Abstract
A long-standing question in community ecology is what determines the identity of species that coexist across local communities or metacommunity assembly. To shed light upon this question, we used a network approach to analyse the drivers of species co-occurrence patterns. In particular, we focus on the potential roles of body size and trophic status as determinants of metacommunity cohesion because of their link to resource use and dispersal ability. Small-sized individuals at low-trophic levels, and with limited dispersal potential, are expected to form highly linked subgroups, whereas large-size individuals at higher trophic positions, and with good dispersal potential, will foster the spatial coupling of subgroups and the cohesion of the whole metacommunity. By using modularity analysis, we identified six modules of species with similar responses to ecological conditions and high co-occurrence across local communities. Most species either co-occur with species from a single module or are connectors of the whole network. Among the latter are carnivorous species of intermediate body size, which by virtue of their high incidence provide connectivity to otherwise isolated communities playing the role of spatial couplers. Our study also demonstrates that the incorporation of network tools to the analysis of metacommunity ecology can help unveil the mechanisms underlying patterns and processes in metacommunity assembly.
Keywords: co-occurrence network, metacommunity structure, modularity analysis, body size, trophic position
1. Introduction
It has long been recognized that the identity of locally coexisting species results from the interaction between factors affecting their immigration and persistence in local communities [1–4]. The relative importance of these, however, will probably vary with scale and with how different individuals interact with the dominant landscape pattern [5–8], which in turn is modulated by species' functional traits. Body size and trophic behaviour are considered key functional traits affecting metacommunity structure as they affect species’ potential to reach local patches, the strength of environmental filters and the result of biotic interactions within local patches [9,10]. For example, owing to their contrasting dispersal abilities and resource requirements, large consumers would probably be more sensitive to landscape-level factors, while individuals of smaller body size could be more affected by local environment ([11,12] but see [10]).
The expectation that large-size species play a main role in the cohesion of communities is supported by their larger niche breath [13–15], their apex insertion in local communities [16,17] and their movement among communities [18]. Similarly, McCann and co-authors considering allometric trends with body size suggested that large consumers act as connectors among local communities [9,17–18]. Indeed, available data suggest that the increase in energetic demand with body size, coupled with larger movement abilities [9] and relaxation in gape limitation [19], typically determine an expansion in prey diversity as size increases [20–22,18], trophic position [22], environmental tolerance [15,23] and the range of energy sources used [17,22]; and thus the coupling of local communities by large consumers [18]. Different studies point to this coupling as a main determinant of metacommunity stability [9,18,24–26]. Intermediate body size species, however, could also play a role in metacommunity cohesion. Intermediate body size species could occur in more patches because of their reduced resource requirement per unit of habitat area [27–29] and because of their ability to use a wider range of trophic positions [19]; which allows them to sustain viable populations where larger or smaller species would not. Notwithstanding the possibility that under some scenarios other mechanisms may become more important [10,30–32], the main expectation is that as a species body size increases so does its potential to become a connector within a metacommunity system, and this would peak at intermediate or large body sizes.
In summary, species with different traits are expected to have different roles within metacommunities. In particular, we hypothesize that: (i) small body size individuals at lower trophic levels will tend to form highly linked subgroups—e.g. local aggregations—and (ii) larger and/or intermediate body size individuals at higher trophic levels could play the role of spatial couplers of such subgroups, thus providing cohesion to the metacommunity [9,18]. Recent advances in network theory allow translating the two hypotheses into specific predictions of topological co-occurrence at the metacommunity level, and thus a formal evaluation of these hypotheses.
Modularity in ecological networks has increasingly been used to detect spatial aggregation of species in compartments of organisms sharing similar ecological attributes [33–37]. In co-occurrence networks, modules correspond to subsets of species whose probability of co-occurring across local communities is higher as compared with the probability of co-occurring with other species of the metacommunity. These modules could represent guilds [38], trophic motifs [18], co-evolved plants and pollinators [33] or related species [35]. Further, the relative number of interactions with other species of their module, other modules and the whole network, allows the estimation of their topological roles within metacommunities [33,39–41]. These roles are directly related to the above proposed hypotheses. Specifically, intermediate and/or large mobile consumers are expected to operate as network hubs (e.g. connecting the whole network), connectors (e.g. connecting different modules) or module hubs (e.g. connecting their module). On the other hand, the larger dependence of small species on local conditions suggests that they could play a peripheral role in co-occurrence patterns.
