Abstract
Background:
Little is known about the tumour suppressive proteins and the underlying mechanisms that suppress colon cancer progression. Homeodomain-containing transcription factor HOXB9 plays an important role in embryogenesis and cancer development. We here aim to uncover the potential role of HOXB9 in the regulation of colon adenocarcinoma progression including epithelial-to-mesenchymal transition.
Methods:
HOXB9 expression in colon adenocarcinoma cells and patients was analysed by western blot and immunohistochemistry separately. Correlation between HOXB9 expressions with patients' survival was assessed by Kaplan–Meier analysis. HOXB9-regulated target gene expression was determined by RNA sequencing in HOXB9-overexpressing colon adenocarcinoma cells.
Results:
Elevated HOXB9 expression was identified in well-differentiated colon adenocarcinoma patients and was associated with a better overall patients' survival. Overexpression of HOXB9 inhibited colon adenocarcinoma cell growth, migration, invasion in vitro and tumour growth, liver as well as lung metastases in nude mice; whereas silencing HOXB9 promoted these functions. HOXB9 promoted colon adenocarcinoma differentiation via a mechanism that stimulates mesenchymal-to-epithelial transition, involving downregulation of EMT-promoting transcriptional factors including Snail, Twist, FOXC2 and ZEB1 and upregulation of epithelial proteins including E-cadherin, claudins-1, -4, -7, occludin and ZO-1.
Conclusions:
HOXB9 is a novel tumour suppressor that inhibits colon adenocarcinoma progression by inducing differentiation. Elevated expression of HOXB9 predicts a longer survival in colon adenocarcinoma patients.
Keywords: HOXB9, colon adenocarcinoma, differentiation, mesenchymal-to-epithelial transition, metastasis
HOXB9 is a member of homeobox-containing (HOX) transcriptional factor family that encompasses 39 genes in human and is classified into four different clusters, including HOX A, B, C and D (Abate-Shen, 2002). HOXB9 is expressed in the mammary gland of adult mice and loss-of-function mutations in HOXB9 gene cause hypoplasia of the mammary gland after pregnancy (Chen and Capecchi, 1999). In addition to their critical roles in development, increasing numbers of studies demonstrated that HOX family genes are associated with tumorigenicity (Chen and Sukumar, 2003; Grier et al, 2005; Shah and Sukumar, 2010) and progression including tumour growth and angiogenesis (Chen and Sukumar, 2003; Rhoads et al, 2005; Winnik et al, 2009; Sun et al, 2013). HOXB9 was known to induce tumour angiogenesis, invasion and lung metastasis in breast cancer (Hayashida et al, 2010), as well as being an important prognostic factor for clinical outcomes of breast cancer patients (Chiba et al, 2012; Morgan et al, 2012; Seki et al, 2012; Shrestha et al, 2012). HOXB9 was also found to induce epithelial-to-mesenchymal transition (EMT), a process that is associated with tumour invasion and resistance to chemotherapeutic drugs and radiations by accelerating DNA damage responses (Nguyen et al, 2009; Hayashida et al, 2010). HOXB9 induction of EMT is mediated by activation of the Wnt signalling pathway (Hayashida et al, 2010). Recently, a report showing that decreased expression of HOXB9 is related to a poor overall survival in patients with gastric carcinoma, identifying an opposite role of HOXB9 in cancer (Sha et al, 2013), suggested that HOXB9 may play a diverse role during cancer progression under various circumstances. Therefore, it is of interest to uncover the many faces of HOXB9 in human cancers.
Colon cancer is one of the leading malignancies worldwide and is the fourth cause of death in cancer patients. Although early diagnoses and therapeutic strategies have been well established for colon cancer patients, invasion, metastasis and recurrence of the disease are still challenging. As for molecules associated with prognosis in colon cancer, the silent mating-type information regulation 2 homologue 1 (SIRT1) has been found associated with good prognosis in colorectal cancer (Jung et al, 2013). CALU and CDH11 are candidate stromal biomarkers of prognostic significance in colon cancer as well (Torres et al, 2013). However, there are no studies regarding the clinical relevance of HOXB9 expression with colon adenocarcinoma progression in human.
In this study, we examined the expression of HOXB9 in 63 cases of colon adenocarcinoma with corresponding normal tissues as control and established the relationship between HOXB9 expression and clinicopathologic features. We also evaluated the prognostic role of HOXB9 in colon adenocarcinoma patients. Further, we investigated the functional role of HOXB9 in the regulation of colon adenocarcinoma progression both in vitro and in vivo. Finally, we identified that HOXB9 is a novel tumour suppressive transcriptional factor that inhibits tumour growth and metastasis in a mouse xenograft model of colon adenocarcinoma via a mechanism of inducing mesenchymal-to-epithelial transition, the opposite process of EMT.
Materials and methods
Ethics
The Ethics Committee of Peking University Health Science Center has approved for mouse experiments (permit number: LA2011-73), and for using human colon adenocarcinoma patient tumour tissues (permit number: ZRLW-5) in the present study. The procedures for handling mice and human tumour specimens were in accordance with the ethical standard of the Helsinki Declaration of 1975, and the revised version in 1983. We also referred to the procedures from Workman et al (2010).
Patient tumour samples
Tumour tissue sections (n=63) were obtained from colon adenocarcinoma patients who underwent surgery at Peking University Health Science Center between July 2006 and September 2007. Normal colon tissue samples (n=63) were obtained from the same colon cancer patient cohort, which were at least 2 cm apart from the tumours. Survival was measured for patients from the time of surgery, with death emerged as the end point. Overall survival was analysed until August 2012. The median observation time for overall survival was 47 months (range, 3–73 months). In total, 27 patients died during the follow-up; the remaining patients were alive and censored. Of note, patients included in this study were not treated by neoadjuvant and adjuvant therapies. The clinicopathological characteristics of patients with colon adenocarcinoma were summarised in Table 1.
Table 1. Clinicopathological characteristics of patients with colon adenocarcinoma (n=63).
| Characteristic | Value | % |
|---|---|---|
| Age median (range), years |
70 (24–90) |
|
|
Gender | ||
| Male | 34 | 53.97% |
| Female |
29 |
46.03% |
|
Tumour Nodes Metastases category | ||
| T1 | 2 | 3.17% |
| T2 | 3 | 4.76% |
| T3 | 49 | 77.78% |
| T4 | 9 | 14.29% |
| N0 | 40 | 63.49% |
| N1 | 19 | 30.16% |
| N2 | 4 | 6.35% |
| M0 | 61 | 96.83% |
| M1 |
2 |
3.17% |
|
American Joint Committee on Cancer category | ||
| I | 3 | 4.76% |
| II | 37 | 58.73% |
| III | 21 | 33.33% |
| IV |
2 |
3.17% |
|
Differentiation | ||
| Low differentiation | 13 | 20.63% |
| Moderate differentiation | 37 | 58.73% |
| High differentiation | 8 | 12.70% |
| Tubular like | 30 | 47.62% |
| Sieve like |
33 |
52.38% |
|
Survival status | ||
| Alive | 36 | 57.14% |
| Dead | 27 | 42.86% |
Histology of all tumour specimens and regional lymph nodes was confirmed with haematoxylin–eosin staining according to the International Union against Cancer Tumour Nodes Metastases classification. AJCC is a classification system developed by the American Joint Committee on Cancer for describing the extent of disease progression in cancer patients.
