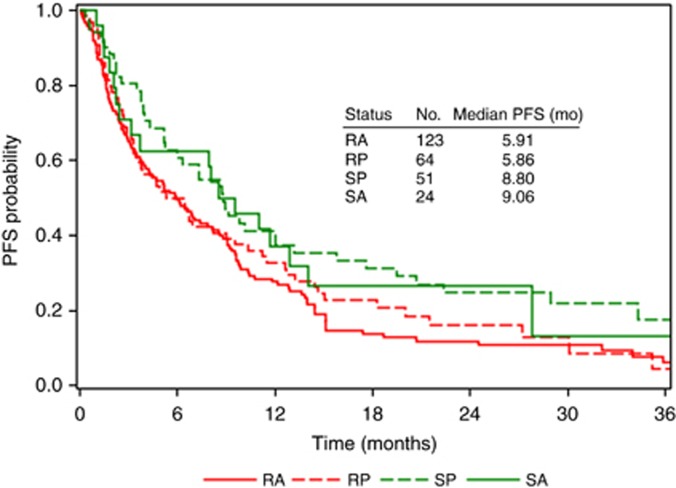
Assay tumour responses differ by therapy, and cross-drug resistance was evident in this population. Of 262 tumours, 123 (47%) were identified as non-sensitive to all tested therapies (RA), whereas 24 (9%) were defined as sensitive to all tested therapies (SA); the remainder (44%) were sensitive to at least one therapy. For those patients showing a heterogeneous pattern of response, 51 were treated with sensitive therapies (SP), and 64 were treated with non-sensitive therapies (RP). The median PFS was 9.1, 8.8, 5.9 and 5.9 months for SA, SP, RP and RA, respectively, with significant difference between SA+SP vs RA+RP, as previously shown (HR=0.67, 95% CI=0.50–0.91, P=0.009; Rutherford et al, 2013), as well as approximately 3 months difference between SA vs RA and between SP vs RP, but without meaningful differences between SP vs SA or between RP vs RA (Figure 3).
Figure 3.
PFS for patients displaying homogeneous vs heterogeneous assay responses. Patients were classified into four groups based on their calculated multiple drug response index (MDRI) and administered clinical treatment: sensitive to all assayed therapies (MDRI=100%) and treated with a sensitive therapy (SA), sensitive to some therapies (0<MDRI<100%) and treated with a sensitive therapy (SP), non-sensitive to all therapies (MDRI=0) and treated with a non-sensitive therapy (RA), and non-sensitive to some therapies (0<MDRI<100%) and treated with a non-sensitive therapy (RP).
The impact of cross-drug response on PFS was further assessed based on MDRI using the Cox model. In univariate analysis, patients whose tumours were sensitive to more therapies (i.e., higher MDRI) experienced better prognoses, as expected (HR=0.96, 95% CI=0.92–0.99, P=0.034). However, MDRI was no longer significant in multivariate analysis (HR=1.02, 95% CI=0.95–1.09, P=0.629), whereas the association between assay result for administered therapy and PFS remained evident (HR=0.60, 95% CI=0.36–1.02, P=0.057; Table 2).
Table 2. Univariate and multivariate analyses of the association of PFS with chemoresponse assay results.
|
Univariate analysis |
Multivariate analysis |
|||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
Age | ||||
| Increasing per 10 years |
1.09 (0.97–1.23) |
0.160 |
1.07 (0.93–1.24) |
0.349 |
|
ECOG PS | ||||
| 1 or 2 vs 0 |
1.24 (0.94–1.65) |
0.136 |
1.12 (0.80–1.57) |
0.518 |
|
Histology | ||||
| Serous vs others |
1.29 (0.97–1.72) |
0.081 |
1.37 (0.97–1.95) |
0.076 |
|
Tumour grade | ||||
| 3 vs 1 or 2 |
1.14 (0.80–1.64) |
0.462 |
1.12 (0.77–1.62) |
0.562 |
|
Platinum sensitivity status | ||||
| Plat sensitive vs plat resistant |
0.59 (0.45–0.76) |
<0.001 |
0.64 (0.47–0.87) |
0.005 |
|
MDRI | ||||
| Increasing each ten percent |
0.96 (0.92–0.99) |
0.034 |
1.02 (0.95–1.09) |
0.629 |
|
Chemoresponse assay results | ||||
| Sensitive vs non-sensitive | 0.67 (0.50–0.91) | 0.009 | 0.60 (0.36–1.02) | 0.057 |
Abbreviations: CI=confidence interval; ECOG=Eastern Cooperative Oncology Group; HR=hazard ratio; PFS=progression-free survival; PS=performance status; MDRI=multiple drug response index.
