Abstract
Pyrazinamide (PZA) is a frontline anti-tuberculosis drug that plays a crucial role in the treatment of both drug-susceptible and multidrug-resistant tuberculosis (MDR-TB). PZA is a prodrug that is converted to its active form, pyrazinoic acid (POA), by a nicotinamidase/pyrazinamidase encoded by the pncA gene, the mutation of which is the major cause of PZA resistance. Although RpsA (ribosomal protein S1, involved in trans-translation) has recently been shown to be a target of POA/PZA, whole-genome sequencing has identified mutations in the panD gene encoding aspartate decarboxylase in PZA-resistant strains lacking pncA and rpsA mutations. To gain more insight into a possible new target of PZA, we isolated 30 POA-resistant mutants lacking mutations in pncA and rpsA from M. tuberculosis in vitro, and whole-genome sequencing of 3 mutants identified various mutations in the panD gene. Additionally, sequencing analysis revealed that the remaining 27 POA-resistant mutants all harbored panD mutations affecting the C-terminus of the PanD protein, with PanD M117I being the most frequent mutation (24/30, 80%). Conditional overexpression of panD from M. tuberculosis, M. smegmatis or E. coli, or of M. tuberculosis mutant PanD M117I, all conferred resistance to POA and PZA in M. tuberculosis. β-alanine and pantothenate, which are downstream products of PanD, were found to antagonize the antituberculosis activity of POA. In addition, the activity of the M. tuberculosis PanD enzyme was inhibited by POA at therapeutically relevant concentrations in a concentration-dependent manner but was not inhibited by the prodrug PZA or the control compound nicotinamide. These findings suggest that PanD represents a new target of PZA/POA. These results have implications for a better understanding of this peculiar persister drug and for the design of new drugs targeting M. tuberculosis persisters for improved treatment.
Keywords: Pyrazinamide, pyrazinoic acid resistance, mode of action, panD, aspartate decarboxylase
INTRODUCTION
Pyrazinamide (PZA) is an important first-line tuberculosis (TB) drug used in combination with other TB drugs for the treatment of both drug-susceptible TB and multidrug-resistant tuberculosis (MDR-TB).1,2 PZA is a peculiar persister drug that acts only on dormant non-growing persisters and has poor activity against growing Mycobacterium tuberculosis.3 Its high activity against persister bacteria is responsible for PZA's unique sterilizing activity, which shortens the TB treatment period from 9–12 months to 6 months.4 Because of its indispensible sterilizing activity, all new TB regimens in clinical development include PZA.3
PZA is a prodrug that requires activation to its active form, pyrazinoic acid (POA), by an M. tuberculosis pyrazinamidase/nicotinamidase enzyme encoded by the pncA gene.5 Mutations in pncA are the major mechanism of PZA resistance in M. tuberculosis.4,6 Recently, we identified ribosomal protein S1 (RpsA, Rv1630), a vital protein involved in trans-translation, as a target of PZA.7 Mutations in rpsA have been found in some PZA-resistant clinical isolates lacking pncA mutations.7,8,9,10 However, some PZA-resistant strains do not have mutations in either the pncA or rpsA genes,3,9,11 indicating the presence of a possible new resistance mechanism or target of PZA.
Recently, we identified a new gene, panD encoding aspartate decarboxylase and involved in β-alanine biosynthesis, mutations in which are associated with PZA resistance in M. tuberculosis.12 PanD is involved in the synthesis of pantothenate (vitamin B5), which in turn is required for the synthesis of coenzyme A (CoA), a molecule that is at the center of all energy metabolism and allows carbohydrates, fats, and proteins to be burned as energy sources. However, how panD mutations cause PZA resistance and how PZA might interfere with pantothenate and CoA function are unclear. In an attempt to shed light on possible new targets of PZA, in this study, we isolated mutants of M. tuberculosis resistant to POA, the active form of PZA, and characterized mutations potentially involved in POA resistance. Whole-genome sequencing of select POA-resistant mutants without pncA or rpsA mutations together with targeted sequencing mapped all the mutations in the panD gene. Our biochemical and genetic studies suggest that PanD is a new target involved in PZA action and resistance.
MATERIALS AND METHODS
Isolation of spontaneous POA-resistant mutants of M. tuberculosis
A single colony of M. tuberculosis strain H37Ra was cultured at 37 °C in 7H9 medium. At 2∼3 weeks, the culture reached an optical density (OD600) of 0.6∼1.0 (approximately 1×106 to 1×108 colony-forming units (CFU)/mL) and was plated onto 7H11 agar plates containing 100 to 500 µg/mL POA (pH 6.8) or 25 to 200 µg/mL POA (pH 5.7) and also plated on a 7H11 plate for CFU counting. Resistant colonies were picked and subcultured on 7H11 plates containing the same POA concentration at the same pH for rescreening. POA-resistant mutants were stocked and subjected to DNA extraction for further analysis by whole-genome sequencing or by targeted panD sequencing as described below.