Here, we use a network approach to infer species roles in the metacommunity structure of a desert ecosystem in the Atacama Desert, Chile. Starting with the incidence matrix of species across local communities, we built a unipartite species network such that any two species that co-occur, more frequently than expected by chance, in local communities were linked. Using this positive co-occurrence network, we: (i) carried out a modularity analysis and classified species into different topological roles; and (ii) tested for the importance of trophic status, body size, numerical abundance and incidence of the species as the biological attributes that could determine these roles. The results herein reported identify modularity as a main component of species co-occurrences networks and, body size and trophic position as chief determinants of the ecological role of species within metacommunities.
2. Material and methods
(a). Study site and sampling
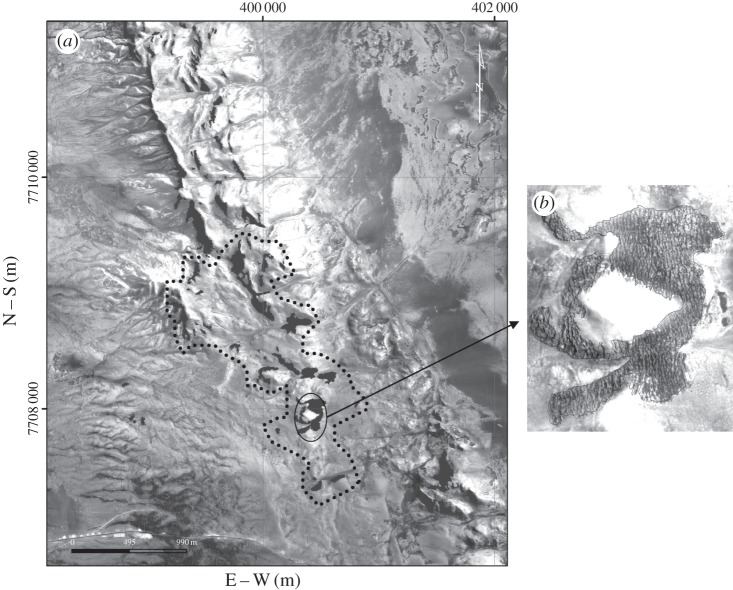
We analysed a metacommunity located in the Atacama Desert, northern Chile (20°29′ S–20°26′ S) one of the most arid ecosystems in the world (average precipitation per year < 2 mm between 1905 and 2001) [42]. This ecosystem is sustained by the humidity provided by fog that, as it is moved inland by the westerly winds, interacts with the terrain giving rise to the development of isolated patches consisting of parallel vegetated bands of the bromeliad, Tillandsia landbeckii (see [43] for a model that explains the emergence of the banded vegetation pattern observed in local communities).
We sampled animal species in 31 local communities (i.e. patches) within the system (figure 1). These communities are mainly composed of arthropods (arachnida and insecta), except for one species of lizard, Phrynosaura reichei and one species of gecko, Phyllodactyllus gerrhopygus (see the electronic supplementary material, table S1, for a list of the species). In each patch, we used a transect sampling scheme where each transect contained a variable number of pitfall traps of 500 cm3 according to the patch area. Pitfall traps have a good performance monitoring active organisms providing confident estimations of species pool and local incidences, when sites are sampled under the same standardized protocol [44].
Figure 1.
Aerial photograph of the study area in the Atacama Desert (20°29′ S–20°26′ S), north of Chile. (a) The dotted black line encloses the 31 patches sampled in this study. (b) A patch internal structure.
A total of 1518 pitfall traps were equi-distantly distributed in each transect and checked twice during 30 days before removal. This represents, to our knowledge, the largest survey of an Atacama Desert metacommunity so far performed. All the animals were collected, preserved in 90% ethanol and identified to the finest possible taxonomic resolution by a specialist (see the electronic supplementary material, table S1, for a list of the species). The length and width of each individual caught was measured under a stereoscopic zoom microscope to estimate its biovolume, calculated as the body length × width2. The following biological attributes were estimated for each species: (i) biovolume, defined as the arithmetic mean of the biovolume of all collected individuals of the species found in the metacommunity, (ii) numerical abundance, as the number of individuals found in the metacommunity, and (iii) incidence as the proportion of occurrences of the species in the metacommunity. Species were classified as carnivorous or herbivorous following González et al. [45].