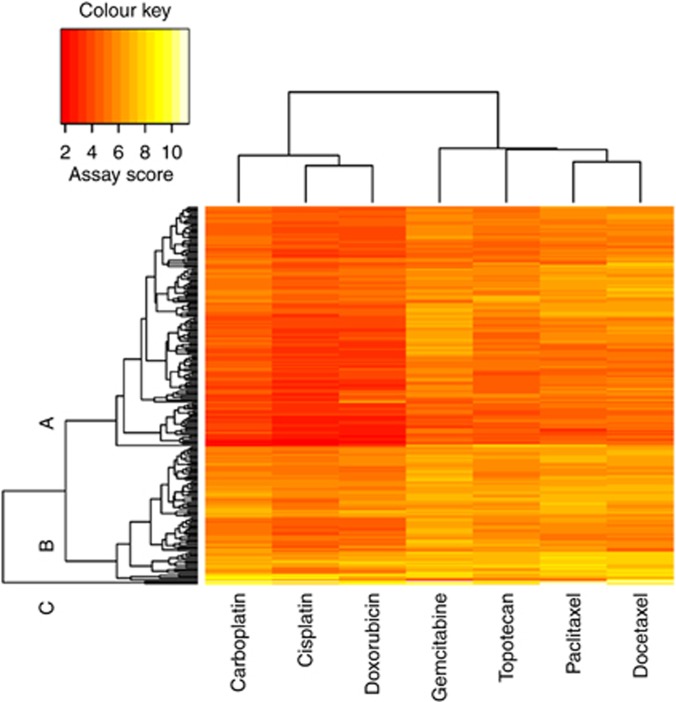
Chemoresponse assay scores for the seven single-agent therapies were further evaluated using two-dimensional clustering analysis. Therapies are represented along the x axis, with carboplatin, cisplatin and doxorubicin clustering together and paclitaxel, docetaxel, topotecan and gemcitabine clustering together. Along the y axis, patients were classified into three clusters (A, B and C), consisting of 63%, 36% and 1% of the study population, respectively (Figure 4). Cluster C was excluded from the analysis owing to the limited number of patients in this group. There is no evidence that this unsupervised clustering was significantly associated with either clinical characteristics or patient outcome (data not shown). These results, together with the data shown in Figure 3 and Table 2, indicate this assay is likely not prognostic of outcome independent of treatment, like some clinical factors (e.g., age, stage, grade, etc.). Rather, the assay's prognostic value (as observed in the mismatch analysis) is likely attributed to cross-drug response (i.e., correlation of assay results for randomly selected therapies with assay result for clinically administered therapy), as anticipated.
Figure 4.
Two-dimensional clustering analysis based on chemoresponse assay data for single-agent treatments. Therapies are represented along the x axis, and patients are represented along the y axis. Red indicates lower assay score (i.e., sensitive), and yellow indicates higher assay score (i.e., resistant).
Discussion
The development and validation of predictive markers has become increasingly vital to improving patient outcomes, especially in the face of an increasing number of available therapies. The current study investigated the association of specific treatment outcomes in 262 prospectively accrued persistent and recurrent EOC patients with assay results blinded to the investigator and patient. Using several approaches, the predictive value of a chemoresponse assay was evaluated and supported.
