Abstract
Pertussis is an infectious respiratory disease of humans caused by the gram-negative pathogen Bordetella pertussis. The use of acellular pertussis vaccines (aPs) which induce immunity of relative short duration and the emergence of vaccine-adapted strains are thought to have contributed to the recent resurgence of pertussis in industrialized countries despite high vaccination coverage. Current pertussis vaccines consist of antigens derived from planktonic bacterial cultures. However, recent studies have shown that biofilm formation represents an important aspect of B. pertussis infection, and antigens expressed during this stage may therefore be potential targets for vaccination. Here we provide evidence that vaccination of mice with B. pertussis biofilm-derived membrane proteins protects against infection. Subsequent proteomic analysis of the protein content of biofilm and planktonic cultures yielded 11 proteins which were ≥three-fold more abundant in biofilms, of which Bordetella intermediate protein A (BipA) was the most abundant, surface-exposed protein. As proof of concept, mice were vaccinated with recombinantly produced BipA. Immunization significantly reduced colonization of the lungs and antibodies to BipA were found to efficiently opsonize bacteria. Finally, we confirmed that bipA is expressed during respiratory tract infection of mice, and that anti-BipA antibodies are present in the serum of convalescent whooping cough patients. Together, these data suggest that biofilm proteins and in particular BipA may be of interest for inclusion into future pertussis vaccines.
Keywords: biofilms, BipA, Bordetella pertussis, proteomics, vaccination
INTRODUCTION
The gram-negative bacterium Bordetella pertussis is the main causative agent of whooping cough or pertussis. During epidemics in the pre-vaccination era, pertussis was attributable for up to 13% of all-cause child mortality (≤10 years).1 While this percentage has dropped significantly following the introduction of widespread vaccination in the 1950s, pertussis has resurged since the 1990s2,3 and is now estimated to (re-)infect 40 million people each year,3,4,5 resulting in approximately 195 000 deaths worldwide, mainly in children.6
Although acellular pertussis vaccines (aPs) are highly effective in protecting infants from developing severe pertussis, immunity already wanes after 5–10 years 7,8 and aPs may not effectively reduce circulation of B. pertussis in the population.9 This may have facilitated the evolution of antigenically divergent strains and, more recently, the emergence of a lineage (designated the P3 lineage) that produces higher amounts of several virulence factors, including pertussis toxin (Ptx).10,11 Alarmingly, within this P3 lineage, strains that do not express pertactin (Prn), a component of most aPs, have emerged in the last six years and are now becoming increasingly dominant.12,13,14 Clearly, there is a need for novel antigens for inclusion in pertussis vaccines to improve their effectiveness. Here we have focused on biofilms as a potential source of protective antigens.
B. pertussis is able to produce biofilms in the respiratory tract and this process has been shown to contribute to successful infection.15,16,17,18 In general, biofilm-associated bacteria differ significantly from their planktonic counterparts 19 and studies with B. pertussis have shown that ∼10% of the proteome is differentially expressed between these conditions.20 Most B. pertussis proteins that interact directly with the host are regulated by the two-component Bordetella master virulence regulatory system BvgAS which transduces environmental signals such as temperature, nicotinic acid and sulfate, to gene regulation (reviewed in Decker et al.21). Based on the activity of this system, the Bvg+, Bvgi, and Bvg− phase have been defined. In the Bvg+ phase, almost all virulence factors are expressed.10,21 The Bvgi phase, characterized by the expression of the Bordetella intermediate protein A (BipA), has been suggested to be involved in transmission and biofilm formation.22,23,24 Bvg+ and Bvgi phase bacteria have been shown to be virulent in mice, whereas Bvg−-locked bacteria, which cannot switch to the Bvg+ or Bvgi phase, are unable to establish infection.24,25
Here we examined the ability of B. pertussis biofilms to protect against pertussis using a mouse model. Having confirmed the protective potential of biofilms, we then comprehensively analyzed the biofilm antigenic protein composition and compared it to in vitro planktonic grown bacteria. Finally, we evaluated whether immunization with a single biofilm protein, BipA, was able to protect mice against respiratory infection with B. pertussis and examined the expression of bipA during infection.
MATERIALS AND METHODS
Ethics statement
Animal experiments were approved by the Radboudumc Committee for Animal Ethics and conducted in accordance with the relevant Dutch legislation. Bacterial strains were collected by regional Medical Microbiology Laboratories from patients suspected of whooping cough and sent to the National Institute for Public Health and the Environment in the context of routine surveillance (as required by law). The strains were sent to the National Institute for Public Health and the Environment for confirmation of clinical diagnosis, species determination and subtyping. Strictly anonymized patient information was included, which was limited to age, sex and postal code. For this type of surveillance, ethical evaluation or patient consent are not required. The strains have been used in previous studies.10
Bacterial strains and growth conditions
For flow cytometry and quantitative polymerase chain reaction analysis, B. pertussis strains were grown under Bvg+, Bvgi and Bvg− conditions as described previously.10 For modulation of the BvgAS regulatory system, magnesium sulfate was added to cultures at a final concentration of 5 mM and 50 mM to induce Bvgi and Bvg− conditions, respectively. In the absence of additional sulfate, the concentration of free sulfate is 0.02 mM, thereby inducing Bvg+ conditions. For proteomics, B. pertussis was grown under planktonic and biofilm conditions in chemically defined THIJS medium26 essentially as described previously.20 Briefly, planktonic cultures were grown overnight in chemically defined THIJS medium supplemented with 0.2 mg/mL Heptakis-cyclodextrin (Sigma-Aldrich Chemie B.V., Zwijndrecht, The Netherlands) and then re-inoculated into pre-warmed medium at an optical density at 620 nm (OD620) of 0.075. Planktonic cultures were harvested by centrifugation (15°C, 8000g, 20 min) at mid-exponential (17 h) and stationary (40 h) growth phases, washed with phosphate-buffered saline (PBS) and snap-frozen in liquid nitrogen. Biofilm cultures were carried out by using glass column reactors (diameter: 6 cm, h=45 cm; Industria Vidriera Argentina, SA, Buenos Aires, Argentina) packed with flat polypropylene beads (300 g; diameter: 4.1 mm, h=1.5 mm, with an average density of 0.901 g/cm3; Pentroken, Buenos Aires, SA, Argentina), which served as growth support. The bioreactors were inoculated with 200 mL of a planktonic culture of B. pertussis B1917 (OD620 of 1.0) grown in THIJS medium and incubated for 5 h at 37°C to allow bacterial attachment to the beads. Then, the suspension was drained to remove unattached cells and 200 mL of fresh THIJS medium was added to each column. Bioreactors were incubated aerobically (0.1 L/min) at 37°C and the THIJS medium was refreshed at 24-h intervals. For proteomic analysis, biofilm cells were harvested after 72 h, when the biofilm achieved its mature structure. Beads were washed three times with sterile PBS buffer to remove unattached cells. Sessile cells were gently detached from beads by slight agitation in PBS buffer, pelleted by centrifugation 8000g for 20 min at 15°C, washed twice and snap-frozen in liquid nitrogen. Sessile cell pellets were stored at −80°C and freeze-dried for protein isolation.