Immunohistochemistry
Colon adenocarcinoma patient tumour tissue sections were formalin-fixed and paraffin-embeded. Deparaffinisation and hydration were performed and followed by abolishing endogenous peroxidase activity using 0.3% hydrogen peroxide for 30 min and microwaved for antigen retrieval in 10 mM sodium citrate buffer (pH 6.0) for 20 min. HOXB9 antibody was used at 2 μg ml−1 in these experiments, and was incubated at 4 °C overnight. Then PV9000 2-step plus Poly-HRP anti-mouse/rabbit IgG detection system (Zhong Shan Jin Qiao, Beijing, China) was applied. The streptavidin–biotin–peroxidase method was used for detection, and diaminobenzidine was applied as substrate (ChemMate Detection Kit, DAKO, Glostrup, Denmark). Haematoxylin was used for counterstaining. Negative controls were performed by omitting the primary antibody.
Assessment of immunohistochemistry
Two pathologists evaluated all immunostainings independently and a consensus justification based on discussion was recorded. The assessment was classified into four grades: no reactivity marked as 0, faint reactivity as 1+, moderate reactivity as 2+ and strong reactivity as 3+. We defined expression level of 3+ as high expression and the others as low expression.
Expression vectors, cell culture and gene transfections
HOXB9 full-length cDNA was cloned from a human placenta cDNA library using primers: 5′-ATCGAATTCATTTCTGGGACGCTTAGC-3′ (forward primer) and 5′-ATTCTCGAGGGTCTTTGATTAGACAGCAC-3′ (reverse primer). PCR product was cloned into PCRII vector, and then further cloned into pCMV10-3 × Flag vector (Sigma, Oakville, ON, Canada) using EcoRI–XhoI sites. Colon adenocarcinoma cell lines SW480, SW620, HCT116, RKO, LST and SW1116 were purchased from ATCC (Rockefeller, MD, USA) or Cell Collection Center of Peking Union Medical School in China. Breast cancer cell lines T47D, Hs578T and SUM159 were also purchased from ATCC in the USA. These cell lines were cultured in RPMI1640 medium (Invitrogen, Carlsbad, CA, USA) with 10% FBS and 50 μg ml−1 gentamycin. Cells were grown in 75-cm2 culture flasks or 60-mm2 dishes at 37 °C in humidity with 5% (v/v) CO2. Media were changed every 2 days. For establishment of stable clones that expressed Flag and Flag-HOXB9 separately, HCT116 cells were transfected with Flag-HOXB9 and empty vector using Lipofectamine. A total of 24 h after transfection, cells were passaged and G418 was added at final concentration of 500 μg ml−1. Mix clone and single clones that expressed HOXB9 were all selected for functional studies as indicated in the text.
Western blot analysis
PBSTDS lysis buffer with the presence of protease cocktail inhibitor (Roche Diagnostics, Bazel, Switzerland) was used to extract total cell lysates. In total, 20% of 5 × SDS-loading buffer was added into 80% of cell lysates, then resolved by 10% SDS–PAGE gel and blotted onto PVDF membranes (pore size 0.45 μm, Millipore, Billierica, MA, USA). The primary antibodies anti-HOXB9 mouse monoclonal antibody (1 : 1000 dilution; Clone M01, Abnova, Taipei, Taiwan), anti-HOXB9 rabbit polyclonal antibody (1 : 1000 dilution; Santa Cruz, Dallas, TX, USA) anti-claudin-1, claudin-7, mouse monoclonal antibody (1 : 1000 dilution; Millipore, USA), anti-E-cadherin rabbit monoclonal antibody (1 : 1000 dilution; Epitomics, Burlingame, CA, USA), anti-Flag-monoclonal antibody (1 : 10 000 dilution; Clone M2, Sigma) and occludin monoclonal antibody (1 : 1000 dilution; Sigma) were incubated with the membranes separately under rotation. After thorough washing, membranes were further incubated with corresponding secondary antibodies recognising either rabbit or mouse Ig (Jackson Laboratories, Bar Harbor, ME, USA). Finally, the bands were visualised by the enhanced chemoluminescence (Pierce, Appleton, WI, USA).
Cell motility and cell invasion assays
Costar Transwell chambers (Coring, NY, USA) with 8 μm pore size were used to perform the cell migration assay. HCT116 cells stable transfected with Flag-BAP (bacterial alkaline phosphatase), Flag-HOXB9 were seeded on the upper surface of the Transwell. After 6-h incubation in migration buffer (RPMI1640, 2 mM CaCl2, 1 mM MgCl2, 0.2 mM MnCl2 and 0.5% BSA) at 37 °C in humidified 5% CO2, the Transwell membranes were fixed with 4% formaldehyde for 15 min and stained by crystal violet for 10 min. At last, six microscopic fields were randomly chosen for counting the migrated cells.
The invasion assay was performed by adding Matrigel to the upper surface of the Transwell before adding cells and incubated for 12 h.
RNA interference
Sequences of RNA interference (RNAi) oligonucleotides used for knockdown of endogenous HOXB9 were designed as follows: HOXB9 siRNA1: 5′-CCCTTCAATTTGTAGACTCTT-3′, HOXB9 siRNA2: 5′-CTCCTCAATCTGAGTGAGAGA-3′.
Scramble siRNA: 5′-UUCUCCGAACGUGUCACGU-3′.
In vivo xenograft tumour growth and metastasis experiments
Balb/c nude mice were implanted subcutaneously into the flank with 1 × 106 cells, which are stably overexpressing HOXB9 and the control vector separately. Tumours were measured for their sizes every 3 days and continued to record for the indicated time. Tumours were sectioned at day 28th when they reached to approximately 1 cm in diameter and the mice were continued to maintain. Tumours were taken out and weighted. At day 56th, mice were killed using euthanasia. Tumour recurrence in situ and metastasis in remote organs were measured.
Quantitative PCR
Quantitative PCR assays were performed to detect the expression of HOXB9, E-cadherin, Vimentin, Twist and ZEB1 mRNA in Flag-HOXB9 HCT116 and Flag HCT116 cells. In brief, total cellular mRNAs were isolated by Trizol (Invitrogen), and 2 μg of total RNA was reverse-transcribed using M-MLV reverse transcriptase (Promega, Madison, WI, USA). Then PCR was performed using Taq PCR MasterMix (TIANGEN, Beijing, China) with the settings as: 94 °C 2 min; 94 °C 30 s, 60 °C 30 s, 72 °C 30 s, for 30 cycles; 72 °C 5 min. The primers for qPCR were as follows: for human HOXB9, forward: 5′-CCATTTCTGGGACGCTTAGCA-3′, reverse: 5′-TGTAAGGGTGGTAGACGGACG-3′ for actin, forward: 5′-CTGAGCGTGGCTACTCCTTC-3′, reverse: 5′-GCCATCTCGTTCTCGAAGTC-3′ for human E-cadherin, forward: 5′-CTGGGCTGGACCGAGAGA-3′, reverse: 5′-GAAGGTCAGCAGCTTGAACCA-3′ for human Vimentin, forward: 5′-GACGCCATCAACACCGAGTT-3′, reverse: 5′-CTTTGTCGTTGGTTAGCTGGT-3′ for human Twist, forward: 5′-GTCCGCAGTCTTACGAGGAG-3′, reverse: 5′-GCTTGAGGGTCTGAATCTTGCT-3′ for human ZEB1, forward: 5′-GATGATGAATGCGAGTCAGATGC-3′, reverse: 5′-ACAGCAGTGTCTTGTTGTTGT-3′. Relative fold changes in qPCR were determined by the ΔΔCt method.