The analyses conducted herein address common clinical concerns regarding the predictive properties of a chemoresponse assay (Markman, 2011), including the ability of the assay to discern specific therapies that are likely to be more effective among the multiple clinically relevant treatment options. Specifically, the ability of a chemoresponse assay to identify treatment-specific sensitivity has been questioned given the clinical and biological factors that may limit the ability of an assay to be predictive of patient response. For example, since cross-drug response to different therapies is a common phenomenon in chemoresponse assays and it is thought that patients can exhibit multidrug resistance, there is concern that these factors may limit the predictive ability of an assay. Furthermore, it is hypothesised that the improved outcome observed for patients treated with assay-sensitive therapies may be driven by the inherent biology of the tumour represented by the chemoresponse profile (i.e., homogeneous sensitivity vs homogeneous resistance to all therapies), suggesting that the assay may simply reflect the inherent biological-based tumour behaviour and, hence, patient prognosis. The studies described herein attempt to address each of these concerns.
First, the match/mismatch analysis assesses whether cross-drug response and correlated assay results across different therapies limit the predictive ability of the assay. This simple approach, evaluating the ability of the assay to predict patient outcome based on the assay result for the clinically administered therapy (match) as compared with a randomly selected treatment assay result for the same patient (mismatch), showed that the association of outcome and assay result for match was stronger than for mismatch (HR: 0.67 vs 0.81, respectively), indicating that a patient receiving an assay-sensitive therapy would be more likely to experience a better outcome than a patient treated empirically.
Next, the heterogeneity of assay response for a given patient to multiple therapies was evaluated, showing that nearly half (44%) of the patients included in the study demonstrated heterogeneous assay response to all therapies tested (i.e., were sensitive to at least one therapy tested, but not all), whereas the remainder were homogeneously sensitive (9%) or non-sensitive (47%). Comparison of PFS for heterogeneous patients vs those displaying homogeneous responses demonstrated that improved outcome in assay-sensitive patients is unlikely due to the inherent (presumably homogeneous) chemosensitivity of a tumour. Further, multivariate analysis demonstrated that an increased or decreased percentage of assay-sensitive results across all therapies tested was not associated with patient outcome when the matched assay result was included. Therefore, although tumour sensitivity or resistance to multiple therapies is evident in a portion of recurrent EOC patients, improved outcome was associated with clinical administration of a sensitive therapy, regardless of whether that tumour displayed homogeneous or heterogeneous assay responses.
Finally, unsupervised clustering analysis, based on assay results for multiple single-agent therapies, was neither associated with common clinical factors nor with patient outcome, further supporting that this assay is not just reflective of intrinsic tumour biology or common clinical factors normally associated with tumour prognosis (i.e., not only prognostic).
With rapid advancements in cancer research and the number of new therapies being developed, clinical interest lies in identifying predictive markers for personalised therapy. Although many molecular-based biomarkers have been clinically accepted (Cobleigh et al, 1999; Slamon et al, 2001; Amado et al, 2008; Karapetis et al, 2008; Mok et al, 2009; Van Cutsem et al, 2009; Rosell et al, 2012) and are supported by regulatory bodies' approvals and inclusion in standardized treatment paradigms for other cancer types (Allegra et al, 2009; Burstein et al, 2010; Keedy et al, 2011; National Comprehensive Cancer Network, 2013b, 2013c, 2013d), unfortunately, to date, no predictive markers have been clinically validated for use in EOC. Although numerous clinical factors (e.g., age, stage, grade, histology) are used extensively in making treatment decisions especially in the recurrent EOC setting, these factors are limited in their ability to discern specific therapies that are likely to be more effective in second-line treatment and beyond.