Escherichia coli OverExpress C41(DE3) (Lucigen, Middleton, WI, USA), used for rBipA expression, was grown in Luria Bertani medium containing appropriate antibiotics (ampicillin, kanamycin and/or chloramphenicol at a concentration of 50 µg/mL) at 37°C.
Proteomics analysis
Bacterial pellets from 10 mL of planktonic (mid-exponential or stationary) or biofilm cultures were lysed by sonication and used for the isolation of cytosolic and membrane protein fractions using the ReadyPrep Protein Extraction Kit Membrane I (Bio-Rad Laboratories, Hercules, CA, USA). The obtained soluble and insoluble fractions, respectively containing the cytosolic and membrane (-associated) proteins were purified using the ReadyPrep 2-D clean-up Kit (Bio-Rad Laboratories). Finally, protein pellets were dissolved in 8 M urea in 10 mM Tris-HCl pH 8.0 and subjected to in-solution digestion and C18 reversed phase nano flow liquid chromatography-mass spectrometry/mass spectrometry analysis as described in Supplementary Information. For vaccination experiments, membrane proteins were diluted in PBS as described below.
Recombinant BipA production
The entire DNA sequence of the bipA (BP1112) gene lacking the N-terminal signal sequence (amino acids 1–20) and membrane spanning domain (amino acids 21–66) was codon optimized for expression in E. coli, synthesized by GenScript (GenScript USA Inc. Piscataway, NJ, USA), and cloned into pET28-TEVsite vector (modified from pET28a vector; Novagen, Madison, WI, USA) to generate an N-terminally His6-tagged BipA protein (rBipA). rBipA expression from the pET28-TEVsite vector was induced in E. coli OverExpress C41(DE3) (Lucigen) cultured to OD600=0.6–0.7 at 37°C by addition of 1 mM isopropyl β-d-1-thiogalactopyranoside for 3 h. Induced cultures were harvested by centrifugation (4000g for 15 min at 4°C) and enzymatically lysed by resuspending the pellet in Bacterial Protein Extraction Reagent (Thermo Scientific, Waltham, MA, USA) containing 25 U/mL benzonase nuclease (Novagen), 100 µg/mL lysozyme, and protease inhibitors (Roche, Mannheim, Germany), and incubating for 30 min at room temperature. The cells were disrupted further by sonicating the bacterial solutions (30 s pulse for 5 min at an intensity of 70%). Cell lysates were centrifuged (4000g for 30 min at 4°C) and pellets containing inclusion bodies dissolved in a buffer containing 20 mM sodium phosphate, 500 mM NaCl, 4 M urea and 20 mM imidazole and used for protein purification. His6-tagged rBipA was purified on a AKTA FPLC system using affinity chromatography on a HisTrap FF crude 1 mL column prepacked with Ni Sepharose 6 Fast Flow (GE Healthcare, Uppsala, Sweden). Bound rBipA was eluted in a single step elution with a buffer containing 20 mM sodium phosphate, 500 mM NaCl, 4 M urea and 500 mM imidazole. Fractions containing rBipA were pooled and protein concentrations were determined using the 2-D Quant Kit according the manufactures protocol (GE Healthcare, Uppsala, Sweden). rBipA was refolded by rapid 50-fold dilution in 10 mM Benzamidine, 1 mM EDTA, 100 mM NaCl and 50 mM Tris-HCL (pH 8.8) as described.27
Protein samples obtained after isopropyl β-d-1-thiogalactopyranoside induction, cell lysis, and purification were analyzed on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gradient gels using Colloidal Coomassie staining and immune blotting. Colloidal Coomassie staining was performed as previously described.28 Briefly, sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels were incubated for 3 h in staining solution consisting of 5% aluminum sulfate 14–18 hydrate, 10% ethanol, 0.02% of Coomassie Brilliant Blue G-250 and 8% phosphoric acid. Then, gels were destained by incubation for 30 min in 2% phosphoric acid and 10% ethanol and overnight incubation in water. For immune blotting, proteins were transferred to polyvinylidene difluoride membranes, blocked with PBS with 0.1% Tween-20, 2% skim milk powder and 1% bovine serum albumin (BSA) (1 h at room temperature) and incubated overnight with a 1∶2000 diluted Penta-His Antibody (Qiagen, Hilden, Germany) at 4°C. After washing three times for 5 min in PBS with 0.1% Tween-20, the membrane was incubated for 1 h at room temperature with a goat-anti-mouse light chain Horseradish peroxidase (HRP) antibody and washed again in a similar manner. Finally, immunoblots were developed using an enhanced chemiluminescence substrate system (GE Healthcare, Uppsala, Sweden).
Vaccination
Individual groups of naïve, female 6–8 week old BALB/c mice (Charles River, Leiden, The Netherlands) were immunized subcutaneously on day 0 and 14 with 5 µg rBipA or 1 µg purified membrane proteins derived from planktonic or biofilm bacteria, mixed 1∶1 in 1.3% alum adjuvant (Sigma-Aldrich Chemie B.V. Zwijndrecht, The Netherlands). As a negative control, mice were immunized with a similar ratio of PBS and Alhydrogel, or with PBS alone. In order to compare efficacy to current aPs, one group of mice was immunized with 1/50th of the human dose (equals 1.16 µg of B. pertussis protein) of the commercial hexavalent acellular pertussis vaccine Infanrix (25 µg Ptx, 25 µg filamentous hemagglutinin (FHA), 8 µg pertactin; GSK, Rixensart, Belgium). On day 35, mice were challenged by intranasal infection with B. pertussis strain B1917 (2×107 colony forming units (CFU) in a total volume of 40 µL). On day 38 and 42, the bacterial load in the nasopharynx (upper respiratory tract) and the lungs (lower respiratory tract) was determined as described previously.29 Serial dilutions of the nasal lavages (NLs) and lung homogenates were plated onto Bordet Gengou agar plates and incubated at 37°C for 3–4 days, after which the bacterial load was determined. Serum samples were collected on days 0, 28, 38 and 42.
In vivo transcriptional analysis
Groups of 4 female BALB/c mice (6–8 weeks old) were infected intranasally with B. pertussis strain B1917 as described above. Mice were sacrificed after seven days, after which bacteria were collected from the lungs through a bronchoalveolar lavage with PBS and RNA was stabilized with 2 volumes of RNA Protect Bacteria Reagent (Qiagen). Total RNA was extracted using the RNeasy Mini Kit (Qiagen) and contaminating genomic DNA was removed by DNase treatment (Ambion, Carlsbad, CA, USA). Bacterial RNA was enriched and amplified using the MICROBEnrich (Ambion) and SensationPlus FFPE Amplification (Affymetrix, Santa Clara, CA, USA) Kits, respectively. Enriched RNA was reverse transcribed using the SuperScript One-Cycle cDNA Kit (Invitrogen, Carlsbad, CA, USA) and used for quantitative real-time PCR analysis. To determine relative expression levels, ΔCt values were calculated by subtracting the Ct value of the recA (BP2546) household gene from the Ct value of the target gene.30 The transcription data are expressed as 40−ΔCt value, with 40 representing the number of PCR cycles as detection limit. A 40−ΔCt value of 40 indicates that the gene is expressed at equal levels as recA, while lower values correspond to lower expression.