RNA deep sequencing
Cellular total RNAs were extracted and RNA deep sequencing analyses were performed at the Biodynamic Optical Imaging Center, Peking University.
Statistical analysis
Methods including χ2, Fisher's exact test and Spearman's correlation coefficient were used to analyse the correlation between HOXB9 expression and clinicopathological characteristics of patients with colon adenocarcinoma. All statistical analyses were performed using SPSS version 14.0 (New York, NY, USA). Survival analysis was undertaken using Kaplan–Meier analysis, and curves were compared using the log-rank test. Results were considered statistically significant at the level of P⩽0.05.
Results
Elevated expression of HOXB9 predicts longer overall survival for patients with colon adenocarcinoma
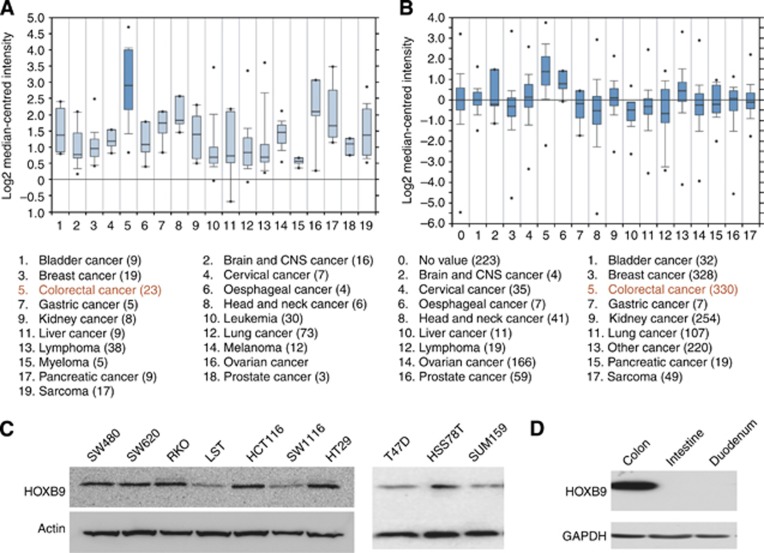
By reanalysing Oncomine Wooster Cell line data set, we discovered that the expression level of HOXB9 in cell lines from 19 different cancer types and found that cell lines from colon cancer expressed the highest level of HOXB9 among cell lines from other cancer types examined (Figure 1A). Furthermore, we also examined the expression level of HOXB9 in 17 different types of cancer patients in an Oncomine Bittner Multi-cancer data set and found that colon adenocarcinoma expressed the highest level of HOXB9 among cancer patients examined (Figure 1B). Other cancer types including pancreas, prostate and ovary cancers expressed relatively higher level of HOXB9 as well (Figure 1B). In addition, we confirmed the expression of HOXB9 protein in colon adenocarcinoma cell lines (Figure 1C). Interestingly, we also found that HOXB9 is highly expressed in colon but not expressed in intestine and duodenum in mouse (Figure 1D), indicating that HOXB9 might play an important role in the maintenance of the normal structure and function of colonic epithelia. Taken together, these data indicated that HOXB9 is expressed at both RNA and protein levels in cell lines and tumour tissues from both colon adenocarcinoma and normal colon, suggesting that HOXB9 may play a role in colon adenocarcinoma progression.
Figure 1.
Expression of HOXB9 in colon adenocarcinoma cell lines and patients. (A) HOXB9 expression in Wooster Cell line data set of oncomine: colorectal cancer cells displayed the highest expression of HOXB9 among 19 types of cancer cell lines included. (B) HOXB9 expression in Bittner Multi-cancer data set of oncomine: colorectal cancer showed the highest expression of HOXB9 in patients among 17 different types of cancer. (C) Left panel: HOXB9 protein levels in colon adenocarcinoma cell lines; right panel: HOXB9 protein levels in breast cancer cell lines. Protein levels of HOXB9 in cell lines were analysed by western blot probed by an anti-HOXB9 monoclonal antibody. Actin was used as loading control. (D) HOXB9 expression in normal colon, intestine and duodenum in mouse. Mouse colon, intestine and duodenum were dissected and tissues were homogenised and quantified. Equal amount of each tissue was used for western blot analysis. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as loading control.
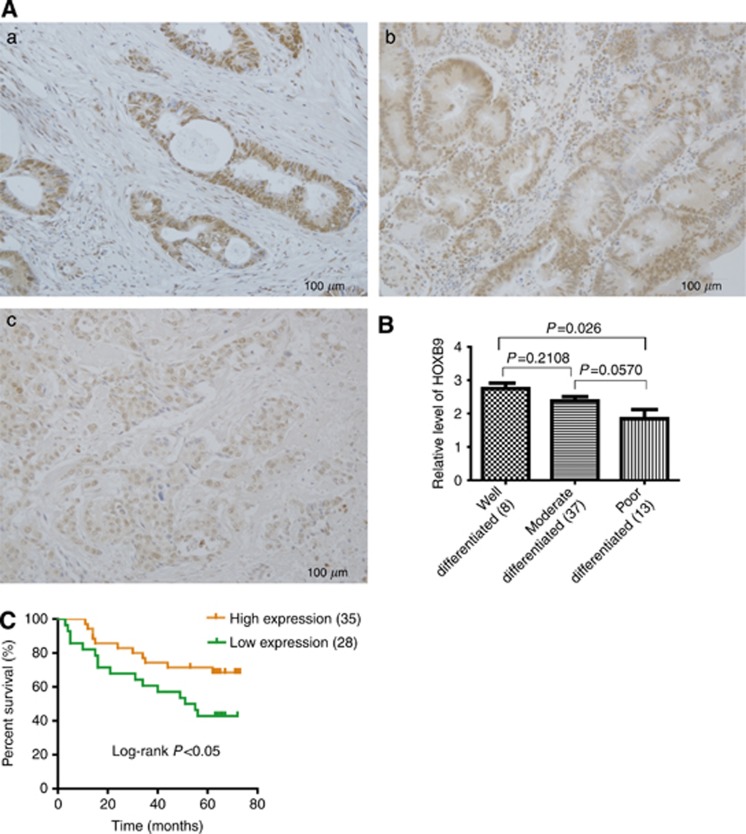
We continued to examine the HOXB9 expression profile in three typical differentiation types of colon adenocarcinoma that include well-, moderate- and poorly differentiated tumours. Interestingly, we found that HOXB9 expression is higher in well-differentiated tumours (Figure 2Aa) than in that of moderate- and poorly differentiated ones (Figure 2Ab and c). Furthermore, the well-differentiated types in the patients were significantly different from the poorly differentiated tumours in terms of HOXB9 expression (Figure 2B, P=0.026), whereas the other two pairs are not statistically significant. However, in Spearman's correlation coefficient test (Table 2), the expression level of HOXB9 correlated with the degree of differentiation in colon adenocarcinomas (Spearman's correlation test=0.26595; P=0.0436), providing further evidence that HOXB9 does play a role in colon adenocarcinoma differentiation.