Whole-genome sequencing
Genomic DNA was isolated from the POA-resistant mutants and subjected to whole-genome sequencing using the Illumina HiSeq 2000 machine (Illumina, Inc., San Diego, CA, USA) as previously described.12 Paired-end sequencing libraries were barcoded and constructed from the genomic DNA of each strain with insert sizes of approximately 300 base pairs (bp) using TruSeq DNA Sample Preparation kits (Illumina, Inc., San Diego, CA, USA) according to manufacturer's instructions. For each strain, 1.5 G–3.0 G bases (345 to 690-fold genome coverage) were generated after the barcodes were trimmed. High-quality data were aligned with the reference sequence of M. tuberculosis H37Ra (NC_009525) using SOAPaligner. We used the M. tuberculosis H37Ra genome sequence13 as a reference strain for sequence comparison with the POA-resistant mutants derived from M. tuberculosis H37Ra. Only reads where both ends aligned to the reference sequence were used for single nucleotide variant (SNV) and insertion and deletion (InDels) analysis. SNVs and InDels ranging from 1 to 5 bp were sorted and called at minimum reads of 10. Synonymous mutations and PE/PPE mutations within coding sequences were not included in the final analysis to focus on mutations that are most likely to be involved in POA resistance.
PCR and DNA sequencing of the panD gene
The panD primers (panD_F: 5′TCA ACG GTT CCG GTC GGC TGC T3′ and panD_R: 5′TAT CCG CCA CTG CTG CAC GAC CTT3′) were used to amplify a 650-bp PCR product that contains the whole panD gene from POA-resistant M. tuberculosis mutants as described previously.12 The nucleotide sequences were analyzed by using Sequencher software (Gene Codes Corporation, Ann Arbor, MI, USA) to identify possible mutations in panD.
panD conditional overexpression and POA susceptibility testing
The panD gene was amplified by PCR from the M. tuberculosis parent strain H37Ra, the POA-resistant mutants, M. smegmatis and E. coli using the primers (panDRvF 5′-GCG CTT AAT TAA GAA GGA GAT ATA CAT ATG TTA CGG ACG ATG CTG AAG-3′ and panDRvR 5′-ATC AAG CTT CTT CGG ATT CGG TGC GTA TCC G-3′ panDMsF 5′-GCG CTT AAT TAA GAA GGA GAT ATA CAT ATG CAA AGA GTG CTG CTG GCA-3′ panDMsR 5′-ATC AAG CTT GTG AAC TCA CTG GTT GGG TTG C-3′ and panDEcF 5′-GCG CTT AAT TAA GAA GGA GAT ATA CAT ATG ATT CGC ACG ATG CTG CAG G-3′ panDEcR 5′-ATC AAG CTT AAC GCC GTG AAG GCA GCA AA-3′), respectively. The forward primers were designed to include the first start codon of the respective panD gene. The PCR products were digested with the restriction enzymes PacI and HindIII (New England BioLabs, Inc., Ipswich, MA, USA) and ligated to the tetracycline-inducible plasmid pUV15tetORM14 digested with the same enzymes to yield recombinant plasmids. Then, recombinant constructs were confirmed by DNA sequencing and transformed into M. tuberculosis H37Ra as described.15 POA susceptibility testing was performed for different M. tuberculosis strains (wild type, wild type/pUV15tetORM, and wild type/pUV15tetORM +panD) in 7H11 agar and 7H9 liquid medium at pH 6.8 or pH 5.7 as described.16 When required, different concentrations of POA were added. The inducing substrate anhydrous tetracycline (ATc) was added to the plate at a concentration of 50 ng/mL, which did not affect M. tuberculosis growth. Calcium pantothenate or β-alanine was incorporated into 7H11 plates at 0.1, 1, 5, 10 and 100 µM to assess their effects on POA susceptibility. Each strain was printed on 7H11 agar plates in triplicate with a replicator or in 96-well plates in liquid medium at a density of ∼105 CFU/mL to assess the effects of conditional PanD overexpression, pantothenate, and β-alanine on POA/PZA activity.
Overexpression and purification of M. tuberculosis PanD
The M. tuberculosis panD gene was PCR amplified from the genome of type strain H37Rv using the primers: panD F: 5′-TAT CCA TGG GCA TGT TAC GGA CGA TGC TGA AG-3′ and panD R: 5′-TAT CTC GAG TCC CAC ACC GAG CCG GGG GT-3′. The PCR product was digested with NcoI and XhoI and cloned into the pET28a plasmid vector digested with the same enzymes. The correct clones were confirmed by DNA sequencing. The PanD protein was purified as described.17 Briefly, log phase E. coli strain BL21(DE3) containing pET28a-panD was induced with 0.5 mM IPTG at 37 °C for 4 h. The cells were harvested, resuspended in buffer A (10 mM Tris-HCl, pH 7.5, 150 mM NaCl), and lysed by sonication. The PanD protein was purified by using a Ni+2-NTA column (Qiagen, Valencia, CA, USA) and dialyzed against 10 mM Tris, pH 8.0. The protein fractions were eluted with imidazole and analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
PanD enzyme activity assay
PanD activity was assayed by evaluating the conversion of [14C]aspartic acid to β-alanine as previously described.17 [14C]aspartic acid (0.1 mCi/mL, 7.7 GBq/mmol) was kindly supplied by the National Institutes of Health AIDS Reagents Program, Rockville, Md. Briefly, the reaction mixture contained (0.2 mL total volume) phosphate buffer saline, [14C]aspartic acid (0.5 µCi/mL, equivalent to 2.4 µM), and 0.5 µM purified M. tuberculosis PanD enzyme. Incubation was for 3 h and the reaction was stopped by 0.02 mL 50% trichloroacetic acid followed by centrifugation at 12 000 rpm for 5 min and the supernatant was analyzed on a thin layer chromatography (TLC) plate as described.18 For TLC, 2-μL portions of reaction mix were spotted onto a 0.2-mm-thick 10 cm × 10 cm silica G gel 60 plate (Whatman, Inc., Piscataway, NJ, USA), which was developed in 1-butanol, glacial acetic acid and water (4:1:1) for 1 h. The plate was air-dried and exposed to X-ray film for autoradiography. Densitometry was performed on scanned images to obtain an absolute intensity (AI) for each blot and its corresponding control blot. Relative intensity (RI) was calculated by normalizing the experimental AI to the corresponding control AI. One unit of aspartate decarboxylase was defined as the amount of enzyme required to produce 1 mM of β-Ala per hour. The inhibition of PanD activity by POA was investigated with different concentrations of POA (25, 50, 100, and 200 µg/mL) as well as the control compounds PZA and nicotinamide.