Antibody analysis
Protein ELISA
IgG titers against rBipA in mouse and human sera were determined by enzyme-linked immunosorbent assay (ELISA) analysis. Microtiter plates (F96, MaxiSorp; Thermo Scientific, Waltham, MA, USA) were coated with 2 µg/mL rBipA in 50 µL PBS per well overnight at 4°C. Control ELISAs were performed with plates coated with 2 µg/mL Ptx in PBS (Kaketsuken, Kumamoto, Japan). Plates were washed with PBS with 0.05% Tween 20 and then incubated for 1 h with PBS containing 1% BSA. Serial dilutions of mouse or human sera were added to the plates and incubated for 1 h at 37°C. After washing, alkaline phosphatase (AP)-conjugated secondary goat antibody directed to mouse IgG-Fc (Sigma-Aldrich) or human IgG (Southern Biotech, Birmingham, AL, USA) was added for 1 h at 37°C using a 1∶2500 and 1∶1000 dilution, respectively. After washing, 50 µL per well of p-nitrophenyl phosphate (1 mg/mL) in substrate buffer (10 mM diethanolamine and 0.5 mM magnesium chloride, pH 9.5) was added and the absorbance was read at 405 nm. IgG concentrations in mouse serum were determined using absorbance data of a standard curve of 1–64 ng mouse IgG1 (Sigma) on wells coated with 2 µg/mL Rat anti-mouse Ig, kappa light chain (BD Pharmingen, San Jose, CA, USA). Human serum titers were calculated by multiplying the highest dilution with an optical density >0.2 with the corresponding dilution factor, as a value >0.2 falls within the linear range of the ELISA curve.
Whole-cell ELISA
Binding of IgG1, IgG2a and IgG2b to whole bacteria was measured using a whole-cell ELISA method adapted from Abdillahi and Poolman.31 Briefly, ELISA plates were coated with Bvg+ mid-log culture of strain B1917 suspended in PBS, washed (PBS containing 0.05% Tween 20), blocked with 1% BSA/PBS, and incubated with serial dilutions of mouse serum. After washing, ELISA plates were incubated with HRP-conjugated rat-anti-mouse IgG1 (1∶50 000 dilution; BD Pharmingen, San Jose, CA, USA), HRP-conjugated rat-anti-mouse IgG2a (BD Pharmingen; 1∶10 000 dilution), or AP conjugated rat-anti-mouse IgG2b (1∶2000 dilution; Southern Biotech, Birmingham, AL, USA) for 1 h at 37°C. 3,3′,5,5′-tetramethylbenzidine substrate was added to each well for the HRP-conjugated antibodies, while p-nitrophenyl phosphate substrate was used for AP-conjugated rat-anti-mouse IgG2b. Optical density was then measured on an ELISA plate reader (Tecan Infinite F50) at 450 and 405 nm for 3,3′,5,5′-tetramethylbenzidine and p-nitrophenyl phosphate substrate, respectively. Antibody subtype concentrations were determined by comparison to standard curves with known concentrations of each IgG subtype.
Opsonization
1% BSA/PBS was used for all dilutions. 106 CFU of Bvg+ and Bvg− B1917 were incubated with serum or NL samples from vaccinated mice for 30 min at 4°C. Bacteria were fixed in 2% paraformaldehyde and surface-bound IgG or IgA was detected using anti-mouse IgG-Fc or IgA α-chain-FITC-conjugated antibodies (Sigma-Aldrich) on a BD LSRII flow cytometer (Biosciences, San Jose, CA, USA). The amount of surface-bound antibodies was expressed in arbitrary units as a fluorescence index (FI), calculated by multiplying the geometric mean fluorescence intensity by the percentage of FITC-positive bacteria.32 Data were analyzed using FlowJo version 7.6.5 (TreeStar Inc, Ashland, OR, USA).
Statistical analyses
A two-tailed Mann–Whitney U test was used for comparison of bacterial load in NL and lung homogenate samples between PBS-vaccinated mice and rBipA or membrane protein-vaccinated mice. A Kruskal–Wallis test followed by a Dunns post-hoc test (α=5%) was used for comparison of antibody-mediated opsonization by serum and NL sample between PBS-vaccinated mice and rBipA-vaccinated mice. A one-tailed Wilcoxon signed-rank test was used to determine whether ELISA measured IgG levels were significantly above the detection limit. All statistical analyses were performed using the GraphPad Prism software program, version 5.0, where P<0.05 was considered significant.
RESULTS
Membrane-associated proteins derived from biofilms or planktonic grown cells confer protection against respiratory tract infection in mice
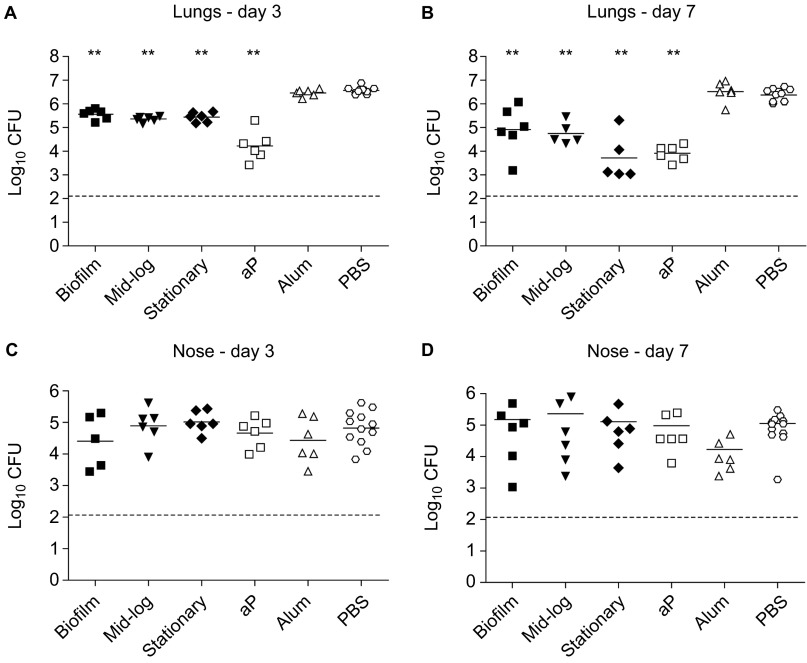
To evaluate the potential of biofilm-grown bacteria to confer protection against respiratory tract infection in mice, we cultured a B. pertussis clinical isolate (B1917, isolated in 2000) under biofilm-forming and planktonic conditions. This particular strain belongs to the P3 lineage which emerged in the 1980s and currently predominates in many vaccinated populations.33 For planktonic conditions, bacteria were grown to mid-log and stationary phase. Following growth, bacteria were harvested and cytosolic and membrane (-associated) proteins were extracted.34 The membrane fractions were adjuvanted with Alhydrogel and the equivalent of 1 µg of protein was used to vaccinate female BALB/c mice 2× by subcutaneous injection, with 2-week intervals. In parallel, groups of mice were vaccinated with a currently licensed three-component aP (Infanrix) or mock-vaccinated with PBS or adjuvant only. Immunized mice were then challenged by intranasal infection with strain B1917, after which the lung and nose bacterial load was determined three and seven days later. Mice vaccinated with aP showed a significant reduction (>220-fold) in lung CFUs at day 3 and day 7 (Figures 1A and 1B). At three days following infection, significant protection was observed in the lungs of mice immunized with proteins derived from planktonic as well as biofilm-grown bacteria (Figure 1A). After seven days, protection was more pronounced, with 29-fold, 43-fold and 170-fold reduction in bacterial load in mice immunized with biofilm, mid-log and stationary proteins, respectively (Figure 1B). The differences in protection induced by the three membrane fractions were not significant. Of note, we observed that none of the vaccinated mice, including those vaccinated with aP, were protected against infection with B. pertussis in the upper respiratory tract (Figures 1C and 1D). Taken together, these data suggest that immunization with biofilms can induce protection against infection.