Figure 2.
Elevated expression of HOXB9 predicts longer overall survival for patients with colon adenocarcinoma. (A) HOXB9 differentially expressed in colon adenocarcinoma patients with different degree of differentiation. For patient samples: (a) well differentiated; (b) moderate differentiated; (c) poorly differentiated. (B) Semi-quantitative analysis of HOXB9 levels in colon adenocarcinoma patients with different degree of differentiation, in which HOXB9 expressed higher in well-differentiated adenocarcinoma than the moderate- or poorly differentiated ones. Statistical analysis was performed and the difference between well- and poorly differentiated is significant (P=0.026), whereas the differences between well- and moderate-differentiated tumours as well as moderate- and poorly differentiated tumours were not significant (Figure 2B, P=0.2108 and 0.0570 separately). (C) Two groups of colon adenocarcinoma patients with HOXB9 high or low expression were determined by Kaplan–Meier analysis with a log-rank at P<0.05, showing that elevated expression of HOXB9 correlates to a better overall survival for colon adenocarcinoma patients.
Table 2. Relationship between HOXB9 expression and various clinicopathological factors in patients with colon adenocarcinoma (n=63).
| Characteristic | Low-expression group | High-expression group | Statistic method | P | ||
|---|---|---|---|---|---|---|
| |
28 |
44.44% |
35 |
55.56% |
|
|
| Sex |
|
|
|
|
χ2 |
0.63 |
| Male |
16 |
25.40% |
18 |
28.57% |
|
|
| Female |
12 |
19.05% |
17 |
26.98% |
|
|
|
Tumour Nodes Metastases category | ||||||
| T1-2 | 1 | 1.59% | 4 | 6.35% | Fisher's exact test | 0.3707 |
| T3-4 | 27 | 26.98% | 31 | 65.08% | ||
| N0 | 16 | 25.40% | 24 | 38.10% | χ2 | 0.5456 |
| N1-2 | 11 | 17.46% | 12 | 19.05% | ||
| M0 | 26 | 41.27% | 35 | 55.56% | χ2 correction for continuity | 0.3768 |
| M1 |
2 |
3.17% |
0 |
0.00% |
|
|
|
American Joint Committee on Cancer | ||||||
| I | 0 | 0.00% | 3 | 4.76% | Fisher's exact test 0.2092 | 0.3294 |
| II | 17 | 26.98% | 20 | 31.75% | ||
| III | 9 | 14.29% | 12 | 19.05% | Kruskal–Walls test 0.1811 | |
| IV |
2 |
3.17% |
0 |
0.00% |
Spearman's correlation coefficient −0.12490 |
|
|
Differentiation | ||||||
| Poorly differentiation | 9 | 14.29% | 4 | 6.35% | Spearman's correlation coefficient 0.26595 | 0.0436* |
| Moderate differentiation | 17 | 26.98% | 20 | 31.75% | Fisher's exact test 0.1432 | |
| Well differentiation | 2 | 3.17% | 6 | 9.52% | Kruskal–Walls test 0.1332 | |
| Tubular like | 8 | 12.70% | 22 | 34.92% | χ2 | 0.0068* |
| Sieve like |
20 |
31.75% |
13 |
20.63% |
|
|
|
Survival status | ||||||
| Alive | 12 | 19.05% | 24 | 38.10% | χ2 | 0.0404* |
| Dead |
16 |
25.40% |
11 |
17.46% |
|
|
| *Significant P-value, P⩽0.05 | ||||||
Methods including χ2, Fisher's exact test and Spearman's correlation coefficient were used to analyse the correlation between HOXB9 expression and clinicopathological characteristics of patients with colon adenocarcinoma. AJCC is a classification system developed by the American Joint Committee on Cancer.
In a cohort study, we examined HOXB9 expression in patients using immunohistochemistry analysis and found that HOXB9 is highly expressed in more than half (55.56%) of the 63 colon adenocarcinoma patients, and the rest of patients showed low HOXB9 expression (Table 2). As demonstrated in Table 2, HOXB9 expression was seen higher in well-differentiated tubular-like colon adenocarcinoma patients than the poorly differentiated sieve-like ones (P=0.0068). In AJCC category, grade IV colon adenocarcinoma has lower HOXB9 expression than grade I (P=0.05), and the level of HOXB9 expression correlated with patients' survival rate. Alive group has higher expression of HOXB9 than the dead group (P<0.05). However, HOXB9 expression has no obvious relationship with the TNM category, age, and gender of colon adenocarcinomas (P=0.1759). In agreement with Figure 2A and B, the tubular-like colon adenocarcinomas, which are on the higher order of differentiation than the sieve-like ones, expressed higher level of HOXB9 (Tables 2, P=0.0068). Importantly, in a Kaplan–Meier analysis, we found that elevated expression of HOXB9 corresponded to a longer patient survival (Figures 2C, P<0.05). This finding suggested that HOXB9 expression level was of prognostic value for colon adenocarcinoma patients, in which elevated expression of HOXB9 may predict a favourable outcome for the patients. However, the functional role of HOXB9 in colon adenocarcinoma progression remained to be elucidated.