RESULTS
POA-resistant M. tuberculosis mutants had PanD mutations
To identify possible new targets of POA, we plated M. tuberculosis H37Ra on 7H11 plates containing POA (25–200 µg/mL at pH 5.7 or 200–700 µg/mL at pH 6.8). Through two more rounds of screening, we were able to isolate 30 mutants resistant to both POA and PZA. DNA sequencing revealed that these 30 POA-resistant mutants did not have the mutations in pncA and rpsA known to cause PZA resistance. Surprisingly, whole-genome sequencing analysis of 3 POA-resistant mutants revealed they had various point mutations (M117I, C deletion at nucleotide position 393, V138A mutations) in the panD gene (Table 1). In view of this finding, the remaining 27 POA-resistant mutants were all subjected to panD DNA sequencing, and remarkably, the results showed that they all had PanD mutations (Table 1). Interestingly, all the mutations occurred in the C-terminus of the 139 amino acid long M. tuberculosis PanD protein within amino acid residues 117–138 (Figure 1). When compared with the wild-type PanD protein, the majority of the mutants (24 of 30 or 80%) had the M117I mutation, and the remaining 6 had the mutations L136R, V138G, V138E, V138A, E126* and a codon shift at 131 (Table 1 and Figure 1). Sequence alignment of the PanD proteins from M. tuberculosis, M. smegmatis, and E. coli indicates that the C-terminal region of M. tuberculosis PanD where all the PanD mutations occurred is variable and confers sequence specificity (Figure 1).
Table 1. panD mutations identified in POA-resistant mutants of M. tuberculosis.
| Strain | Nucleotide change | Amino acid change |
|---|---|---|
| Mycobacterium tuberculosis H37Ra POAR37 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR38 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR48 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR54 | G351A | M117I# |
| Mycobacterium tuberculosis H37Ra POAR55 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR56 | T407G | L136R |
| Mycobacterium tuberculosis H37Ra POAR57 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR59 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR64 | Del 393G | 131codon shift# |
| Mycobacterium tuberculosis H37Ra POAR68 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR71 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR72 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR73 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR76 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR88 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR89 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR90 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR92 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR102 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR106 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR108 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR125 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR126 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR133 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR135 | T413G | V138G |
| Mycobacterium tuberculosis H37Ra POAR136 | G376T | E126* |
| Mycobacterium tuberculosis H37Ra POAR137 | T413A | V138E |
| Mycobacterium tuberculosis H37Ra POAR140 | T413C | V138A# |
| Mycobacterium tuberculosis H37Ra POAR147 | G351A | M117I |
| Mycobacterium tuberculosis H37Ra POAR156 | G351A | M117I |
indicates the three POA-resistant mutants that were sequenced by whole-genome sequencing.
indicates a stop codon.
Figure 1.

Alignment of the PanD amino acid sequences of M. tuberculosis (MTB), M. smegmatis (MSG) and E. coli (ECO). The conserved residues are boxed. The C-terminal sequence without crystal structure is highlighted in yellow. The asterisks indicate the mutant residues identified in the POA-resistant M. tuberculosis mutants.
panD overexpression caused POA/PZA resistance in M. tuberculosis
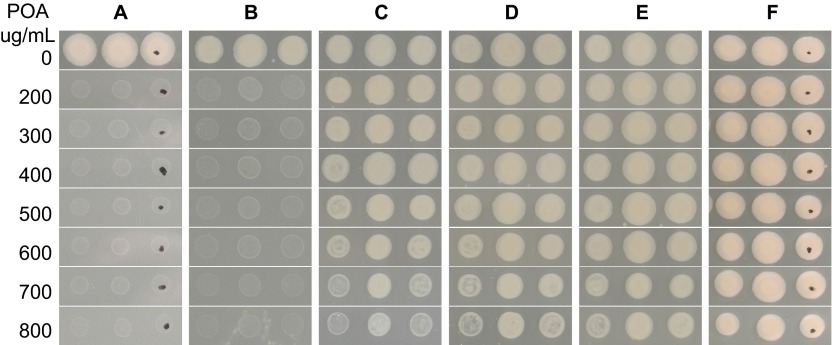
To confirm PanD as a target of PZA, we overexpressed panD genes from M. tuberculosis in M. tuberculosis strain H37Ra and tested their PZA and POA susceptibility. Overexpression of the panD gene from wild-type M. tuberculosis, mutant POAR54 (nucleotide change G351A; amino acid change M117I), mutant POAR140 (nucleotide change T431C; amino acid change V138A), or the panD genes from M. smegmatis or E. coli caused POA resistance in M. tuberculosis H37Ra using the tetracycline-inducible vector pUV15tetORs14 in the presence of ATc (50 ng/mL) inducing agent (Figure 2). In contrast, the recombinant strains were sensitive to POA (<300 µg/mL, pH 6.8) without the inducing agent ATc (Figures 2A–2C). POA-resistant M. tuberculosis mutants with the M117I mutation grew in the absence or presence of ATc induction (Figure 2D) because this is the mutant alone and is not transformed with a panD expression construct. It is interesting to note that expression of either wild-type H37Rv panD or mutant panD (G351A, M117I and T431C, V138A) from M. tuberculosis conferred POA resistance to M. tuberculosis (POA>700 µg/mL, pH 6.8) (Figures 2B and 2C), as did panD from M. smegmatis and E. coli (Figures 2E and 2F),. The vector control remained susceptible to POA with or without ATc induction (Figure 2G). In addition, conditional overexpression of the above constructs caused a three-fold increase in the MIC of PZA for M. tuberculosis from 25 µg/mL in the uninduced and control strains to 75 µg/mL (pH 5.7) in the ATc-induced strains.