(b). Co-occurrence network
A total of 17 667 individuals (76% herbivores and 24% carnivorous) were used to build the species co-occurrence network. In this network, species that co-occurred more often than expected by chance were linked. To determine significant co-occurrences, we propose a novel null model based on the abundance and incidence of species in the metacommunity. Thus the co-occurrence network was created following three steps: (i) estimation of the probability of finding an individual of a given species in a given patch, (ii) estimation of the probability of species co-occurrence and the expected co-occurrence along all the patches and all pair of species, and (iii) detection of significant deviations in the co-occurrence pattern.
(i) The probability of observing a species i in a patch j was estimated as follows:
where N is the total number of individuals observed in the metacommunity without considering species identity, mi is the number of individuals of species i in the metacommunity and nj is the number of individuals observed in the patch j without considering species identity. The large parenthesis indicates the calculus of a combinatory.
(ii) The probability of two species i and q co-occurring in a single patch j was estimated as Pi,q;j = Pij × Pqj and the expected number of co-occurrences along all patches was estimated as follows:
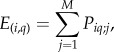 |
where M is the total number of patches.
(iii) The variance in the number of co-occurrences of species i and q was estimated as follows:
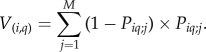 |
Finally, for each pair of species, a Z-value was calculated as follows:
where Obsiq is the number of co-occurrences of species i and q observed across all patches.
The significance of the links was evaluated with a Z-test [46]. Co-occurrence of species with Z-values larger than 2, corresponding to a 95% confidence interval (CI), were considered significant. The co-occurrence matrix was constructed using the complete list of species in rows and columns putting a one for significant positive co-occurrence (Z > 2) and a value of zero otherwise. This Z-value, which comes from a standardized normal distribution, is different from the z-value that defines the topological role of species (see below).
(c). Modularity analysis
Modularity refers to the degree to which a network is organized into sets of nodes with more connections within them than among them [47]. In our positive co-occurrence network, a module is a group of species that co-occur more than expected by chance among themselves than with other species in the metacommunity. To detect such structure, we used a recently developed module-detecting algorithm based on simulated annealing (SA) [39]. This is a stochastic optimization technique that identifies modules in a graph by maximizing a function of modularity [48]. Compared to alternative approaches, SA has the advantage that the number and size of modules are determined by the network itself and not by the researcher; thus allowing for the possibility that no good division of the network might exist [47]. Herein, the modularity function used was the one proposed by Newman & Girvan [49] for a unipartite graph. To test its significance, we compared the estimated modularity on the empirical network with the 99% percentiles of the distribution of modularity in 2000 random networks with the same degree distribution—i.e. conserving the original distribution of edges per species. Random networks were generated reassigning edges randomly among species, retaining only those that did not include a species connected to itself.
(d). Topological roles of the species
Once a significant modular organization was detected, we estimated the topological role of species based on their membership in a module [39]. The role of species i has two components. The first one is defined as the standardized within-module degree z, which is the number of links that species i has to other species in the same module [33], and is defined as follows:
where kis is the number of links k of the species i to other species in the same module s, while 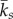 and SDks are the average and the standard deviation of within-module links of species in s. The second component is the among-module connectivity, c or the participant coefficient (sensu [39]). This is a measure of the number of links of species i with species in other modules [33] normalized by the degree of the species i (ki) and is estimated as follows:
and SDks are the average and the standard deviation of within-module links of species in s. The second component is the among-module connectivity, c or the participant coefficient (sensu [39]). This is a measure of the number of links of species i with species in other modules [33] normalized by the degree of the species i (ki) and is estimated as follows:
where kit is the number of links from species i to the other species in the module t (including i's own module). Finally, the roles of each species are depicted in a zc-parameter space that was divided in four regions following the criteria of Olesen et al. [33] in setting z and c thresholds. Note that the threshold values which define the topological roles were defined by [39], as corresponding to z = 2.5 and c = 0.62. The definition of the threshold value of z = 2.5 corresponds to a 99% CI, while c = 0.62 refers to a node that has at least 60% of its links within the module [39]. Using these threshold values, four roles could be identified (see also figure 2b). Peripherals or specialists are species that have z ≤ 2.5 and c ≤ 0.62, that is they have few links and most of them go to other species within their module. Module hubs are species that have z > 2.5 and c < 0.62, that is they are species with many links and most of them to species in their own module. Connectors, on the other hand, are species that have low z ≤ 2.5 but high c > 0.62, and they are key to linking several modules in the network. Finally, network hubs or super-generalist species, are the ones that have both a high z > 2.5 and c > 0.62 and in consequence they could act as module hubs and connectors at the same time.