HOXB9 inhibits colon adenocarcinoma cell growth, migration and invasion in vitro
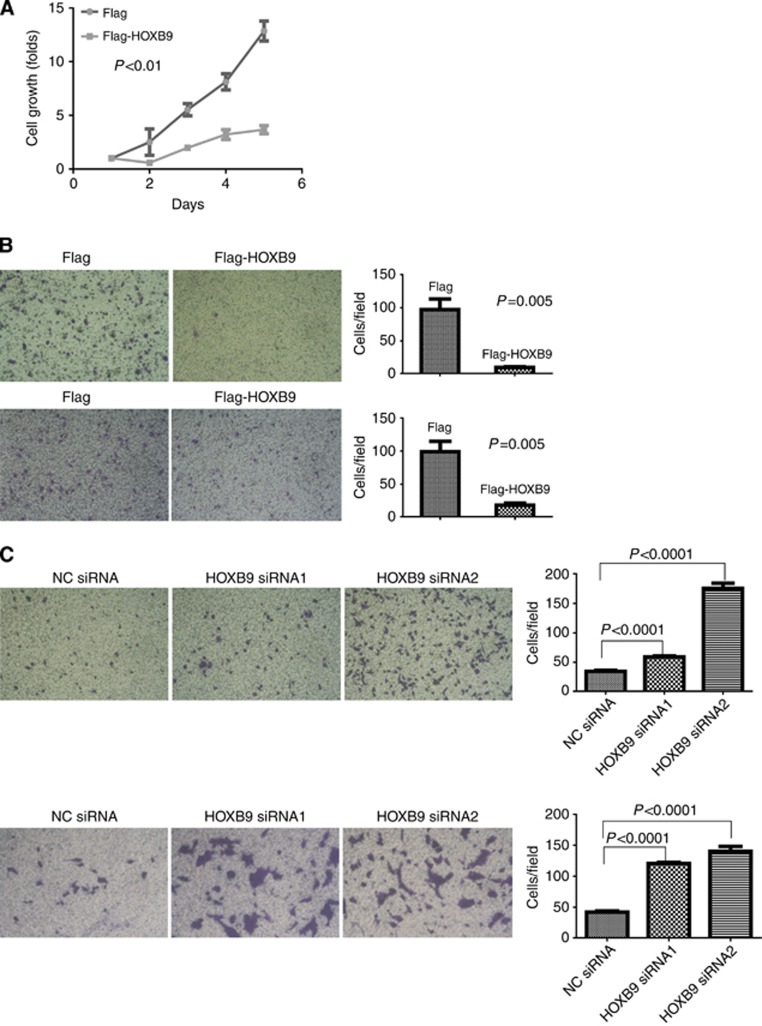
Given that the aforementioned data demonstrated that elevated HOXB9 expression predicts longer colon adenocarcinoma patient survival, suggesting that HOXB9 may play a tumour suppressive role. To this end, we examined the role of HOXB9 in the regulation of colon adenocarcinoma progression. In a gain-of-function experiment, overexpression of HOXB9 in HCT116 cells inhibited cell growth (Figure 3A), migration (Figure 3B upper panels) and invasion on Matrigel (Figure 3B lower panels). In contrast, in a loss-of-function experiment by knocking down the endogenous expression of HOXB9 using small interference RNAs (siRNAs), colon adenocarcinoma cells HCT116 were rescued from the inhibitory effect of HOXB9 and re-gained the capacities for cell migration (Figure 3C upper panels) and invasion (Figure 3C lower panels). These data indicated that the existence of endogenous HOXB9 exerts an important effect that suppresses cell growth, migration and invasion in colon adenocarcinoma.
Figure 3.
HOXB9 inhibits colon cancer cell growth, migration and invasion. (A) Cell growth assay. HOXB9 effect on colon adenocarcinoma cell growth was measured using WST-1 method in HCT116 cells stably expressing Flag-HOXB9 or Flag. (B) Gain-of-function experiments. Upper panels: HOXB9 effect on colon adenocarcinoma cell migration was determined in a Transwell assay by comparing HCT116 cells stably expressing Flag-HOXB9 or Flag (the left two panels), with quantification on the right panel. Lower panels: HOXB9 effect on colon adenocarcinoma cell invasion in Matrigel was examined using HCT116 cells stably expressing Flag-HOXB9 or Flag (the left two panels). The cell invasion assay was quantified and displayed on the right panel. (C) Loss-of-function experiments. Upper panels: the requirement of HOXB9 in colon adenocarcinoma cell migration was examined in HCT116 cells transiently transfected with two HOXB9 siRNAs separately, controlled by scramble siRNA (the left three panels). The migratory cells were quantified as shown on the right panel. Lower panels: the effect of HOXB9 on colon adenocarcinoma cell invasion was examined in HCT116 cells transiently transfected with two HOXB9 siRNAs separately, controlled by scramble siRNA (the left three panels). Quantification of cell invasion was shown on the right panel. Student's t-tests were applied for analysing the difference between the two cell populations in growth, migration and invasion assays, and in each case the difference was regarded as significant with p⩽0.05.
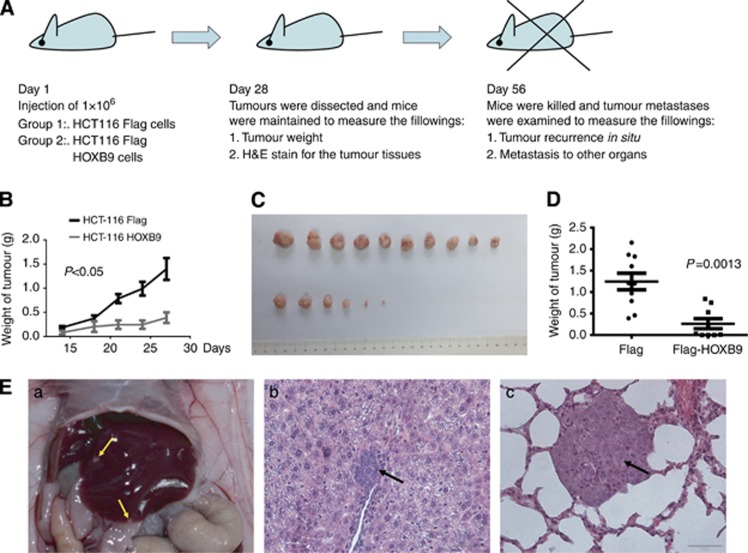
HOXB9 inhibits colon adenocarcinoma growth and metastasis in vivo in a mouse model
To answer the question that whether HOXB9 plays a similar inhibitory role in vivo as in vitro, we examined the potential blockade of HOXB9 on tumour growth and metastasis in a nude mouse xenograft experiment as designed in Figure 4A. Overexpression of HOXB9 was identified to inhibit tumour growth in xenograft nude mice (Figure 4B–D). When tumours grew with a size of approximately 1 cm in diameter, they were removed by surgery. Interestingly, tumours derived from the HOXB9-expressing HCT116 cells displayed a low or no capability to metastasise to the remote organs including liver and lung of the nude mouse; whereas tumours derived from the control HCT116 cells demonstrated a high capability to metastasise to the liver and lung (Figure 4E, left: liver metastasis for a gross view, middle: a histology for liver metastasis, and right: a histology for lung metastasis). In addition, the capability of HOXB9 in blockade of tumour formation, recurrence and metastasis in nude mice was summarised in Table 3 from a representative experiment. These data strongly suggested that HOXB9 inhibited colon adenocarcinoma cell growth and metastasis to remote organs in nude mice. Taken together, these data indicated that elevated expression of HOXB9 exerted a suppressive role for colon adenocarcinoma cell progression both in vitro and in vivo. Thus, these functional experiments well interpreted the fact that elevated expression of HOXB9 correlated with a better patients' overall survival in colon adenocarcinomas.
Figure 4.
HOXB9 suppresses tumour growth and metastasis in a mouse xenograft model. (A) Design of mouse xenograft experiment. (B) Tumour growth curve. (C) Tumour formation at day 28, which were dissected and photographed. (D) Average of tumour weights at day 28. (E) For HCT116-Flag control cells: (a) liver metastasis; (b) histology of live metastasis; (c) histology of lung metastasis. Please refer to Table 3 for summary of tumour formation, recurrence in situ and metastasis to remote organs for Flag HCT116 or Flag-HOXB9 HCT116 cells in the nude mouse model.