Figure 2.

M. tuberculosis became resistant to POA when panD expression was induced by ATc. Each strain was printed in triplicate with a replicator at a density of ∼105 CFU/mL. (A) WT/pUV15tetORs-panDRv (expressing panD from M. tuberculosis H37Rv) without (− sign) and with (+ sign) ATc, (B) WT/pUV15tetORs-panDG351A (expressing panD mutant) without and with ATc, (C) WT/pUV15tetORs-panDT431C (expressing panD mutant) without and with ATc, (D) POAR 156 (M117I) without and with ATc, (E) WT/pUV15tetORs-panDMS (expressing panD from M. smegmatis) without and with ATc, (F) WT/pUV15tetORs-panDEC (expressing panD from E. coli) without and with ATc, (G) WT/pUV15tetORs vector control without and with ATc.
Pantothenate and β-alanine antagonized POA/PZA activity against M. tuberculosis
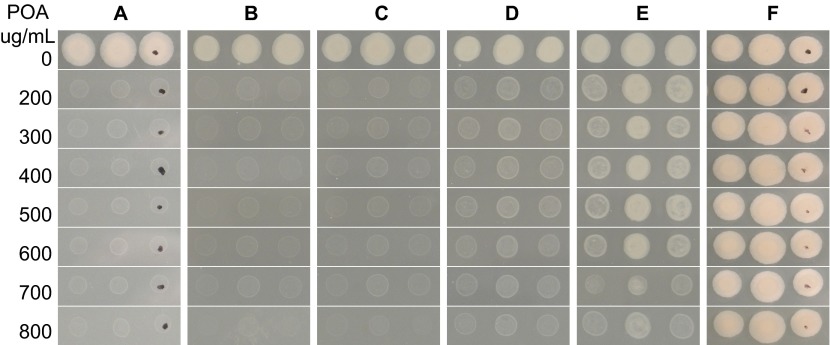
Because PanD (aspartate decarboxylase) is involved in converting L-aspartic acid to β-alanine, a precursor for pantothenate and coenzyme A biosynthesis, we wondered if supplementation with β-alanine or pantothenate might render M. tuberculosis resistant to POA. To test this, pantothenate, β-alanine, and different amino acids that served as controls, were incorporated into 7H11 agar containing different concentrations of POA (200–800 µg/mL pH 6.8) followed by inoculation with M. tuberculosis H37Ra. The results showed that the parent strain and the mutant strain were sensitive and resistant to POA, respectively, on 7H11 plates containing different concentrations of POA (Figures 3A and 3B), but susceptibility to POA dramatically decreased when pantothenate or β-alanine (Figures 3C and 3D) was added to the medium at 0.1 mM. However, other amino acids such as L-alanine, L-aspartate, L-valine and L-glutamate had no effect on POA activity (Figures 3E–3H). This finding suggests that pantothenate and β-alanine, which are the downstream products of PanD, specifically antagonize the antituberculosis activity of POA and that POA inhibits pantothenate or β-alanine synthesis by blocking PanD activity in M. tuberculosis.
Figure 3.
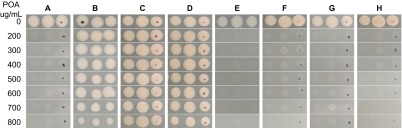
Antagonism of POA's activity against M. tuberculosis by pantothenate and β-alanine. (A) M. tuberculosis parent strain H37Ra on 7H11 agar, (B) a POA-resistant mutant with a PanD mutation (M117I), (C) parent strain on 7H11 agar containing 0.1 mM calcium pantothenate, (D) parent strain with 0.1 mM β-alanine, (E) parent strain with 0.1 mM L-alanine, (F) parent strain with 5 mM calcium L-aspartate, (G) parent strain with 10 mM L-valine, (H) parent strain with 10 mM glutamate.