Figure 2.
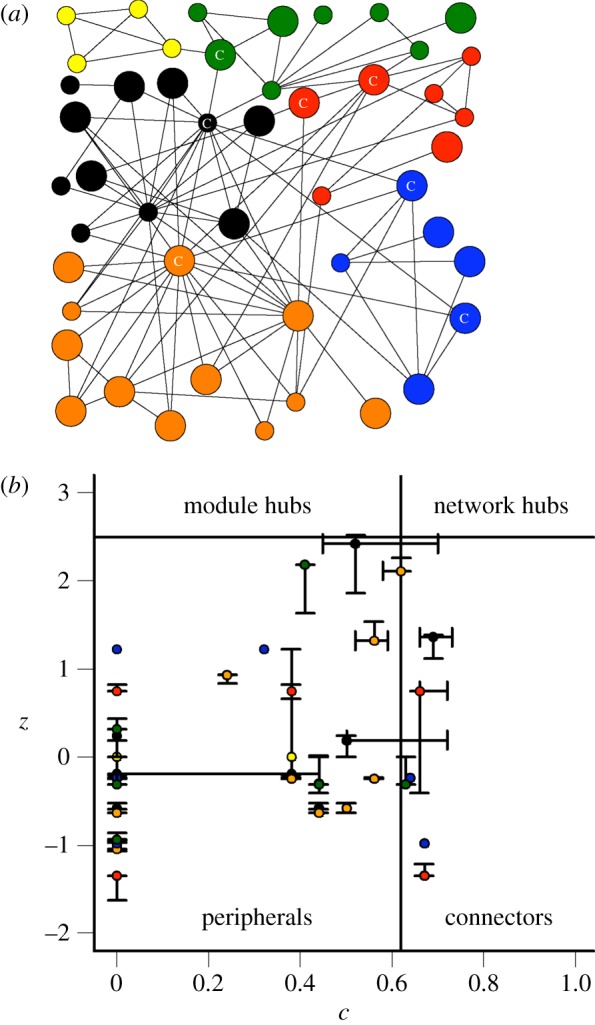
Co-occurrence network structure and species roles. (a) Co-occurrence network showing the modular structure of the metacommunity (48 nodes and 103 links). The small circles correspond to herbivorous species, whereas the large ones correspond to carnivorous species. The circles with a ‘C’ indicate which species are identified as connectors. (b) zc parameter space showing the distribution of species according to their network roles. The threshold value of z = 2.5 and c = 0.62 used here were heuristically determined by Guimera & Amaral [39]. The bars at each node correspond to the 95% CI from 200 estimations of species roles. In both cases, the different colours refer to the six modules found. (Online version in colour.)
The stochastic nature of our module detection algorithm implies the existence of variability in the assignment of species to modules, and consequently, in species role identification. To check for the robustness of our results, we performed 200 sets of independent modularity analyses. In each analysis, the two components of the role of each species, z and c, were retained and a 95% CI from the distributions of the 200 z- and c-values for each species was constructed. Finally, a logistic regression was used to evaluate if the probability of occupying a certain role (1) or the other three roles (0) could be associated with trophic status (carnivores or herbivores), biovolume, quadratic biovolume, numerical abundance per species and incidence of the species, which were used as predictor variables. The quadratic expression of biovolume was included because theoretical expectation [28] and previous results [12] indicate a main role of intermediate-size species. Thus, four possible models—one for each role—were used to describe the association between a role and the predictor variables. In all cases, models were generated using a best subset procedure and the best model for each role was that with the lowest Akaike's information criterion (AIC) [50]. Differences in AIC values between models greater than two units were considered statistically significant [51]. When several models presented AIC values with less than two units of difference, the model with larger R2 values and/or significant parameters was selected.