Clinical validation of prognostic and predictive markers requires study designs that differ from those employed in traditional drug trials (Institute of Medicine, 2011). Clinical validation for prognostic markers is more straightforward, as only the clinical outcomes between marker-positive and marker-negative patients are compared within a cohort of uniformly treated patients. If a marker is significantly associated with outcome when the effects of other possible confounding factors are controlled, then the marker is considered to be prognostic. In contrast, the clinical studies required to demonstrate the clinical validation of predictive markers are more complex. Often, data from previously conducted randomized drug trials are employed to conduct such validations if archived tissue is available and appropriate for the marker. This strategy may be advantageous in certain instances where outcomes require prolonged follow-up to achieve a meaningful difference in survival.
Clinical study designs for markers have been extensively discussed and reviewed in the literature (Simon and Maitournam, 2004; Sargent et al, 2005; Freidlin et al, 2010; Tajik et al, 2013). The biomarker-stratified design, which uses the marker status to guide analysis but not to assign treatment, is considered to be more efficient regarding required study sample size, can be used when the number of markers and treatment choices is limited, and can investigate a marker's predictive properties. However, this approach is not directly compatible with chemoresponse assays where a larger number of therapies (multiple markers) are evaluated simultaneously (e.g., the response profile of each therapy is considered to be a separate marker). The biomarker-strategy design, in which patients are randomized to two arms (assay-directed vs empirical treatment), is a reasonable approach for evaluating multiple markers and has been recommended for evaluation of chemoresponse assays (Blue Cross and Blue Shield Association, 1995, 2000, 2002; Schrag et al, 2004; Burstein et al, 2011). However, this design is inefficient in its use of patients (i.e., large sample size not logistically feasible) and may not clearly distinguish between better treatments in the marker arm vs the standard of care arm (Freidlin et al, 2010; Buyse et al, 2011; Ziegler et al, 2012; Buyse and Michiels, 2013; Center for Medical Technology Policy, 2013). Furthermore, this approach has been shown to be subject to a treating physician ‘learning effect' bias where, across the study duration, empirically selected treatments became more similar to those indicated in the assay-informed arm, as described by Cree et al (2007).
Over the last two decades, numerous assessments have called for more thorough evaluation of the validation and justification of chemoresponse assays for clinical use (Blue Cross and Blue Shield Association, 1995, 2000, 2002; Schrag et al, 2004; Burstein et al, 2011). The study design uniformly recommended in these assessments is based on the biomarker-strategy design, specifically an interventional (i.e., treatment assigned by marker), randomized, two-arm (assay-directed vs empiric treatment assignment) marker study design. Although the design proposed in these assessments can theoretically address a few markers simultaneously and has been historically referenced as the ‘gold standard', it would require a larger number of patients, would be subject to the physician bias ‘learning effect' and, yet, still not able to cleanly evaluate the predictive marker elements. Also, importantly, alternate approaches to evaluating both predictive and prognostic marker elements can be executed on a more achievable sample size (Wieand, 2005). As a result, the biomarker-strategy approach is not the only design that can effectively validate a predictive marker and may not be the preferred approach (Center for Medical Technology Policy, 2013).
The match/mismatch analysis method employed in this study represents a promising alternate approach to validating a chemoresponse assay, in that it has an achievable sample size, retains the ability to assess both prognostic and predictive marker properties and is appropriate for use with multiple markers (Simon and Maitournam, 2004; Maitournam and Simon, 2005; Simon, 2005; Mandrekar and Sargent, 2009, 2010; Simon, 2010; Center for Medical Technology Policy, 2013). This is especially important in recurrent EOC where numerous, clinically-equivalent therapies are available for use and enrolment of a large sample size is not logistically feasible. Furthermore, this approach has been previously recommended for assessment of chemoresponse assays and is expected to be able to evaluate whether the assay is predictive of response (Wieand, 2005). This method does lack the ability to assess each therapy/marker individually due to prohibitive sample size requirements, which increases with each individual therapy/marker evaluated.
In both the current analyses and the prior analysis reported by Rutherford et al, the association of assay result with clinical outcome was evaluated using two strata (sensitive vs non-sensitive), with the thresholds for each treatment predefined in an external reference sample, independent of clinical outcome. Analysis of quantitative data, such as a receiver operating characteristic (ROC) curve, may further illustrate assay performance; however, due to the individualised nature of establishing assay treatment concentrations and response category thresholds for each distinct treatment, assay results cannot be directly compared across different treatments (i.e., the assay result range varies across individual treatments).