Effects of different concentrations of β-alanine or pantothenate on POA susceptibility
To identify the minimum concentration of β-alanine or pantothenate that antagonizes POA activity, M. tuberculosis strain H37Ra was plated on 7H11 plates containing various concentrations of β-alanine or pantothenate (0, 0.1, 1, 5, 10 and 100 µM). The results showed that the parent strain H37Ra was susceptible to 200 µg/mL POA (Figure 4A) and 0.1 µM β-alanine did not appear to antagonize POA activity (Figure 4B). The minimum concentration of β-alanine that showed obvious POA antagonism was 1 µM (Figure 4C), and the antagonism increased with the concentration of β-alanine (Figures 4C–4F). Interestingly, there was not much variation in the level of β-alanine antagonism across the range of 1–100 µM as all concentrations caused resistance to 600–800 µg/mL POA (Figures 4C–4F). Similarly, we evaluated the effect of pantothenate on POA susceptibility and found that concentration required to cause resistance to POA was an order of magnitude higher for pantothenate (10 µM) than β-alanine (1 µM) (Figure 5).
Figure 4.
Effect of β-alanine concentration on POA susceptibility. M. tuberculosis H37Ra on 7H11 agar with no β-alanine (A), 0.1 µM β-alanine (B), 1 µM β-alanine (C), 5 µM β-alanine (D), 10 µM β-alanine (E), and 100 µM β-alanine (F).
Figure 5.
Effect of pantothenate concentration on POA susceptibility. M. tuberculosis H37Ra on 7H11 agar with no calcium pantothenate (A), 0.1 µM calcium pantothenate (B), 1 µM calcium pantothenate (C), 5 µM calcium pantothenate (D), 10 µM calcium pantothenate (E), and 100 µM calcium pantothenate (F).
POA but not the prodrug PZA inhibits the activity of the M. tuberculosis PanD enzyme
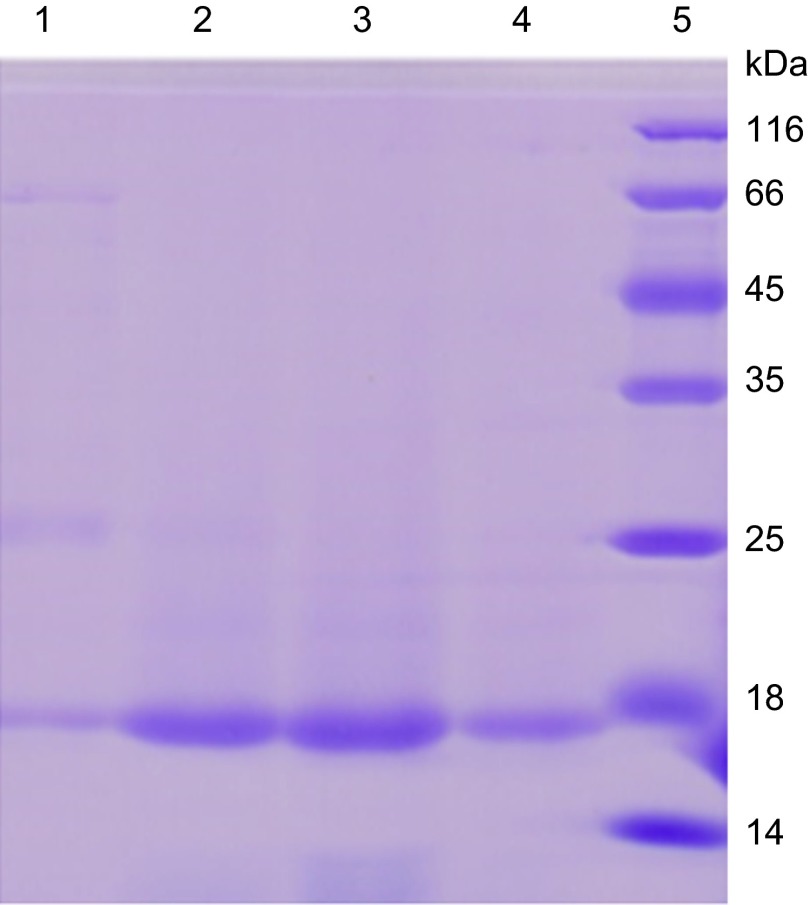
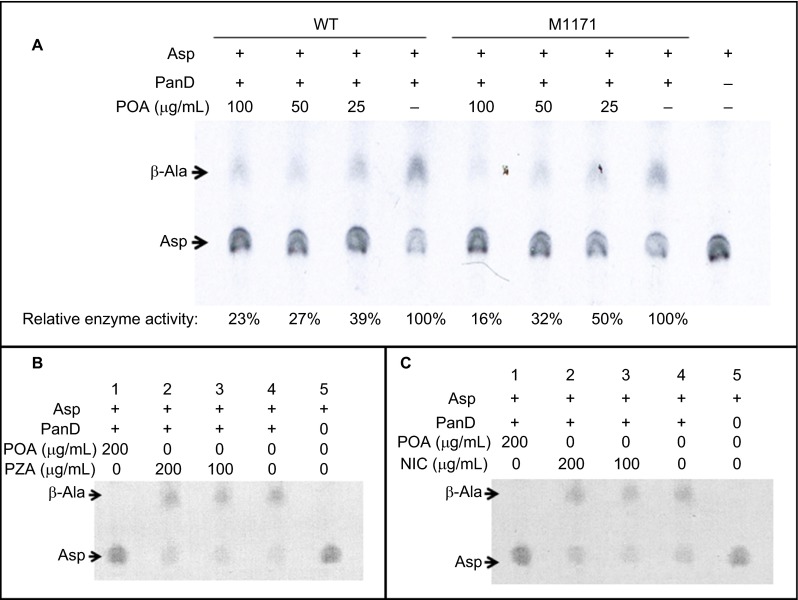
PanD (aspartate decarboxylase) catalyzes the formation of β-alanine from aspartic acid in the reaction L-aspartate + H+ → β-alanine + CO2. To assess if POA can inhibit the enzymatic activity of PanD, we overexpressed and purified the wild-type M. tuberculosis PanD protein, which appeared as a 16 kD band on SDS-PAGE (Figure 6). The purified PanD protein had activity of 107.8 U/mg and was used for a PanD enzymatic assay in the presence or absence of POA. The PanD M117I mutant was similarly overexpressed and purified and was found to have slightly reduced enzymatic activity (76.3 U/mg). The wild-type M. tuberculosis PanD hydrolyzed aspartic acid (Asp) to β-alanine (β-Ala) in the absence of POA (Figure 7A, Lane 4 from the left). Importantly, POA at the physiologically relevant concentrations of 100, 50, or 25 µg/mL inhibited the activity of M. tuberculosis PanD in a concentration-dependent manner with 23%, 27%, or 39% residual enzymatic activity as shown by the increasing accumulation of the substrate aspartate and the decreasing amount of the product β-alanine (Figure 7A). The M117I PanD mutant enzyme was also inhibited by POA (100, 50, 25 µg/mL) to varying degrees with 16%, 32%, and 50% residual enzymatic activity, except that the M117I mutant enzyme appeared to be slightly less inhibited by POA than the wild-type enzyme was at 50 and 25 µg/mL (Figure 7A). POA at 200 µg/mL completely inhibited the activity of the wild-type PanD enzyme as shown by the lack of conversion of aspartate to β-alanine (Figures 7B and 7C, first lane from left). In contrast, neither the prodrug PZA nor nicotinamide when used as controls at 100 or 200 µg/mL inhibited the enzymatic activity of PanD (Figures 7B and 7C) as expected.