Figure 1.
Immunization with membrane protein fractions derived from biofilm and mid-log and stationary planktonic cultures. Naive adult female BALB/c mice were subcutaneously immunized with 1 µg membrane proteins. Negative controls consisted of adjuvant only (Alum) and PBS, while an acellular vaccine (aP) containing 0.5 µg Ptx, 0.5 µg FHA and 0.16 µg Prn was used as a positive control. Subsequently, mice were infected intranasally with 2×107 CFU of B. pertussis strain B1917. The bacterial load in the lungs and nose was quantified three (A and C) and seven (B and D) days after challenge. Each symbol represents one mouse. Horizontal lines represent the mean. Dashed lines indicate the lower limit of detection. **P<0.005 relative to PBS mice; two-tailed Mann–Whitney U test. FHA, filamentous hemagglutinin.
Proteomic analysis of B. pertussis grown under biofilm or planktonic conditions
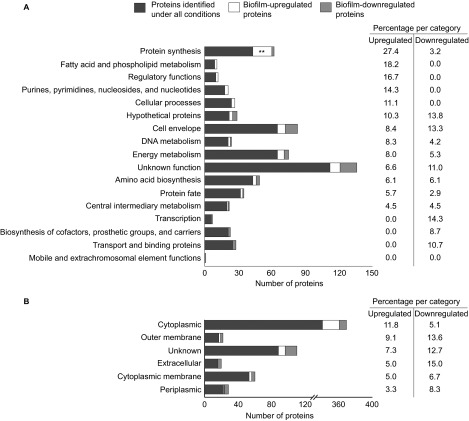
As our initial results implied that biofilms contain protective antigens, we compared the proteome of cytosolic and membrane protein fractions of the mid-log, stationary and biofilm cultures by mass spectrometry, as described in Supplementary Information. This approach identified a total of 749, 729 and 825 proteins in the extracts from mid-log, stationary and biofilm cultures, respectively, with an overlap of 645 proteins (88%). This analysis covered 21%–24% of the predicted total of 3449 protein-coding open reading frames in the B. pertussis genome.35 IDEAL-Q36 was then used to identify proteins which were differentially regulated in biofilms (Supplementary Information). This method normalizes and quantifies comparable peptides between parallel runs of different samples.36 Proteins that were ≥three-fold more or less abundant under biofilm (either cytoplasmic or membrane fraction) conditions compared to either mid-log or stationary conditions, were defined as biofilm-upregulated and biofilm-downregulated proteins, respectively. This strategy yielded 60 biofilm-upregulated proteins. Aggregation based on function and predicted subcellular localization showed that these proteins were significantly enriched for (ribosomal) proteins involved in protein synthesis (N=17, 27.4%, P=7.7×10−4; Figure 2 and Supplementary Table S1). Additionally, 48 proteins were identified which were downregulated in biofilms. While this set of proteins was not significantly enriched for any specific functional classes, 17 of these proteins have previously been identified as Bvg-activated genes in strain B1917,10 including the virulence factors Vag8, FimX, and eight type III secretion system (T3SS) proteins (Supplementary Table S1).
Figure 2.
Functional clustering of proteins identified with proteomics. 645 proteins were identified under all three growth conditions (biofilm, planktonic mid-log and planktonic stationary), of which 60 and 48 proteins were found to be upregulated or downregulated, respectively, in biofilms relative to planktonic conditions. The identified proteins were grouped by functional categories (A) and PSORTb-predicated subcellular localization (B). The relative frequencies of biofilm-up- or -downregulated proteins, compared to the total number of annotated proteins identified under all conditions for each functional class, are listed on the right-hand side. Asterisks indicate statistically significant enrichment of biofilm-regulated proteins in a certain class as determined by Fisher's exact test. **P<0.005.
Vaccine candidate antigen selection
We focused on the 11 proteins that were upregulated in biofilms compared to both mid-log and stationary planktonic conditions, as these potentially represent protein antigens specifically expressed in biofilms. These proteins were ranked based on protein abundance,37,38 as well as predicted surface accessibility (i.e., outer membrane or extracellular)39 (Table 1). Protein abundance was estimated for each fraction and growth condition by emPAI score,37,38 a method which provides an approximate protein ranking based on the number of observed peptides per protein relative to the number of observable peptides (Supplementary Table S2). Based on these criteria, BipA was identified as the most abundant surface-associated biofilm protein.
Table 1. Biofilm-associated proteins and vaccine candidate selection criteria.
| General information | Fold changea | Protein abundance (emPAI values)b | Predicted localizationc | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cytosol | Membrane | Cytosol | Membrane | ||||||||||
| ORF | Gene | Protein name | BF/ML | BF/ST | BF/ML | BF/ST | BF | ML | ST | BF | ML | ST | |
| BP1112 | bipA | Bordetella intermediate protein A | 5.41 | 4.63 | 4.71 | 7.43 | 1.19 | 0.21 | 0.36 | 3.36 | 0.61 | 0.38 | Om |
| BP0878 | Phospholipase | — | — | 3.76 | 3.74 | — | — | — | 0.32 | — | — | Om | |
| BP0038 | gloA | Lactoylglutathione lyase | 12.48 | 9.00 | 9.00* | 3.13* | — | — | — | — | — | — | C |
| BP0782 | Exported protein | — | — | 3.90 | 13.56 | — | — | — | — | — | — | Un | |
| BP0901 | Oxidoreductase | 5.73 | 7.33 | — | — | 0.07 | — | — | — | — | — | C | |
| BP1315 | Universal stress family protein | 3.81 | 7.89 | 3.03* | 2.27* | 1.67 | 1.25 | 0.81 | 1.71 | 1.11 | 1.64 | C | |
| BP1519 | fba | Fructose-bisphosphate aldolase | 3.83 | 11.15 | −1.33 | −1.32 | 4.96 | 2.16 | 1.12 | 1.61 | 1.47 | 1.95 | C |
| BP2039 | Amidase | 3.62 | 9.89 | — | — | — | — | — | — | — | — | C | |
| BP2782 | Lipoprotein | — | — | 10.64 | 15.84 | — | — | — | 1.18 | 0.80 | 0.93 | Un | |
| BP2873 | tctC | Tripartite tricarboxylate transporter family receptor | 2.52 | 3.80 | 3.12 | −1.79 | 0.37 | 0.33 | 0.27 | 0.32 | 0.11 | — | P |
| BP2896 | Hydrolase | — | — | 4.19 | 6.81 | — | — | — | — | — | — | C | |
Abbreviations: BF, biofilm; C, cytoplasmic; ML, mid-log; Om, outer membrane; ORF, open reading frame; P, periplasmic; ST, stationary; Un, unknown.