Table 3. Comparison for the ability of tumour formation, recurrence and metastasis between the control Flag HCT116 and the Flag-HOXB9 HCT116 cells in nude mice.
| Groups | Flag HCT116 (No. mice=10) (%) | Flag-HOXB9 HCT116 (No. mice=9) (%) |
|---|---|---|
| Number of cells injected subcutaneously |
1 × 106 |
1 × 106 |
| Tumour formation on day 28th |
10/10 (100%) |
6/9 (66.7%) |
| Surgery on day 28th |
All alive |
One dead of unknown reason |
| Recurrence in situ |
3/10 (30%) |
1/8 (12.5%) |
| Metastasis to the liver |
7/10 (70%) |
3/8 (37.5%) |
| Metastasis to the lung | 5/10 (50%) | 3/8 (37.5%) |
Balb/c nude mice were implanted subcutaneously into the flank with 1 × 106 cells, which are stably overexpressing HOXB9 and the control vector separately. The ability of tumor formation was significantly decreased by HOXB9 stably expression (P=0.0016). Tumours were dissected on day 28th when the diameter of the tumours reached to approximately 1 cm and the mice were continued to maintain. After another 28 days growth (on day 56th), mice were killed using euthanasia, and tumor recurrence and metastasis were measured.
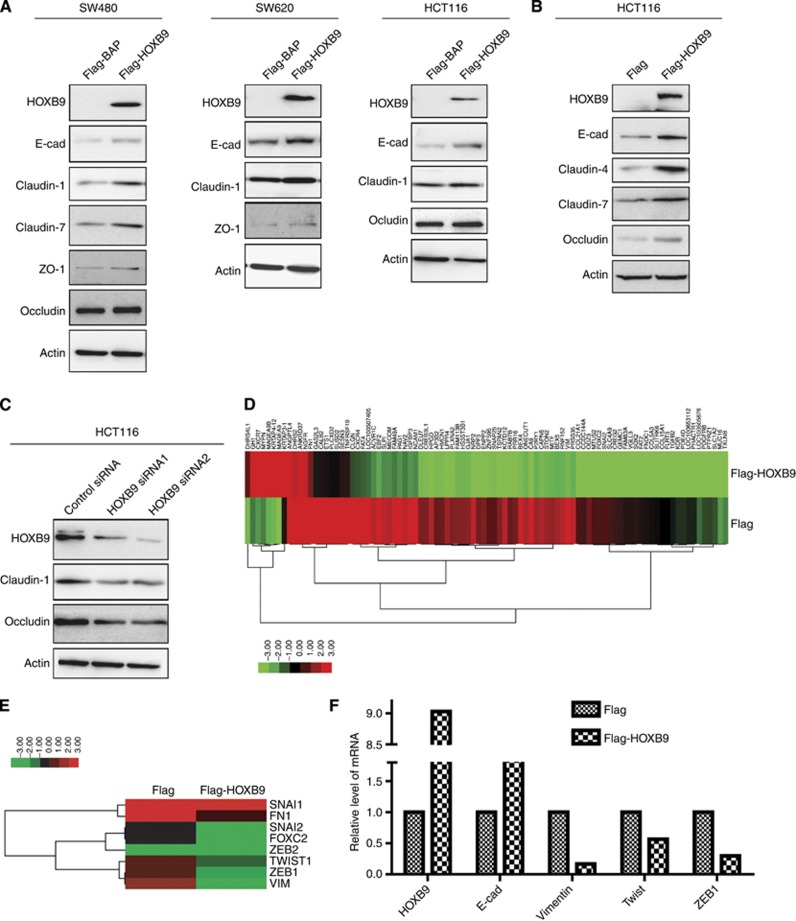
HOXB9 promotes colon adenocarcinoma differentiation by upregulating cellular junction protein expressions—a mechanistic interpretation
The aforementioned findings have demonstrated that HOXB9 functions as a tumour suppressive transcriptional factor in colon adenocarcinoma cells and its expression correlated with differentiation patterns and overall survival in colon adenocarcinoma patients. However, the mechanism accounting for these inhibitory effects of HOXB9 remained unknown. To this end, we examined changes of a panel of cell differentiation markers in colon adenocarcinomas. In a gain-of-function experiment, we found that transient expression of HOXB9 upregulated cellular junction proteins E-cadherin, claudin-1, claudin-7, ZO-1 and occludin in SW480 cells (Figure 5A left), E-cadherin, claudin-1 and ZO-1 in SW620 cells (Figure 5A middle), and E-cadherin, claudin-1 and occludin in HCT116 cells (Figure 5A right). Similarly, E-cadherin, claudin-4 and -7, and occludin were found upregulated in HCT116 cells stably expressing HOXB9 (Figure 5B). In contrast, in a loss-of-function experiment using HOXB9 siRNAs transient transfection, we found that claudin-1 and occludin were both downregulated in HCT116 cells (Figure 5C). These data together suggested that ectopic expression of HOXB9 promoted colon adenocarcinoma cell differentiation by driving the cells toward an epithelial phenotype, which may involve a mesenchymal-to-epithelial transition (MET) process. To identify the global effect of HOXB9 expression on the colon adenocarcinoma cell phenotypic changes, we performed RNA deep sequencing analyses (Supplementary Table 1). As Figure 5D indicated, genes with 16-fold changes under the HOXB9 regulation were displayed. Furthermore, selective gene expressions which are related to EMT including SNAI1, SNAI2, FOXC2, ZEB1 ZEB2, Twist, FN1 and VIM were shown in the heatmap presentation (Figure 5E). The HOXB9 effect on global expressions of the EMT-related markers was further confirmed in a selective panel of key genes in a quantitative PCR assay (Figure 5F).
Figure 5.
HOXB9 promotes colon adenocarcinoma cell differentiation by upregulating cellular junction protein expressions. (A) In the gain-of-function experiment, transient expression of HOXB9 upregulated adherens junction protein E-cadherin and tight junction proteins claudin-1, claudin-7, occludin and ZO-1 in SW480 cells (left); E-cadherin, claudin-1 and ZO-1 in SW620 cells (middle) and E-cadherin, claudin-1, claudin-7 and occludin in HCT116 cells (right). (B) In HCT116 cells, stable expression of HOXB9 upregulated E-cadherin, claudin-4, claudin-7 and occludin. (C) In the loss-of-function experiment, stably knocking down of HOXB9 in HCT116 cells led to downregulation of claudin-1 and occludin. (D) HOXB9-regulated genes with 16-fold changes were presented in a heatmap for displaying gene expression profile. (E) Important EMT-related genes SNAI1, SNAI2, FOXC2, ZEB1 ZEB2, Twist, FN1 and VIM were selected to plot in a heatmap presentation, plotted showing that EMT-related transcriptional factors are drastically downregulated, so do mesenchymal markers FN1 and VIM. (F) Confirmation of changes in gene expression for selective key genes in Flag HCT116 or Flag-HOXB9 HCT116 cells. Epithelial marker E-cadherin, mesenchymal marker Vimentin and EMT-stimulating transcriptional factors Twist and ZEB1 were measured by quantitative PCR in the cells expressing HOXB9. Data were expressed as relative expression of mRNAs.