Figure 6.
Overexpression and purification of M. tuberculosis PanD in E. coli. Lane 1–4: PanD protein, which appears as a 16-kD band, eluted by 50, 100, 200, 400 mM imidazole, respectively. Lane 5: Protein molecular weight marker (116, 66, 45, 35, 25, 18, 14 kD).
Figure 7.
Concentration-dependent inhibition of M. tuberculosis PanD by POA but not by PZA or nicotinamide. The rightmost lane is a 14C-aspartic acid control without PanD or POA addition. (A) POA at 25, 50, or 100 µg/mL significantly inhibited the enzymatic activity of both wild-type and M117I PanD in a concentration-dependent manner as shown by the reduced conversion of aspartate (Asp) to β-alanine (β-Ala). At 200 µg/mL, POA completely inhibited the enzymatic activity of wild-type PanD (panels B and C, first lane from left). In contrast, neither PZA as a prodrug (B) nor nicotinamide (NIC) (C) as a structural analog control at 100 and 200 µg/mL had an apparent effect on PanD's enzymatic activity.
DISCUSSION
PZA resistance in M. tuberculosis is most commonly caused either by mutations in the pncA gene encoding the PZase required to convert the PZA prodrug to POA5 or (occasionally) by mutations in the target gene rpsA encoding ribosomal protein S1 involved in trans-translation.7 We recently identified mutations in a third gene, panD, (encoding an aspartate decarboxylase involved in pantothenate (vitamin B5 synthesis) in five PZA-resistant strains when PZA was used to select resistant mutants.12 In this study, to shed new light on possible new targets of PZA, we isolated mutants of M. tuberculosis resistant to POA, the active form of PZA. Interestingly, all 30 POA-resistant mutants had mutations in the C-terminus of the panD gene (Figure 1 and Table 1). This surprising finding suggests that panD mutations are closely associated with POA resistance and that the C-terminus of the PanD protein may be involved in POA binding.
Previous attempts to isolate POA-resistant M. tuberculosis mutants have failed,6 and this has hindered the identification of the targets of PZA/POA. It is possible that the previous studies used very low pH (pH 5.6) such that POA-resistant mutants could not survive the combined lethal action of low pH and POA because the level of resistance conferred by target mutations such as PanD mutations may be too low to counteract the combined action of POA and acidic pH. Indeed, when the pH of the medium was lowered to pH 5.6, the POA-resistant mutants obtained at pH 6.8 became sensitive to POA at 25–50 µg/mL (data not shown). The isolation of POA-resistant mutants in this study most likely succeeded because a less acidic pH (pH 6.8, that of the 7H11 medium) was used. At this pH (pH 6.8), the parent M. tuberculosis strain H37Ra was susceptible to 200 µg/mL POA (Figure 2A), allowing POA-resistant mutants to be isolated at 200–800 µg/mL POA readily in this study. This has made it possible to characterize the genetic basis of POA resistance. Because POA is the active form of the drug PZA, the finding that all the POA-resistant mutants had panD mutations (Table 1) suggests that PanD is most likely to be a target of POA/PZA.