Clinical validations for chemoresponse assays, which simultaneously assess multiple markers/therapies, must be carefully considered. Through further analysis of a prospective study and by using several analytical approaches, the current study further evaluated the clinical value of a chemoresponse assay. The results provide reasonable evidence that this assay is a predictive marker, with the capacity to discern specific therapies that are likely to be more effective, and women with recurrent EOC may benefit from assay-informed therapy selection.
Acknowledgments
We thank Shuguang Huang for his critical review of the manuscript. This study was sponsored by Precision Therapeutics, Inc. DJS received compensation for consulting work from Precision Therapeutics, Inc. CT, MJG and SLB are paid employees of Precision Therapeutics, Inc. TCK, MAP and RLC have no financial disclosures to make relevant to the work described.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- Blue Cross and Blue Shield Association 1995. Nonclonogenic cytotoxic drug resistance assay. Technology Evaluation Center Assessment Program 10(22).
- Blue Cross and Blue Shield Association 2000. Chemotherapy sensitivity and resistance assays. Technology Evaluation Center Assessment Program 15(11). [PubMed]
- Blue Cross and Blue Shield Association 2002. Chemotherapy sensitivity and resistance assays. Technology Evaluation Center Assessment Program 17(12).
- Burstein HJ, Mangu PB, Somerfield MR, Schrag D, Samson D, Holt L, Zelman D, Ajani JA, American Society of Clinical Oncology American society of clinical oncology practice guideline update on the use of chemotherapy sensitivity and resistance assays. J Clin Oncol. 2011;29:3328–3330. doi: 10.1200/JCO.2011.36.0354. [DOI] [PubMed] [Google Scholar]
- Burstein HJ, Prestrud AA, Seidenfeld J, Anderson H, Buchholz TA, Davidson NE, Gelmon KE, Giordano SH, Hudis CA, Malin J, Mamounas EP, Rowden D, Solky AJ, Sowers MR, Stearns V, Winer EP, Somerfield MR, Griggs JJ, American Society of Clinical Oncology American Society of Clinical Oncology clinical practice guideline: update on adjuvant endocrine therapy for women with hormone receptor-positive breast cancer. J Clin Oncol. 2010;28:3784–3796. doi: 10.1200/JCO.2009.26.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyse M, Michiels S. Omics-based clinical trial designs. Curr Opin Oncol. 2013;25:289–295. doi: 10.1097/CCO.0b013e32835ff2fe. [DOI] [PubMed] [Google Scholar]
- Buyse M, Michiels S, Sargent DJ, Grothey A, Matheson A, de Gramont A. Integrating biomarkers in clinical trials. Expert Rev Mol Diagn. 2011;11:171–182. doi: 10.1586/erm.10.120. [DOI] [PubMed] [Google Scholar]
- Center for Medical Technology Policy 2013. Evaluation of clinical validity and clinical utility of actionable molecular diagnostic tests in adult oncology.