Collectively, HOXB9 exerts a tumour suppressive effect on colon adenocarcinoma progression by inhibition of master EMT transcriptional factors ZEB1 and Twist that lead to upregulation of the epithelial markers and promotes differentiation in colon adenocarcinomas.
Discussion
HOXB9 has been known to play a very important role in embryonic development and in the progression of a number of malignancies, including breast and lung cancers (Nguyen et al, 2009; Hayashida et al, 2010). Sharply contrast to the previous studies that HOXB9 induces tumour invasion and metastasis in breast and lung cancers, on the opposite we identified in this report that HOXB9 suppressed tumour growth, invasion and metastasis to the lung and liver in a mouse xenograft model of colon adenocarcinomas. Furthermore, we also found that elevated HOXB9 expression correlated with longer patient survival, indicating that HOXB9 is a tumour suppressive transcriptional factor in colon adenocarcinomas. In lines with our results, Sha et al (2013) found that decreased expression of HOXB9 is related to poor overall survival in gastric carcinoma patients. In addition, silencing of HOXB9 was linked to extrathyroidal extension and advanced pathologic stage of papillary thyroid carcinoma, hinting that HOXB9 may suppress invasion in thyroid tumour (Kim et al, 2012). Taken together, these results strongly suggest that HOXB9 plays a diverse and even an opposite role in different types of cancer. The fact that HOXB9 promotes or suppresses tumour progression may be attributed to the tissue origin and the microenvironment of the tumours.
HOXB9 was found to activate cellular proliferation and differentiation in leukaemic cells (Ohnishi et al, 1998). However, no indication of HOXB9 regulation on differentiation was found in solid tumours. We presented in this report that elevated level of HOXB9 promoted cell differentiation in colon adenocarcinomas via a mechanism called MET, a process that drives mesenchymal cells to epithelial cells. This important finding implies that raising the levels of transcriptional factor HOXB9 or its targets in colon adenocarcinoma patients may represent a way of developing new therapeutic strategies or drugs that block colon adenocarcinoma invasion and metastasis. Inducing differentiation has been applied to treat a variety of cancers (Liu et al, 2000). Malignant cells can be converted to a benign phenotype accompanying with reduced tumour cell proliferation and increased apoptosis (Spira and Carducci, 2003). In colon adenocarcinomas, differentiation is associated with therapeutic outcome and patient survival. Tubular-like colon adenocarcinomas are usually well differentiated with lower tumour grade, whereas sieve-like adenocarcinomas are poorly differentiated with higher tumour grade. Microsatellite instability (MSI) is a mode of genomic instability resulting in colorectal cancer development and is associated with poor differentiation, proximal location and failure of chemotherapy (Heinimann, 2013). Therefore, poor differentiation of a tumour in the context of MSI is not an adverse prognostic factor, suggesting that HOXB9-induced colon adenocarcinoma differentiation may not be beneficial to the patients. The relationships between the HOXB9 expression profile and the status of MSI or microsatellite stability (MSS) in colon adenocarcinomas are interesting questions that warrant future investigations.
In the present study, we identified that HOXB9 is elevated in well-differentiated colon adenocarcinoma patients (Figure 2A–C), and ectopic expression of HOXB9 inhibits colon adenocarcinoma cell growth and invasion. The underling mechanism for these functions may be achieved by upregulation of a panel of epithelial proteins including E-cadherin, tight junction proteins claudins, occludin and ZO-1 (Figure 5A and B). Tight Junctions are multiprotein complexes including occludin, claudins and junctional adhesion molecules. Tight Junctions seal neighbouring cells and control the paracellular pathways and restrict the diffusion of apical and basolateral membrane components. Tight Junctions also play a role in cell differentiation and proliferation (Schneeberger and Lynch 2004; Aijaz et al, 2006; Furuse and Tsukita 2006; Balda and Matter 2009; Harten et al, 2009). Our finding that HOXB9 inducing expressions of tight junction protein shed light on the hope that enhancing the level of HOXB9 in colon adenocarcinoma cells may help to drive the de-differentiated cells to differentiate back to epithelial cells that hold low growth and invasion potentials. Our RNA deep sequencing results from HOXB9-overexpressing HCT116 cells showed that important EMT-stimulating transcriptional factors including Snail, Slug, Twist, ZEB1 and TCF-7 have been greatly downregulated by HOXB9 expression (Figure 5F). As a consequence, the expression of mensenchymal markers including vimentin and fibronectin were markedly reduced (Figure 5E and F). These data strongly indicated that MET did happen in colon adenocarcinoma cells that express HOXB9.
Of note, HOXB9 expression level was not correlated with the sequence of colon adenocarcinoma in the TNM category (Table 2). The reason could be the inadequate numbers of patients included in the cohort applied. However, we did observed a correlation between grades I and IV in the AJCC category (P=0.05, not shown in Table 2). Therefore, the relationship between HOXB9 expression level and the progression of colon adenocarcinomas remains to be elucidated using a larger patient cohort in the future. However, HOXB9 expression level was found correlated with the tumour differentiation patterns classified either by well-, moderate- or poorly differentiated colon adenocarcinomas or by the tubular-like or sieve-like colon adenocarcinomas (Table 2). Furthermore, we found that HOXB9 expression level was raised in patients who were alive and patients with elevated HOXB9 expression displayed a longer overall survival (Table 2 and Figure 2C). Therefore, these data suggested that detection of HOXB9 level may help to assess the therapeutic outcomes and HOXB9 may be of prognostic value for colon adenocarcinoma patients. In addition, the detailed molecular mechanisms accounting for HOXB9 suppressive effects on colon adenocarcinoma cell growth, migration, invasion and metastasis deserve for further investigations.
In summary, we demonstrated that HOXB9 is a novel tumour suppressive protein that inhibits tumour cell growth, migration, invasion and metastasis in colon adenocarcinomas via a primary mechanism of inducing cell differentiation by promoting mesenchymal-to-epithelial transition. Elevated expression of HOXB9 anticipates a favourable outcome in colon adenocarcinoma patients. Therefore, targeting HOXB9 may give insight of generating new strategies for colon adenocarcinoma therapeutics.