3. Results
(a). Modularity analysis
The modularity analysis showed that the co-occurrence network (48 nodes and 103 links) was significantly more modular (modularity = 0.49; p < 0.001) than expected for a random graph (figure 2a), suggesting the presence of six modules. The number of species in modules ranged from 4 to 12 species (see figure 2a) and, as expected for a modular organization, the mean connectance of a module was higher (0.23 ± 0.14) than the connectance of the entire network (0.05). In general, all modules were dominated by a few abundant species. Carnivores and herbivores were present in all modules, except in one module composed only of four herbivorous species (see figure 2a).
(b). Species role in the metacommunity
Our results show that species were classified as either peripherals or connectors (i.e. z < 2.5 in both cases), implying that no module or network hub species are present in the analysed metacommunity; hence, most of the variation was associated with the c-axis (see figure 2b). In particular, 85% of the species played peripheral roles with most of their links restricted to their own modules. Interestingly, 51% of these nodes had connections only inside their own modules (i.e. c = 0). These are called ultraperipheral species sensu Guimera & Amaral [39]. The remaining 15% of the nodes are connector species, which are important in linking their modules to the rest of the network. All modules have at least one connector species (C), except for the one that did not have carnivorous species (see figure 2a). In general, we found that most of the topological roles assigned by the SA procedure presented narrow 95% CIs supporting the robustness of the method (see figure 2b).
As most of the variation in the topological roles was associated to the c-axis, we could only analyse the probability of being a connector-node or not (i.e. a peripheral species) through the logistic regression, whose results (figure 3) show that only body size and trophic status were associated with the probability of being a connector-node. In particular, we found a humped relationship between the probability of being a connector-node and body size. This relationship was significant and also included the trophic status in the whole model (see the electronic supplementary material, table S2). In consequence, in the co-occurrence network herein studied, connector-nodes are more likely to be carnivorous species with intermediated body size (see figure 3, dotted and dashed line).
Figure 3.
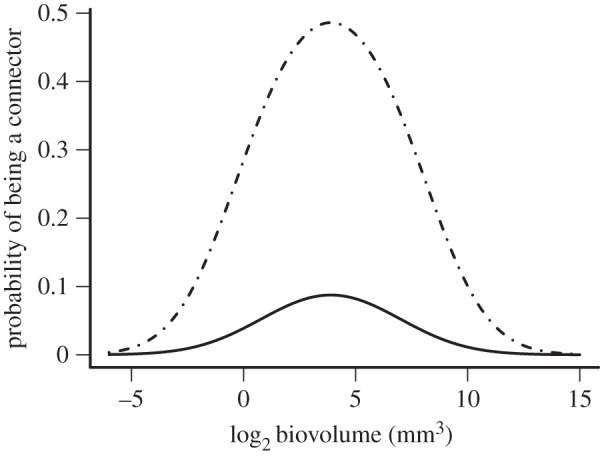
Relationship between the probability of being a connector species and body size. The solid line corresponds to herbivorous species, whereas the dotted and dashed one to carnivorous species. The logistic regression model was statistically significant (see the electronic supplementary material, table S2).
4. Discussion
Community structure has usually been quantified by characterizing species as belonging to guilds, functional groups or taxonomic assemblages [52,53]. The modularity analysis of species co-occurrence networks herein reported clearly identifies subsets of organisms with similar responses to biotic or abiotic conditions and/or mutual interactions that determine their local co-occurrence. These modules could be related to classical categorizations but are not restricted to them [33]. In our system, modules are not composed of species of particular body sizes, taxonomic identity or trophic behaviour. However, our study shows that the connection among modules is related to classical biological traits. In this sense, the modular structure of species occurrences represents an emergent feature of the underlying biological structure.
Closely related to the modular structure of networks is the potential identification of the topological role of species [33,39]. The structure of the co-occurrence network herein analysed, suggests a distribution of roles where only a few species are responsible for the metacommunity level connectivity. This pattern of role distribution [39] has also been described in a wide variety of biological networks including spatial [36,54], mutualistic [33,40,55], food web [35], genetic [34] and metabolic ones [39], supporting the commonality of this network structure. However, few studies have underscored the importance of biological attributes as determinants of species roles (but see [35,55]).