Not significant (P>0.05).
Fold change specifies the protein abundance ratio of the indicated conditions as determined using IDEAL-Q (Supplementary Table S1). ‘—': protein not quantifiable.
emPAI values37 were calculated based on the protein false discovery rate validation method of Weatherly38 (Supplementary Table S2). ‘—': protein was not detectable.
Localization prediction based on PSORTb v3.0.39
BipA expression during infection
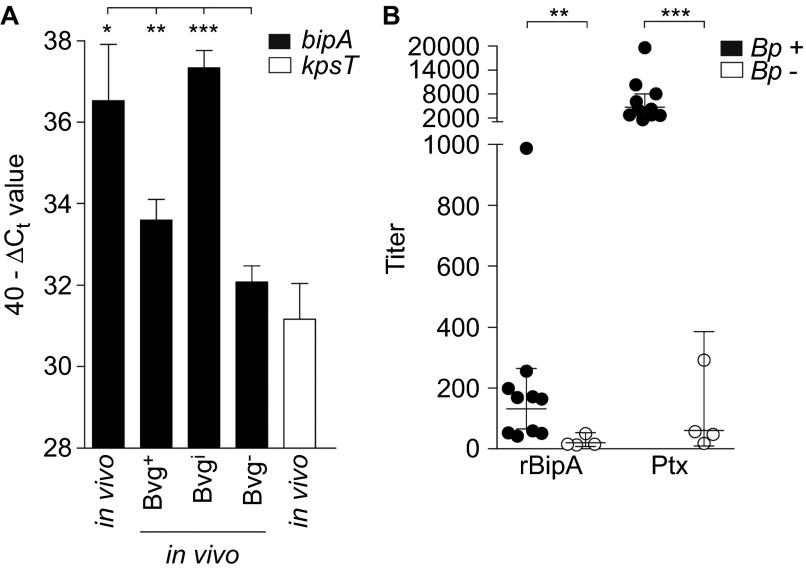
To determine whether BipA is expressed during natural infection, naive adult BALB/c mice were infected intranasally with B. pertussis strain B1917. Seven days after infection, bacteria were isolated from the lungs and gene expression of bipA was analyzed by quantitative polymerase chain reaction and compared to Bvg+, Bvgi and Bvg− in vitro conditions. As a negative control, the kpsT gene encoding a Bvg− protein involved in capsule biosynthesis was included.40 Transcriptional analysis showed that bipA is expressed in the lungs of mice, at a level also observed under virulent in vitro Bvg+ and Bvgi conditions (Figure 3A). Thus, these data indicate that BipA is expressed during respiratory infection in mice.
Figure 3.
bipA expression during infection. (A) Naive adult female BALB/c mice were infected intranasally with B. pertussis strain B1917. After seven days, bacteria were collected through BAL and used for in vivo transcriptional analysis using bipA-specific primers as described in the text. Expression levels were compared to in vitro Bvg+, Bvgi and Bvg− conditions. The transcription data are expressed as 40−ΔCt value, which is a measure of expression relative to the recA gene (ΔCt=Ct target–Ct recA). *P<0.05, **P<0.005, ***P<0.0005 relative to the Bvg− condition; Student's t-test with Welch's correction. (B) Human serum, from 10 confirmed B. pertussis-infected individuals (Bp+) and four non-infected individuals (Bp−) was diluted and incubated on ELISA plates coated with recombinant BipA or Ptx and the titers were determined. Horizontal lines represent the geometrical mean. **P<0.005, ***P<0.0005 relative to Bp- sera; one-tailed Mann–Whitney U test. BAL, bronchoalveolar lavage.
To investigate whether BipA was also expressed during infection of humans, a panel of convalescent sera from individuals who were recently infected with B. pertussis was used to measure rBipA-specific antibody responses. We used serum from individuals with anti-Ptx titers above 400 ELISA Units per milliliter, as this level is strongly indicative of recent infection.41 Sera from humans with undetectable titers of anti-Ptx antibodies were chosen as negative control. Analysis of these serum samples indicated that infection of humans also results in antibodies to BipA (Figure 3B), suggesting that BipA is expressed and immunogenic during infection.
BipA is a protective B. pertussis antigen
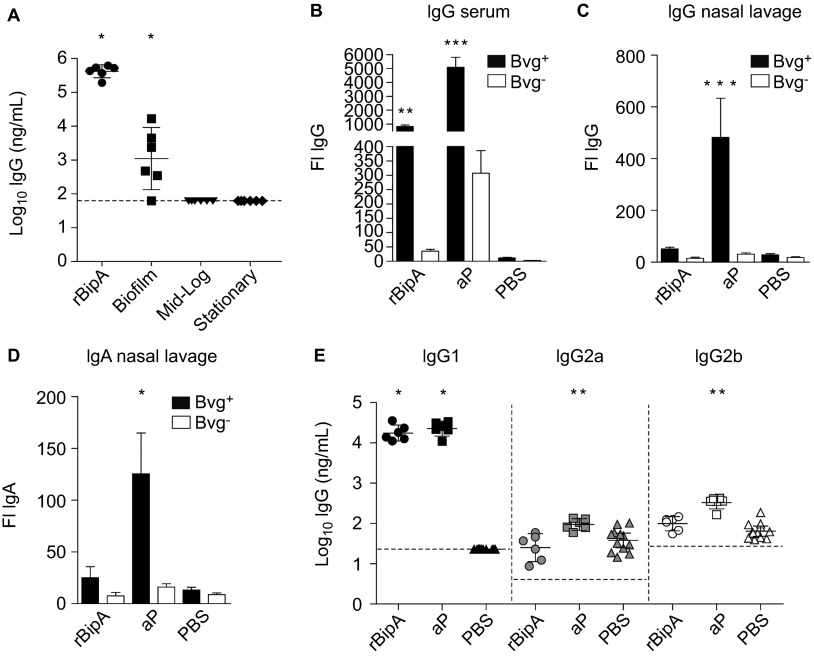
To determine the protective effects of BipA, we constructed a recombinant His-tagged fusion protein of BipA (rBipA). After purification and refolding, the purity of the produced protein was confirmed by Coomassie staining and immunoblotting (Supplementary Figure S1). rBipA was then adjuvanted with Alhydrogel and used as stand-alone antigen (5 µg) to vaccinate female adult BALB/c mice. Mice were immunized 2× in 2-week intervals, after which they were challenged by intranasal infection with B1917. Vaccination with rBipA resulted in a small but significant reduction of bacterial load in the lungs as compared to the PBS-vaccinated mice (Figures 4A and 4B, 5.4 and 3.4-fold at days 3 and 7, respectively) which was not observed in the nasopharynx (Figures 4C and 4D). Thus, these data show that rBipA is able to reduce bacterial load.