Acknowledgments
This work was supported by grants from the Ministry of Science and Technology of China 2013CB910501, 2010CB912203 and 2010CB529402, the National Natural Science Foundation of China 81230051, 30830048, 31170711 and 81321003, the 111 Project of the Ministry of Education, Beijing Natural Science Foundation 7120002 and Peking University grants BMU20120314 and BMU20130364, and a Leading Academic Discipline Project of Beijing Education Bureau to HZ This work was also supported by the National Natural Science Foundation of China Grant 81301802 to JZ.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence. Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Aijaz S, Balda MS, Matter K. Tight junctions: molecular architecture and function. Int Rev Cytol. 2006;248:261–298. doi: 10.1016/S0074-7696(06)48005-0. [DOI] [PubMed] [Google Scholar]
- Balda MS, Matter K. Tight junctions and the regulation of gene expression. Biochim Biophys Acta. 2009;1788:761–767. doi: 10.1016/j.bbamem.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Chen F, Capecchi MR. Paralogous mouse Hox genes, Hoxa9, Hoxb9, and Hoxd9, function together to control development of the mammary gland in response to pregnancy. Proc Natl Acad Sci USA. 1999;96:541–546. doi: 10.1073/pnas.96.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Sukumar S. HOX genes: emerging stars in cancer. Cancer Biol Ther. 2003;2:524–525. doi: 10.4161/cbt.2.5.525. [DOI] [PubMed] [Google Scholar]
- Chiba N, Comaills V, Shiotani B, Takahashi F, Shimada T, Tajima K, Winokur D, Hayashida T, Willers H, Brachtel E, Vivanco MD, Haber DA, Zou L, Maheswaran S. Homeobox B9 induces epithelial-to-mesenchymal transition-associated radioresistance by accelerating DNA damage responses. Proc Natl Acad Sci USA. 2012;109:2760–2765. doi: 10.1073/pnas.1018867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16:181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. The pathophysiology of HOX genes and their role in cancer. J Pathol. 2005;205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- Harten SK, Shukla D, Barod R, Hergovich A, Balda MS, Matter K, Esteban MA, Maxwell PH. Regulation of renal epithelial tight junctions by the von Hippel-Lindau tumor suppressor gene involves occludin and claudin 1 and is independent of E-cadherin. Mol Biol Cell. 2009;20:1089–1101. doi: 10.1091/mbc.E08-06-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashida T, Takahashi F, Chiba N, Brachtel E, Takahashi M, Godin-Heymann N, Gross KW, Vivanco MD, Wijendran V, Shioda T, Sgroi D, Donahoe PK, Maheswaran S. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc Natl Acad Sci USA. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinimann K. Toward a molecular classification of colorectal cancer: the role of microsatellite instability status. Front Oncol. 2013;3:272. doi: 10.3389/fonc.2013.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Hong KD, Jung WY, Lee E, Shin BK, Kim HK, Kim A, Kim BH. SIRT1 expression is associated with good prognosis in colorectal cancer. Korean J Pathol. 2013;47:332–339. doi: 10.4132/KoreanJPathol.2013.47.4.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kim YH, Han JH, Lee KB, Sheen SS, Lee J, Soh EY, Park TJ. Silencing of homeobox B9 is associated with down-regulation of CD56 and extrathyroidal extension of tumor in papillary thyroid carcinoma. Hum Pathol. 2012;43:1221–1228. doi: 10.1016/j.humpath.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Liu TX, Zhang JW, Tao J, Zhang RB, Zhang QH, Zhao CJ, Tong JH, Lanotte M, Waxman S, Chen SJ, Mao M, Hu GX, Zhu L, Chen Z. Gene expression networks underlying retinoic acid-induced differentiation of acute promyelocytic leukemia cells. Blood. 2000;96:1496–1504. [PubMed] [Google Scholar]
- Morgan R, Boxall A, Harrington KJ, Simpson GR, Gillett C, Michael A, Pandha HS. Targeting the HOX/PBX dimer in breast cancer. Breast Cancer Res Treat. 2012;136:389–398. doi: 10.1007/s10549-012-2259-2. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massagué J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Tobita T, Sinjo K, Takeshita A, Ohno R. Modulation of homeobox B6 and B9 genes expression in human leukemia cell lines during myelomonocytic differentiation. Leuk Lymphoma. 1998;31:599–608. doi: 10.3109/10428199809057620. [DOI] [PubMed] [Google Scholar]
- Rhoads K, Arderiu G, Charboneau A, Hansen SL, Hoffman W, Boudreau N. A role for Hox A5 in regulating angiogenesis and vascular patterning. Lymphat Res Biol. 2005;3:240–252. doi: 10.1089/lrb.2005.3.240. [DOI] [PubMed] [Google Scholar]
- Schneeberger EE, Lynch RD. The tight junction: a multifunctional complex. Am J Physiol Cell Physiol. 2004;286:C1213–C1228. doi: 10.1152/ajpcell.00558.2003. [DOI] [PubMed] [Google Scholar]
- Seki H, Sakata M, Takahashi M, Yuko Kitagawa M. HOXB9 expression promoting tumor cell proliferation and angiogenesis is associated with clinical outcomes in breast cancer patients. Ann Surg Oncol. 2012;19:1831–1840. doi: 10.1245/s10434-012-2295-5. [DOI] [PubMed] [Google Scholar]
- Sha S, Gu Y, Xu B, Hu H, Yang Y, Kong X, Wu K. Decreased expression of HOXB9 is related to poor overall survival in patients with gastric carcinoma. Dig Liver Dis. 2013;45:422–429. doi: 10.1016/j.dld.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- Shrestha B, Ansari KI, Bhan A, Kasiri S, Hussain I, Mandal SS. Homeodomain-containing protein HOXB9 regulates expression of growth and angiogenic factors, facilitates tumor growth in vitro and is overexpressed in breast cancer tissue. FEBS J. 2012;279:3715–3726. doi: 10.1111/j.1742-4658.2012.08733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spira AI, Carducci MA. Differentiation therapy. Curr Opin Pharmacol. 2003;3:338–343. doi: 10.1016/s1471-4892(03)00081-x. [DOI] [PubMed] [Google Scholar]
- Sun M, Song CX, Huang H, Frankenberger CA, Sankarasharma D, Gomes S, Chen P, Chen J, Chada KK, He C, Rosner MR. HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci USA. 2013;110:9920–9925. doi: 10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres S, Bartolome RA, Mendes M, Barderas R, Fernandez-Acenero MJ, Pelaez-Garcia A, Peña C, Lopez-Lucendo M, Villar-Vázquez R, de Herreros AG, Bonilla F, Casal JI. Proteome profiling of cancer-associated fibroblasts identifies novel proinflammatory signatures and prognostic markers for colorectal cancer. Clin Cancer Res. 2013;19:6006–6019. doi: 10.1158/1078-0432.CCR-13-1130. [DOI] [PubMed] [Google Scholar]
- Winnik S, Klinkert M, Kurz H, Zoeller C, Heinke J, Wu Y, Bode C, Patterson C, Moser M. HoxB5 induces endothelial sprouting in vitro and modifies intussusceptive angiogenesis in vivo involving angiopoietin-2. Cardiovasc Res. 2009;83:558–565. doi: 10.1093/cvr/cvp133. [DOI] [PubMed] [Google Scholar]
- Workman P, Aboagye EO, Balkwill F, Balmain A, Bruder G, Chaplin DJ, Double JA, Everitt J, Farningham DA, Glennie MJ, Kelland LR, Robinson V, Stratford IJ, Tozer GM, Watson S, Wedge SR, Eccles SA. Guidelines for the welfare and use of animals in cancer research. Br J Cancer. 2010;102:1555–1577. doi: 10.1038/sj.bjc.6605642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.