- Cobleigh MA, Vogel CL, Tripathy D, Robert NJ, Scholl S, Fehrenbacher L, Wolter JM, Paton V, Shak S, Lieberman G, Slamon DJ. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17:2639–2648. doi: 10.1200/JCO.1999.17.9.2639. [DOI] [PubMed] [Google Scholar]
- Coleman RL, Monk BJ, Sood AK, Herzog TJ. Latest research and treatment of advanced-stage epithelial ovarian cancer. Nat Rev Clin Oncol. 2013;10:211–224. doi: 10.1038/nrclinonc.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. Regression models and life-tables. J R Stat Soc B. 1972;34:187–220. [Google Scholar]
- Cree IA, Kurbacher CM, Lamont A, Hindley AC, Love S. A prospective randomized controlled trial of tumour chemosensitivity assay directed chemotherapy versus physician's choice in patients with recurrent platinum-resistant ovarian cancer. Anticancer Drugs. 2007;18:1093–1101. doi: 10.1097/CAD.0b013e3281de727e. [DOI] [PubMed] [Google Scholar]
- Freidlin B, McShane LM, Korn EL. Randomized clinical trials with biomarkers: design issues. J Natl Cancer Inst. 2010;102:152–160. doi: 10.1093/jnci/djp477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grambsch PM, Therneau TM. Proportional hazards tests and diagnostics based on weighted residuals. Biometrika. 1994;81:515–526. [Google Scholar]
- Holloway RW, Mehta RS, Finkler NJ, Li KT, McLaren CE, Parker RJ, Fruehauf JP. Association between in vitro platinum resistance in the EDR assay and clinical outcomes for ovarian cancer patients. Gynecol Oncol. 2002;87:8–16. doi: 10.1006/gyno.2002.6797. [DOI] [PubMed] [Google Scholar]
- Institute of Medicine . Generating Evidence for Genomic Diagnostic Test Development: Workshop Summary. The National Academies Press: Washington, DC, USA; 2011. [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc. 1958;53:457–481. [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- Kaufman L, Rousseeuw PJ. Finding Groups in Data: an Introduction to Cluster Analysis. Wiley: New York, NY, USA; 1990. [Google Scholar]
- Keedy VL, Temin S, Somerfield MR, Beasley MB, Johnson DH, McShane LM, Milton DT, Strawn JR, Wakelee HA, Giaccone G. American Society of Clinical Oncology provisional clinical opinion: epidermal growth factor receptor (EGFR) mutation testing for patients with advanced non-small-cell lung cancer considering first-line EGFR tyrosine kinase inhibitor therapy. J Clin Oncol. 2011;29:2121–2127. doi: 10.1200/JCO.2010.31.8923. [DOI] [PubMed] [Google Scholar]
- Maitournam A, Simon R. On the efficiency of targeted clinical trials. Stat Med. 2005;24:329–339. doi: 10.1002/sim.1975. [DOI] [PubMed] [Google Scholar]
- Mandrekar SJ, Sargent DJ. Clinical trial designs for predictive biomarker validation: one size does not fit all. J Biopharm Stat. 2009;19:530–542. doi: 10.1080/10543400902802458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandrekar SJ, Sargent DJ. Predictive biomarker validation in practice: lessons from real trials. Clin Trials. 2010;7:567–573. doi: 10.1177/1740774510368574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markman M. Counterpoint: chemosensitivity assays for recurrent ovarian cancer. J Natl Compr Canc Netw. 2011;9:121–124. doi: 10.6004/jnccn.2011.0010. [DOI] [PubMed] [Google Scholar]
- Matsuo K, Eno ML, Im DD, Rosenshein NB, Sood AK. Clinical relevance of extent of extreme drug resistance in epithelial ovarian carcinoma. Gynecol Oncol. 2010;116:61–65. doi: 10.1016/j.ygyno.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM. Statistical challenges in the development and evaluation of marker-based clinical tests. BMC Med. 2012;10:52. doi: 10.1186/1741-7015-10-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics Reporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- National Comprehensive Cancer Network 2013. NCCN Clinical Practice Guidelines in Oncology. Ovarian Cancer: Including Fallopian Tube Cancer and Primary Peritoneal Cancer. Version 2.2013.
- National Comprehensive Cancer Network 2013. NCCN Clinical Practice Guidelines in Oncology. Colon Cancer. Version 3.2013. [DOI] [PubMed]
- National Comprehensive Cancer Network 2013. NCCN Clinical Practice Guidelines in Oncology. Breast Cancer. Version 3.2013.
- National Comprehensive Cancer Network 2013. NCCN Clinical Practice Guidelines in Oncology. Non-Small Cell Lung Cancer. Version 2.2013.