We identified two biological attributes—body size and trophic status—as determinants of the connection among modules of co-occurring species. This result gives support to previous findings that emphasize these biological attributes as determinants of a species' ecological role in metacommunity structure [9,18,22,55]. In this sense, it has been proposed that consumers of large body sizes and higher trophic positions, owing to their higher energetic demands and dispersal ability, could operate as connectors or network hubs in the spatial integration of local communities [9,18,55]. However, intermediate body size species could also play the role of connectors. These species have the lowest environmental requirement to persist in a local community [13–15] and could occupy a wider range of trophic positions [19,22]. This is the case observed in the Atacama Desert metacommunity, where the connector roles are occupied by carnivorous species of intermediate body size (see figure 3) as it was also recently observed in a seed dispersal network (see [55]). In a previous study, we showed for this metacommunity that intermediate-size individuals are distributed in all patches including isolated ones, whereas smaller and larger individuals are more abundant in connected patches (see [12]). Congruently, intermediate-size species able to inhabit isolated and connected patches are here playing the role of connectors of different modules integrating the studied metacommunity of the Atacama ecosystem.
In addition to the effects of body size on efficiency [27,28] and constraints [19] in resource utilization, other associations between body size and population parameters should be considered. Species body size is related to abundance [56], incidence [57], species richness [27] and landscape perception [8]. These associations could be involved in the observed role of intermediate body size species as connectors. However, these associations tend to weaken at local scales [56] and this was the pattern herein observed. In the network analysed herein, abundance and incidence were not associated with body size (electronic supplementary material, figure S1), and therefore these variables are not indirectly determining species roles. The larger richness of species at intermediate body size observed here (electronic supplementary material, figure S2) and elsewhere [27], could determine that independently of body size a random selection of connectors will probably be aggregated at intermediate body sizes. The logistic regression analysis allows discrimination between this random expectation and the occurrence of a significant trend. This statistical method has good performance in analyses with low incidence of positive observations as in this study [58,59] and indicates a significant aggregation of connectors in carnivorous of intermediate body size species. This agrees with our previous finding [12] that carnivorous species of intermediate body size concentrate in well-connected patches; thus suggesting that the landscape structure, through its differential effect on species with different body size and trophic status [12,60], is probably a main determinant of the topological role of species. A main emerging challenge in this context is how the ongoing changes in biodiversity and landscape structure by humans' actions could impact on species roles and community cohesion and functioning.
The relevance of modularity in ecological networks has been discussed for a long time, and no consensus has emerged as yet. In food webs, modularity has been typically used to identify recurring significant patterns of predatory–prey species [16]. In pollination networks, modules correspond to subgroups of related species considered as fundamental functional blocks and candidate for coevolutionary units [33,61,62]. More recently, modularity has been applied to biogeographic networks to detect the impact of geological history upon the identity of species present in different areas [37]. In positive co-occurrence networks—as the one presented here—modules comprise a subgroup of species with likely similar environmental requirements as patch area, heterogeneity or connectivity [12]. It seems that the modular structure of communities is consolidating as a pervasive and recurrent property of community organization from local to metacommunity scales. It should be noted that our study describes the co-occurence network of a metacommunity, considering all macroscopic species found in it. This is important, as recent studies have highlighted that patterns in the modular structure of ecological networks, and explanations thereof, are sensitive to the identity of the species included in the analysis [55], hence the methods used to determine the presence of species.