Further evidence that PanD is a target of POA/PZA was provided by the PanD overexpression experiment. Target overexpression is a well-known drug resistance mechanism in bacteria. Our results showed that inducible overexpression of wild-type M. tuberculosis PanD caused an M. tuberculosis strain to be resistant to POA (Figure 2C). In contrast, uninduced strains containing various PanD constructs and the strain that harbored the empty vector were still susceptible to POA, indicating that the induced PanD expression is responsible for the POA resistance. Intriguingly, overexpressing the M. tuberculosis PanD M117I mutant or PanD from either M. smegmatis or E. coli also increased the resistance of M. tuberculosis to POA (Figures 2D–2F). These findings suggest that elevated levels of active PanD enzyme, irrespective of the source, confer POA resistance.
The most direct evidence that PanD is a target of POA/PZA was provided by the finding that POA inhibits the enzymatic activity of PanD (Figure 7A) while the prodrug PZA and the structural analog nicotinamide both fail to do so (Figures 7B and 7C). In addition, the observation that all the PanD mutations from the 30 POA-resistant mutants are localized to the C-terminus of the PanD protein (Table 1 and Figure 1) suggests that POA may bind to this region, and in particular to residue M117 as the most frequent site that is altered in POA-resistant mutants, to inhibit the enzymatic activity of PanD. Moreover, the finding that M117I overexpression confers POA/PZA resistance to M. tuberculosis and that the M117I mutation retains the enzymatic activity of PanD (Figure 7A) suggest that the C-terminal mutations in PanD may affect the binding of POA without eliminating PanD's enzymatic activity. It is interesting to note that the C-terminal region spanning amino acid residues 114–139 of M. tuberculosis PanD, where all the PanD mutations are mapped (Figure 1), could not be determined in the crystal structure of the M. tuberculosis PanD protein.19 It is likely that the C-terminal region of M. tuberculosis PanD, where it shows the most sequence variability (Figure 1), is flexible and dispensable for enzymatic activity but is required for POA binding. Future studies are needed to confirm this possibility.
As a peculiar persister drug that plays a crucial role in shortening TB therapy,3,4 PZA's mode of action is unusual and complex and has attracted considerable recent attention due to increasing interest in developing persister drugs not only for TB20,21 but also for other persistent infections.22 PZA clearly interferes with multiple targets in M. tuberculosis, with activities including membrane potential disruption,23 internal pH acidification,18,24 and trans-translation inhibition.7 In this study, we found new evidence that PanD may represent a target of PZA/POA in M. tuberculosis. PanD converts L-aspartate to β-alanine, which is in turn needed for the synthesis of pantothenate (vitamin B5) and CoA. CoA is a ubiquitous cofactor found in all domains of life that plays a central role in energy metabolism. In addition, M. tuberculosis PanD is a virulence factor25 that could be involved in persistence in vivo. Our finding that POA inhibits PanD's enzymatic activity (Figure 7A) involved in energy metabolism and virulence could help to explain the unique sterilizing activity of PZA in vivo.
An interesting observation of this study was that both β-alanine and pantothenate were able to antagonize POA's activity against M. tuberculosis. This finding suggests that POA inhibits PanD's activity and that supplementation of downstream products such as β-alanine or pantothenate could rescue or antagonize the lethal action of PZA. This raises the intriguing possibility that variations in the levels of β-alanine or pantothenate in the diet of the host might affect the activity of PZA in vivo. Future studies are needed to address this possibility using animal models. Because humans lack the pathway to synthesize pantothenate that is present in various bacteria including M. tuberculosis, PanD could represent an attractive drug target for the development of new drugs targeting persister bacteria. Indeed, there is recent interest in developing new drugs targeting the pantothenate synthesis pathway in M. tuberculosis.26,27,28
In summary, we found that panD mutations are closely associated with POA resistance in POA-resistant mutants lacking pncA and rpsA mutations. Inducible PanD overexpression caused significant resistance to POA in M. tuberculosis. In addition, the antituberculosis activity of POA could be antagonized by β−alanine or pantothenate. POA, but not the prodrug PZA or nicotinamide, inhibited the enzymatic activity of PanD in a concentration-dependent manner. These findings suggest that PanD is a new target of POA/PZA. These results have implications for the development of new drugs targeting pantothenate or CoA biosynthesis to improve TB treatment. Future studies are needed to further our understanding of the genetic, biochemical, and structural basis of PanD inhibition by POA and to assess the potential for food-derived β−alanine and pantothenate to antagonize PZA activity in vivo.
Acknowledgments
This work was supported in part by National Institutes of Health grants AI099512 and AI108535, the Major Project of the Twelfth Five-Year Plan (2013ZX10003008-003 and 2013ZX10003001-002), and the National Natural Science Foundation of China (81101226). We thank Sabine Ehrt (Weill Cornell Medical College, New York, USA) for provision of the Tet-inducible plasmid.