Figure 4.
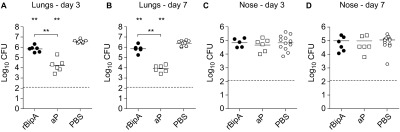
Immunization with rBipA protects against B. pertussis infection. Naive adult female BALB/c mice were subcutaneously immunized with rBipA or controls as described in Figure 1 and infected intranasally with 2×107 CFU of B. pertussis strain B1917. The bacterial load in the lungs and nose was quantified three (A and C) and seven (B and D) days after challenge. Each symbol represents one mouse. Horizontal lines represent the mean. Dashed lines indicate the lower limit of detection. **P<0.005 difference between rBipA and aP and between vaccinated and PBS control mice; two-tailed Mann–Whitney U test.
BipA-specific antibody responses
To determine the role of antibody-mediated protection by rBipA, an ELISA was performed using sera from vaccinated mice. Figure 5A shows that vaccination with rBipA induced high levels of rBipA-specific IgG. Similarly, rBipA-specific antibodies were also detected in serum from biofilm-vaccinated mice, while these were below the detection limit in serum derived from mice vaccinated with planktonic culture-derived membrane proteins (Figure 5A). Next, the ability of serum and NL-derived rBipA-specific antibodies to bind its native epitopes on the surface of Bvg+ and Bvg− B. pertussis was assessed by flow cytometry. We found significant IgG-mediated opsonization of Bvg+ B. pertussis using serum, but not NL, from rBipA-vaccinated mice (Figure 5B). For aP-vaccinated mice, strong opsonization of Bvg+ B. pertussis was observed with serum IgG and NL IgG and IgA (Figures 5B–5D). Both aP- and rBipA-induced antibodies showed significantly reduced binding to Bvg− bacteria, which confirms the low expression of these antigens by Bvg−-phase bacteria. These data show that rBipA vaccination-induced antibodies recognize native BipA epitopes. Since IgG binding alone does not necessarily reflect the quality of the subsequent antibody response,42 the subtype distribution of the opsonizing IgG molecules was also determined using a whole-cell ELISA.31 Analysis of the IgG1, IgG2a and IgG2b response showed that rBipA and aP antigen-specific antibodies were predominantly of the phagocytosis-mediating IgG1 subtype43,44 (Figure 5E).
Figure 5.
Bacterial opsonization by BipA-specific antibodies. (A) Post-immune mouse sera (day 28) were used to determine the amount of total IgG specific for rBipA using an ELISA. Each symbol represents one mouse and the geometrical mean is represented by a horizontal line. Dashed lines indicate the lower limit of detection (93 ng/mL). *P<0.05; one-tailed Wilcoxon signed-rank test. The binding of serum IgG (B) and nasal lavage IgG (C) and IgA (D) antibodies from vaccinated mice to Bvg+ or Bvg− grown B. pertussis B1917 was determined by flow cytometry. Bars represent the geometric mean±95% CI of six individual mice. **P<0.005, ***P<0.0005 relative to PBS group; Kruskal–Wallis test followed by a Dunns post-hoc test (α=5%). (E) To determine the distribution of IgG subtypes, the amount of pertussis-specific IgG1, IgG2a and IgG2b binding to Bvg+ grown whole B1917 cells was assessed. Each symbol represents one mouse and the geometrical mean is represented by a line. Dashed lines indicate the lower limit of detection (23, 4 and 27 ng/mL for IgG1, IgG2a and IgG2b, respectively). For IgG1, a one-tailed Wilcoxon signed-rank test was performed because IgG1 levels of the PBS mice were below the detection limit. IgG2a and IgG2b levels were statistically compared to the PBS mice using a two-tailed Mann–Whitney U test. *P<0.05, **P<0.005. CI, confidence interval; FI, fluorescence index.
DISCUSSION
The recent resurgence of B. pertussis infections is of significant concern. The observation that Prn negative strains are now increasingly being isolated from patients with whooping cough12,13,14 highlights the importance of identifying novel pertussis antigens. All current B. pertussis antigens used in aP vaccines have been derived from planktonic grown bacteria. In this study, we focused on a novel class of proteins which are upregulated in biofilms. Vaccination with planktonic and biofilm-derived proteins may target different steps in infection and broaden immunity. While biofilm formation has been suggested to be important for successful B. pertussis infection,15,16 the ability of biofilm-antigens to protect against infection has not been evaluated.
We showed that mice immunized with membrane proteins derived from biofilm bacteria are protected against respiratory B. pertussis infection and that the degree of protection was not significantly different from mice immunized with planktonic culture-derived membrane proteins (Figure 1). This observation suggests that pertussis biofilms are a potential target for vaccination, similar to other respiratory pathogens such as Pseudomonas aeruginosa.45 To characterize the antigenic composition of a biofilm, we compared the biofilm and planktonic proteomes of a representative strain of the P3 lineage (B1917) which has recently spread worldwide.11
We found 48 proteins which were downregulated in biofilms. While these proteins did not include any of the antigens present in current aPs, a relatively high proportion of these proteins represent Bvg+ phase, virulence-associated proteins.10 This suggests that heterogeneous populations are involved in infections,46 even with respect to virulence-associated proteins. Therefore, inclusion of highly expressed biofilm proteins in new vaccine formulations may enhance the efficacy of pertussis vaccines by broadening the immune response to different phases of infection. Sixty proteins were identified as more abundantly present in biofilms, the majority of which represented ribosomal proteins involved in protein synthesis. However, the majority of these ribosomal proteins were biofilm-upregulated only when compared to stationary growth, and were expressed at similar levels under mid-log conditions (Supplementary Table S1). This suggests that biofilm formation requires high levels of de novo synthesis of proteins, similar to mid-log bacteria.20,47 Of these 60 proteins, only 11 were upregulated when compared to both mid-log and stationary planktonic growth conditions. Moreover, only two of these were predicted to be surface-accessible, i.e. a phospholipase (BP0878) and BipA. Although BP0878 may also have vaccine potential,48 we only assessed the vaccine potential of BipA because it was more abundantly expressed under biofilm conditions. Further, BipA was also found upregulated in Tohama I biofilms relative to planktonic cultures suggesting that expression of BipA is a common signature of Bordetella biofilms.18 BipA shows a large degree of amino-acid sequence identity with the biofilm-associated proteins of Staphylococcus aureus49 and Acinetobacter baumannii,50 which mediate cell-to-cell interactions within biofilms 51 (36% and 33% identity, respectively). Importantly, vaccination with biofilm-associated protein derived from A. baumannii has been shown to protect against infection.52 BipA may play a similar role in B. pertussis biofilm formation during infection. Some clues towards a role for BipA during infection were established in this work. For instance, we showed that during natural infection, BipA was expressed at high levels in the lungs of mice and that infection resulted in BipA-specific serum antibodies in humans (Figure 3). Challenge experiments showed that vaccination with rBipA induced significant protection to lower respiratory tract infection (Figure 4), which was associated with the presence of high amounts of opsonizing, protection-associated IgG1 antibodies43,44 (Figure 5). Significant amounts of rBipA antibodies were also detected in serum from mice vaccinated with biofilm membrane proteins, but not in mice vaccinated with proteins derived from planktonic bacteria, suggesting that BipA is highly expressed in B. pertussis biofilms and immunogenic. An important question is how efficacious BipA will be in natural B. pertussis populations which consist of different strains. The strain used to assess the vaccine efficacy of rBipA, is a representative of the P3 lineage, the members of which have spread worldwide in the last 10–15 years replacing the resident P1 strains.11,53 In many countries, P3 strains are found in frequencies of 60%–100%. Further, immune escape from BipA antibodies seems unlikely, as in a worldwide collection of 343 B. pertussis strains, only one protein variant was found (designated BipA2),53 which differed in only one amino acid from the BipA variant used in this study. Further research is needed to establish the exact mechanisms by which antibodies to biofilms and BipA in particular interfere with infection, and whether protection by BipA can be enhanced through the use of alternative vaccine adjuvants or vaccination routes.