- Pant AC, Diaz-Montes T, Tanner E, Ahmad S, Giuntoli RL, Holloway RW, Bristow RE. Correlation of extreme drug resistant assay results and progression-free survival following intraperitoneal chemotherapy for advanced ovarian cancer. J Chemother. 2010;22:270–274. doi: 10.1179/joc.2010.22.4.270. [DOI] [PubMed] [Google Scholar]
- Peto R, Peto J. Asymptotically efficient rank invariant test procedures (with discussion) J R Stat Soc A. 1972;135:185–206. [Google Scholar]
- Rosell R, Carcereny E, Gervais R, Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R, Pallares C, Sanchez JM, Porta R, Cobo M, Garrido P, Longo F, Moran T, Insa A, De Marinis F, Corre R, Bover I, Illiano A, Dansin E, de Castro J, Milella M, Requart N, Altavilla G, Jimenez U, Provencio M, Moreno MA, Terrasa J, Munoz-Langa J, Valdivia J, Isla D, Domine M, Molinier O, Mazieres J, Baize N, Garcia-Campelo R, Robinet G, Rodriquez-Abreu D, Lopez-Vivanco G, Gebbia V, Ferrera-Delgado L, Bombaron P, Bernabe R, Bearz A, Artal A, Cortesi E, Rolfo C, Sanchez-Ronco M, Drozdowskyi A, Queralt C, de Aquirre I, Ramirez JL, Sanchez JJ, Molina MA, Taron M, Paz-Ares L, Spanish Lung Cancer Group in collaboration with Groupe Francias de Pneumo-Cancerologie and Associazione Italiana Oncologia Toracica Erlotinib versus standard chemotherapy as a first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2012;13:239–246. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- Rutherford T, Orr J, Jr, Grendys E, Jr, Edwards R, Krivak TC, Holloway R, Moore RG, Puls L, Tillmanns T, Schink JC, Brower SL, Tian C, Herzog TJ. A prospective study evaluating the clinical relevance of a chemoresponse assay for treatment of patients with persistent or recurrent ovarian cancer. Gynecol Oncol. 2013;131:362–367. doi: 10.1016/j.ygyno.2013.08.009. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Conley BA, Allegra C, Collette L. Clinical trial designs for predictive marker validation in cancer treatment trials. J Clin Oncol. 2005;23:2020–2027. doi: 10.1200/JCO.2005.01.112. [DOI] [PubMed] [Google Scholar]
- Schrag DL, Garewal HS, Burstein HJ, Samson DJ, Von Hoff DD, Somerfield MR. American Society of Clinical Oncology technology assessment: chemotherapy sensitivity and resistance assay. J Clin Oncol. 2004;22:3631–3638. doi: 10.1200/JCO.2004.05.065. [DOI] [PubMed] [Google Scholar]
- Simon R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J Clin Oncol. 2005;23:7332–7341. doi: 10.1200/JCO.2005.02.8712. [DOI] [PubMed] [Google Scholar]
- Simon R. Clinical trial designs for evaluating the medical utility of prognostic and predictive biomarkers in oncology. Per Med. 2010;7:33–47. doi: 10.2217/pme.09.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Maitournam A. Evaluating the efficiency of targeted designs for randomized clinical trials. Clin Cancer Res. 2004;10:6759–6763. doi: 10.1158/1078-0432.CCR-04-0496. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Tajik P, Zwinderman AH, Mol BW, Bossuyt PM. Trial designs for personalizing cancer care: a systematic review and classification. Clin Cancer Res. 2013;19:4578–4588. doi: 10.1158/1078-0432.CCR-12-3722. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pinter T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- Wieand HS. Chemotherapy sensitivity and response assays: are the ASCO guidelines for clinical trial design too restrictive. J Clin Oncol. 2005;23:3643–3644. doi: 10.1200/JCO.2005.05.321. [DOI] [PubMed] [Google Scholar]
- Ziegler A, Koch A, Krockenberger K, Grosshennig A. Personalized medicine using DNA biomarkers: a review. Hum Genet. 2012;131:1627–1638. doi: 10.1007/s00439-012-1188-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.