The use of pitfall traps to assess the presence of terrestrial invertebrates is the most widely used method, but as most methods, it is not exempted of potential biases. What is important in this context is understanding the biases and assessing their impact upon data and analyses [50]. Fortunately, pitfall performance has been repeatedly evaluated and its potential biases are well understood [44]. Pitfall traps favour the collection of active organisms and are sensitive to vegetation structure, weather conditions and variation in attributes of the traps themselves, such as colour, or time since set up [44]. Vegetation structure is exceptionally homogeneous in this study system (monospecific bands of T. landbeckii), local communities were sampled at the same time and the sites show a small variation in weather conditions, traps were identical, new and only used in the present sampling. As a consequence, a systematic bias in the pitfall performance among local communities does not appear as a main matter of concern in this study. This is particularly important because this study is based on the analysis of the relative abundances of species among local communities. Further, the null model herein introduced also accounts for biases related to species variation in activity level. The model is based on assessing deviations from expected co-occurrences based on the null assumption that observed distributions of individuals among local patches are mutually independent. Finally, it should be noted that neither modules nor the topological role of species was related to species abundance—the metric potentially biased by pitfall trapping. Further, the body size of consumers, which was associated with the topological role of species, was not associated with abundance in our study system (electronic supplementary material, figure S1). In summary, the potential biases related to the use of pitfall traps do not undermine the validity of the results herein presented.
The Atacama Desert is one of the most arid ecosystems in the world with an average precipitation per year of less than 2 mm [42]. Fog advection provides the only reliable source of moisture, which determines a spatial self-organization of the dominant vegetation [43]. In this stressful and isolated ecosystem, where most of the structure is bottom-up regulated, it is, not surprising that medium size species, which are considered less energetically restricted, act as connectors. However, in metacommunities open to the arrival of large predators not limited by local energetic constraints, the larger species should play the connector's role [9,18,22]. Our results highlight the need for additional empirical studies on the determinants of species roles within communities and its variation across ecosystems. This is especially important to foresee the consequences of human-driven global change and particularly those associated to the removal and addition of species [63]. In this context, network approaches could provide valuable insights to improve our mechanistic understanding of these ongoing processes and its consequences [64].
Supplementary Material
This study meets the terms of the ethics committee at the Pontificia Universidad Católica de Chile.
Funding statement
We acknowledge support from FONDAP-FONDECYT 1501–0001 to the Center of Advanced Studies in Ecology and Biodiversity, (ICM) P05–002 and Programa de Financiamiento Basal de Conicyt (PFB-023). A.I.B. would like to thank a postdoctoral fellowship CONICYT—FONDECYT N°3130360 (Chile) and a grant from Comisión Sectorial de Investigación Científica (CSIC) 2011–463 (Uruguay). M.A. thanks the grant FCE 2007–054 and FCE-2–2011–1–7117.
References
- 1.Huffaker CB. 1958. Experimental studies on predation: dispersion factors and predator–prey oscillations. Hilgardia 27, 343–384. [Google Scholar]
- 2.Ricklefs RE. 1987. Community diversity: relative roles of local and regional processes. Science 235, 167–171. ( 10.1126/science.235.4785.167) [DOI] [PubMed] [Google Scholar]
- 3.Brown JH, Kodric-Brown A. 1977. Turnover rates in insular biogeography: effect of immigration on extinction. Ecology 58, 445–449. ( 10.2307/1935620) [DOI] [Google Scholar]
- 4.Kneitel JM, Miller TE. 2003. Dispersal rates affect community composition in metacommunities of Sarracenia purpurea inquilines. Am. Nat. 162, 165–171. ( 10.2307/3473207) [DOI] [PubMed] [Google Scholar]