Membrane proteins extracted from biofilm or planktonic grown cells conferred protection in the mouse lung, while only the biofilm extract induced rBipA antibodies, albeit at lower levels compared to vaccination with rBipA. Clearly, these planktonic fractions harbor other protective antigens. Proteomic analyses of these fractions revealed high expression of many virulence associated factors, including those who have been shown to confer protection in humans (i.e., Ptx, Fim3, Prn and FHA), integral outer membrane proteins and autotransporters (Supplementary Tables S1 and S2). Further fractionation of these samples may elucidate the distinct role of these antigens and reveal additional vaccine candidates.
Taken together, this work provides evidence that vaccination against B. pertussis biofilms induces protection to infection. While further research is still needed to optimize immunity, a strategy by which multiple steps in infection are targeted, including biofilm formation, represents an attractive approach to improve pertussis vaccines. The inclusion of BipA in novel vaccines may thus provide an additional level of protection to current aPs and thereby enhance the efficacy of pertussis vaccination.
Acknowledgments
We thank Maurice van Dael en Jolein Gloerich (Radboud Proteomics Centre, Radboud University Medical Centre, Nijmegen, The Netherlands) for technical assistance regarding proteomics experiments and Fred van Opzeeland, Elles Simonetti and Saskia van Selm (Laboratory of Pediatric Infectious Diseases, Department of Pediatrics, Radboud university medical center, Nijmegen, The Netherlands; Laboratory of Medical Immunology, Department of Laboratory Medicine, Radboud University Medical Center, Nijmegen, The Netherlands) for assistance with the animal experiments. We thank Guy Berbers (Netherlands Centre for Infectious Disease Control, National Institute for Public Health and the Environment) for human sera. Daan de Gouw is supported by Grant 125020001 from the Netherlands Organization of Scientific Research (ZonMw).
Footnotes
Note: Supplementary Information for this article can be found on Emerging Microbes & Infections' website (http://www.nature.com/emi/).
Supplementary Information
Proteomics data of statistically significant biofilm up- and downregulated proteins identified in the cytosolic and membrane fraction of B. pertussis strain B1917
Protein abundance of all proteins identified in the cytosolic and membrane fraction of B. pertussis strain B1917 grown under planktonic and biofilm conditions
References
- Stocks P, Kam MN. On the epidemiology of pertussis cough in London. J Hyg. 1932;32:582–619. doi: 10.1017/s0022172400018301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, McIntyre P, Quinn H, Hueston L, Gilbert GL, McVernon J. Increased population prevalence of low pertussis toxin antibody levels in young children preceding a record pertussis epidemic in Australia. PloS One. 2012;7:e35874. doi: 10.1371/journal.pone.0035874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Greeff SC, de Melker HE, van Gageldonk PG, et al. Seroprevalence of pertussis in The Netherlands: evidence for increased circulation of Bordetella pertussis. PloS One. 2010;5:e14183. doi: 10.1371/journal.pone.0014183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melker HE, Versteegh FG, Schellekens JF, Teunis PF, Kretzschmar M. The incidence of Bordetella pertussis infections estimated in the population from a combination of serological surveys. J Infect. 2006;53:106–113. doi: 10.1016/j.jinf.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Ward JI, Cherry JD, Chang SJ, et al. Bordetella pertussis infections in vaccinated and unvaccinated adolescents and adults, as assessed in a national prospective randomized Acellular Pertussis Vaccine Trial (APERT) Clin Infect Dis. 2006;43:151–157. doi: 10.1086/504803. [DOI] [PubMed] [Google Scholar]
- Black RE, Cousens S, Johnson HL, et al. Global, regional, and national causes of child mortality in 2008: a systematic analysis. Lancet. 2010;375:1969–1987. doi: 10.1016/S0140-6736(10)60549-1. [DOI] [PubMed] [Google Scholar]
- Klein NP, Bartlett J, Rowhani-Rahbar A, Fireman B, Baxter R. Waning protection after fifth dose of acellular pertussis vaccine in children. N Engl J Med. 2012;367:1012–1019. doi: 10.1056/NEJMoa1200850. [DOI] [PubMed] [Google Scholar]
- Witt MA, Katz PH, Witt DJ. Unexpectedly limited durability of immunity following acellular pertussis vaccination in preadolescents in a North American outbreak. Clin Infect Dis. 2012;54:1730–1735. doi: 10.1093/cid/cis287. [DOI] [PubMed] [Google Scholar]
- Warfel JM, Zimmerman LI, Merkel TJ. Acellular pertussis vaccines protect against disease but fail to prevent infection and transmission in a nonhuman primate model. Proc Natl Acad Sci USA. 2014;111:787–792. doi: 10.1073/pnas.1314688110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gouw D, Hermans PW, Bootsma HJ, et al. Differentially expressed genes in Bordetella pertussis strains belonging to a lineage which recently spread globally. PloS One. 2014;9:e84523. doi: 10.1371/journal.pone.0084523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi FR, van Loo IH, van Gent M, et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15:1206–1213. doi: 10.3201/eid1508.081511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, He Q. Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol. 2012;19:1703–1704. doi: 10.1128/CVI.00367-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka N, Han HJ, Toyoizumi-Ajisaka H, et al. Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PloS One. 2012;7:e31985. doi: 10.1371/journal.pone.0031985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawloski LC, Queenan AM, Cassiday PK, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the US. Clin Vaccine Immunol. 2014;21:119–125. doi: 10.1128/CVI.00717-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover MS, Mishra M, Deora R. Extracellular DNA is essential for maintaining Bordetella biofilm integrity on abiotic surfaces and in the upper respiratory tract of mice. PloS One. 2011;6:e16861. doi: 10.1371/journal.pone.0016861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan GP, Love CF, Sukumar N, Mishra M, Deora R. The Bordetella Bps polysaccharide is critical for biofilm development in the mouse respiratory tract. J Bacteriol. 2007;189:8270–8276. doi: 10.1128/JB.00785-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover MS, Sloan GP, Love CF, Sukumar N, Deora R. The Bps polysaccharide of Bordetella pertussis promotes colonization and biofilm formation in the nose by functioning as an adhesin. Mol Microbiol. 2010;77:1439–1455. doi: 10.1111/j.1365-2958.2010.07297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra DO, Conover MS, Arnal L, et al. FHA-mediated cell-substrate and cell-cell adhesions are critical for Bordetella pertussis biofilm formation on abiotic surfaces and in the mouse nose and the trachea. PloS One. 2011;6:e28811. doi: 10.1371/journal.pone.0028811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Serra DO, Lucking G, Weiland F, et al. Proteome approaches combined with Fourier transform infrared spectroscopy revealed a distinctive biofilm physiology in Bordetella pertussis. Proteomics. 2008;8:4995–5010. doi: 10.1002/pmic.200800218. [DOI] [PubMed] [Google Scholar]