References
- Zhang Y, Chang K, Leung C, et al. “ZS-MDR-TB” versus “ZR-MDR-TB”: Improving Treatment of MDR-TB by Identifying Pyrazinamide Susceptibility. Emerg Microbes Infect. 2012;1:e5. doi: 10.1038/emi.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Treatment of tuberculosis: guidelines, Fourth edition Geneva: WHO; 2010. Available at http://www.who.int/tb/publications/2010/9789241547833/en/ (accessed 28 June 2014). [Google Scholar]
- Zhang Y, Shi W, Zhang W, Denis Mitchison. Mechanisms of Pyrazinamide Action and Resistance. Microbiol Spectrum. 2014;2:2.4.03. doi: 10.1128/microbiolspec.MGM2-0023-2013. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mitchison D. The curious characteristics of pyrazinamide: a review. Int J Tuberc Lung Dis. 2003;7:6–21. [PubMed] [Google Scholar]
- Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- Scorpio A, Lindholm-Levy P, Heifets L, et al. Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997;41:540–543. doi: 10.1128/aac.41.3.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Zhang X, Jiang X, et al. Pyrazinamide inhibits trans-translation in Mycobacterium tuberculosis. Science. 2011;333:1630–1632. doi: 10.1126/science.1208813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y, Hu Z, Zhang T, et al. Role of pncA and rpsA gene sequencing in detection of pyrazinamide resistance in Mycobacterium tuberculosis isolates from southern China. J Clin Microbiol. 2014;52:291–297. doi: 10.1128/JCM.01903-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons SO, Mulder A, van Ingen J, Boeree MJ, van Soolingen D. Role of rpsA gene sequencing in diagnosis of pyrazinamide resistance. J Clin Microbiol. 2013;51:382. doi: 10.1128/JCM.02739-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerriegel S, Koser CU, Richter E, Niemann S. Mycobacterium canettii is intrinsically resistant to both pyrazinamide and pyrazinoic acid. J Antimicrob Chemother. 2013;68:1439–1440. doi: 10.1093/jac/dkt042. [DOI] [PubMed] [Google Scholar]
- Alexander DC, Ma JH, Guthrie JL, Blair J, Chedore P, Jamieson FB. Gene sequencing for routine verification of pyrazinamide resistance in Mycobacterium tuberculosis: a role for pncA but not rpsA. J Clin Microbiol. 2012;50:3726–3728. doi: 10.1128/JCM.00620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen J, Shi W, Liu W, Zhang WH, Zhang Y. Mutations in panD encoding aspartate decarboxylase are associated with pyrazinamide resistance in Mycobacterium tuberculosis. Emerg Microbes Infect. 2013;2:e34. doi: 10.1038/emi.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Lu L, Wang B, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3:e2375. doi: 10.1371/journal.pone.0002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrt S, Guo XV, Hickey CM, et al. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 2005;33:e21. doi: 10.1093/nar/gni013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Garbe T, Young D. Transformation with katG restores isoniazid-sensitivity in Mycobacterium tuberculosis isolates resistant to a range of drug concentrations. Mol Microbiol. 1993;8:521–524. doi: 10.1111/j.1365-2958.1993.tb01596.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Permar S, Sun Z. Conditions that may affect the results of susceptibility testing of Mycobacterium tuberculosis to pyrazinamide. J Med Microbiol. 2002;51:42–49. doi: 10.1099/0022-1317-51-1-42. [DOI] [PubMed] [Google Scholar]
- Chopra S, Pai H, Ranganathan A. Expression, purification, and biochemical characterization of Mycobacterium tuberculosis aspartate decarboxylase, PanD. Protein Expr Purif. 2002;25:533–540. doi: 10.1016/s1046-5928(02)00039-6. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Scorpio A, Nikaido H, Sun Z. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol. 1999;181:2044–2049. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan G, Chopra S, Ranganathan A, Swaminathan K. Crystal structure of uncleaved L-aspartate-alpha-decarboxylase from Mycobacterium tuberculosis. Proteins. 2006;65:796–802. doi: 10.1002/prot.21126. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yew WW, Barer MR. Targeting persisters for tuberculosis control. Antimicrob Agents Chemother. 2012;56:2223–2230. doi: 10.1128/AAC.06288-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore C, Schleif AC, Bernstein JB, Heilman CA. The role of biomedical research in global tuberculosis control: gaps and challenges. Emerg Microbes Infect. 2012;1:e9. doi: 10.1038/emi.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Persisters, Persistent Infections and the Yin-Yang Model. Emerg Microbes Infect. 2014;3:e3. doi: 10.1038/emi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wade MM, Scorpio A, Zhang H, Sun Z. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J Antimicrob Chemother. 2003;52:790–795. doi: 10.1093/jac/dkg446. [DOI] [PubMed] [Google Scholar]
- Darby CM, Ingolfsson HI, Jiang X, et al. Whole cell screen for inhibitors of pH homeostasis in Mycobacterium tuberculosis. PLoS One. 2013;8:e68942. doi: 10.1371/journal.pone.0068942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandamurthy VK, Wang X, Chen B, et al. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med. 2002;8:1171–1174. doi: 10.1038/nm765. [DOI] [PubMed] [Google Scholar]
- Sharma R, Kothapalli R, Van Dongen AM, Swaminathan K. Chemoinformatic identification of novel inhibitors against Mycobacterium tuberculosis L-aspartate alpha-decarboxylase. PLoS One. 2012;7:e33521. doi: 10.1371/journal.pone.0033521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EL, Southworth K, Ross L, et al. A novel inhibitor of Mycobacterium tuberculosis pantothenate synthetase. J Biomol Screen. 2007;12:100–105. doi: 10.1177/1087057106296484. [DOI] [PubMed] [Google Scholar]
- Kumar A, Casey A, Odingo J, et al. A high-throughput screen against pantothenate synthetase (PanC) identifies 3-biphenyl-4-cyanopyrrole-2-carboxylic acids as a new class of inhibitor with activity against Mycobacterium tuberculosis. PLoS One. 2013;8:e72786. doi: 10.1371/journal.pone.0072786. [DOI] [PMC free article] [PubMed] [Google Scholar]