- Decker KB, James TD, Stibitz S, Hinton DM. The Bordetella pertussis model of exquisite gene control by the global transcription factor BvgA. Microbiology. 2012;158:1665–1676. doi: 10.1099/mic.0.058941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie Y, Mattoo S, Yuk MH. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J Bacteriol. 2004;186:5692–5698. doi: 10.1128/JB.186.17.5692-5698.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbauer KE, Fuchslocher B, Miller JF, Cotter PA. Identification and characterization of BipA, a Bordetella Bvg-intermediate phase protein. Mol Microbiol. 2001;39:65–78. doi: 10.1046/j.1365-2958.2001.02191.x. [DOI] [PubMed] [Google Scholar]
- Vergara-Irigaray N, Chavarri-Martinez A, Rodriguez-Cuesta J, Miller JF, Cotter PA, Martinez de Tejada G. Evaluation of the role of the Bvg intermediate phase in Bordetella pertussis during experimental respiratory infection. Infect Immun. 2005;73:748–760. doi: 10.1128/IAI.73.2.748-760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez de Tejada G, Cotter PA, Heininger U, et al. Neither the Bvg-phase nor the vrg6 locus of Bordetella pertussis is required for respiratory infection in mice. Infect Immun. 1998;66:2762–2768. doi: 10.1128/iai.66.6.2762-2768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalen M, van den IJssel J, Jiskoot W, et al. Rational medium design for Bordetella pertussis: basic metabolism. J Biotechnol. 1999;75:147–159. doi: 10.1016/s0168-1656(99)00155-8. [DOI] [PubMed] [Google Scholar]
- Hijnen M, van Gageldonk PG, Berbers GA, van Woerkom T, Mooi FR. The Bordetella pertussis virulence factor P.69 pertactin retains its immunological properties after overproduction in Escherichia coli. Protein Expr Purif. 2005;41:106–112. doi: 10.1016/j.pep.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Pink M, Verma N, Rettenmeier AW, Schmitz-Spanke S. CBB staining protocol with higher sensitivity and mass spectrometric compatibility. Electrophoresis. 2010;31:593–598. doi: 10.1002/elps.200900481. [DOI] [PubMed] [Google Scholar]
- Cron LE, Stol K, Burghout P, et al. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect Immun. 2011;79:3697–3710. doi: 10.1128/IAI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta CT) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Abdillahi H, Poolman JT. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J Med Microbiol. 1988;26:177–180. [PubMed] [Google Scholar]
- Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun. 2010;78:704–715. doi: 10.1128/IAI.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi FR, Van Der Maas NA, De Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation—two sides of the same coin. Epidemiol Infect. 2014;142:685–694. doi: 10.1017/S0950268813000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Parkhill J, Sebaihia M, Preston A, et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica. Nat Genet. 2003;35:32–40. doi: 10.1038/ng1227. [DOI] [PubMed] [Google Scholar]
- Tsou CC, Tsai CF, Tsui YH, et al. IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol Cell Proteomics. 2010;9:131–144. doi: 10.1074/mcp.M900177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Oda Y, Tabata T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- Weatherly DB, Atwood JA, 3rd, Minning TA, Cavola C, Tarleton RL, Orlando R. A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteomics. 2005;4:762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- Yu NY, Wagner JR, Laird MR, et al. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics. 2010;26:1608–1615. doi: 10.1093/bioinformatics/btq249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neo Y, Li R, Howe J, et al. Evidence for an intact polysaccharide capsule in Bordetella pertussis. Microbes Infect. 2010;12:238–245. doi: 10.1016/j.micinf.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Berbers GA, van de Wetering MS, van Gageldonk PG, Schellekens JF, Versteegh FG, Teunis PF. A novel method for evaluating natural and vaccine induced serological responses to Bordetella pertussis antigens. Vaccine. 2013;31:3732–3738. doi: 10.1016/j.vaccine.2013.05.073. [DOI] [PubMed] [Google Scholar]
- Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez ME, Hellwig SM, Hozbor DF, Leusen J, van der Pol WL, van de Winkel JG. Fc receptor-mediated immunity against Bordetella pertussis. J Immunol. 2001;167:6545–6551. doi: 10.4049/jimmunol.167.11.6545. [DOI] [PubMed] [Google Scholar]
- Hazenbos WL, Heijnen IA, Meyer D, et al. Murine IgG1 complexes trigger immune effector functions predominantly via Fc gamma RIII (CD16) J Immunol. 1998;161:3026–3032. [PubMed] [Google Scholar]
- Digiandomenico A, Warrener P, Hamilton M, et al. Identification of broadly protective human antibodies to Pseudomonas aeruginosa exopolysaccharide Psl by phenotypic screening. J Exp Med. 2012;209:1273–1287. doi: 10.1084/jem.20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- Oosthuizen MC, Steyn B, Theron J, et al. Proteomic analysis reveals differential protein expression by Bacillus cereus during biofilm formation. Appl Environ Microbiol. 2002;68:2770–2780. doi: 10.1128/AEM.68.6.2770-2780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Baird G, Connor KM, Rudge K, Sales J, Donachie W. Vaccination confers significant protection of sheep against infection with a virulent United Kingdom strain of Corynebacterium pseudotuberculosis. Vaccine. 2006;24:5986–5996. doi: 10.1016/j.vaccine.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Cucarella C, Solano C, Valle J, Amorena B, Lasa I, Penades JR. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehfelm TW, Luke NR, Campagnari AA. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol. 2008;190:1036–1044. doi: 10.1128/JB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I, Penades JR. Bap: a family of surface proteins involved in biofilm formation. Res Microbiol. 2006;157:99–107. doi: 10.1016/j.resmic.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Fattahian Y, Rasooli I, Mousavi Gargari SL, Rahbar MR, Darvish Alipour Astaneh S, Amani J. Protection against Acinetobacter baumannii infection via its functional deprivation of biofilm associated protein (Bap) Microb Pathog. 2011;51:402–406. doi: 10.1016/j.micpath.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Bart MJ, Harris SR, Advani A, et al. Global population structure and evolution of Bordetella pertussis and their relationship with vaccination. mBio. 2014;5:e01074. doi: 10.1128/mBio.01074-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomics data of statistically significant biofilm up- and downregulated proteins identified in the cytosolic and membrane fraction of B. pertussis strain B1917
Protein abundance of all proteins identified in the cytosolic and membrane fraction of B. pertussis strain B1917 grown under planktonic and biofilm conditions