- 5.Keitt TH, Urban DL, Milne BT. 1997. Detecting critical scales in fragmented landscapes. Conserv. Ecol. 1, 4 (http://www.consecol.org/vol1/iss1/art4/) [Google Scholar]
- 6.Wiens JA, Schooley RL, Weeks RD., Jr 1997. Patchy landscapes and animal movements: do beetles percolate? Oikos 78, 257–264. ( 10.2307/3546292) [DOI] [Google Scholar]
- 7.Keymer JE, Marquet PA, Velasco-Hernandez JX, Levin SA. 2000. Extinction thresholds and metapopulation persistence in dynamic landscapes. Am. Nat. 156, 478–494. ( 10.1086/303407) [DOI] [PubMed] [Google Scholar]
- 8.Urban DL, Keitt TH. 2001. Landscape connectivity: a graph-theorethic perspective. Ecology 82, 1205–1218. ( 10.1890/0012-9658(2001)082[1205:LCAGTP]2.0.CO;2) [DOI] [Google Scholar]
- 9.McCann KS, Rasmussen JR, Umbanhowar J. 2005. The dynamics of spatially coupled food webs. Ecol. Lett. 8, 513–523. ( 10.1111/j.1461-0248.2005.00742.x) [DOI] [PubMed] [Google Scholar]
- 10.De Bie T, et al. 2012. Body size and dispersal mode as key traits determining metacommunity structure of aquatic organisms. Ecol. Lett. 15, 740–747. ( 10.1111/j.1461-0248.2012.01794) [DOI] [PubMed] [Google Scholar]
- 11.MacArthur R, Levins R. 1964. Competition, habitat selection, and character displacement in a patchy environment. Proc. Natl Acad. Sci. USA 51, 1207–1210. ( 10.1073/pnas.51.6.1207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borthagaray AI, Arim M, Marquet PA. 2012. Connecting landscape structure and patterns in body size distributions. Oikos 121, 697–710. ( 10.1111/j.1600-0706.2011.19548.x) [DOI] [Google Scholar]
- 13.Schoener TW. 1974. Resource partitioning in ecological communities. Science 185, 27–39. ( 10.1126/science.185.4145.27) [DOI] [PubMed] [Google Scholar]
- 14.Brown JH, Maurer BA. 1989. Macroecology: the division of food and space among species on continents. Science 243, 1145–1150. ( 10.1126/science.243.4895.1145) [DOI] [PubMed] [Google Scholar]
- 15.Brandle M, Ohlschlager S, Brandl R. 2002. Range sizes in butterflies: correlation across scales. Evol. Ecol. Res. 4, 993–1004. [Google Scholar]
- 16.Moore JC, de Ruiter P. 2012. Energetic food webs: an analysis of real and model ecosystems. Oxford, UK: Oxford University Press. [Google Scholar]
- 17.Rooney N, McCann KS, Gellner G, Moore JC. 2006. Structural asymmetry and the stability of diverse food webs. Nature 442, 265–269. ( 10.1038/nature04887) [DOI] [PubMed] [Google Scholar]
- 18.McCann KS. 2012. Food webs. Princeton, NJ: Princeton University Press. [Google Scholar]
- 19.Arim M, Bozinovic F, Marquet PA. 2007. On the relationship between trophic position, body mass and temperature: reformulating the energy limitation hypothesis. Oikos 116, 1524–1530. ( 10.1111/j.2007.0030-1299.15768.x) [DOI] [Google Scholar]
- 20.Brose U, et al. 2006. Consumer-resource body-size relationships in natural food webs. Ecology 87, 2411–2417. ( 10.1890/0012-9658(2006)87[2411:CBRINF]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 21.Otto SB, Rall BC, Brose U. 2007. Allometric degree distributions facilitate food-web stability. Nature 450, 1226–1230. ( 10.1038/nature06359) [DOI] [PubMed] [Google Scholar]
- 22.Arim M, Abades S, Laufer G, Loureiro M, Marquet PA. 2010. Food web structure and body size: trophic position and resource acquisition. Oikos 119, 147–153. ( 10.1111/j.1600-0706.2009.17768.x) [DOI] [Google Scholar]
- 23.Ritchie ME. 2010. Scale, heteogeneity, and the structure and diveristy of ecological communities. Princeton, NJ: Princeton University Press. [Google Scholar]
- 24.Pillai P, Gonzalez A, Loreau M. 2012. Metacommunity theory explains the emergence of food web complexity. Proc. Natl Acad. Sci. USA 108, 19 293–19 298. ( 10.1073/pnas.1106235108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gravel D, Canard E, Guichard F, Mouquet N. 2011. Persistence increases with diversity and connectance in trophic metacommunities. PLoS Biol. 6, e19374 ( 10.1371/journal.pone.0019374) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gravel D, Massol F, Canard E, Mouillot D, Mouquet N. 2011. Trophic theory of island biogeography. Ecol. Lett. 14, 1010–1016. ( 10.1111/j.1461-0248.2011.01667.x) [DOI] [PubMed] [Google Scholar]
- 27.Brown JH, Marquet PA, Taper ML. 1993. Evolution of body size: consequences of an energetic definition of fitness. Am. Nat. 142, 573–584. ( 10.1086/285558) [DOI] [PubMed] [Google Scholar]
- 28.Marquet PA, Taper ML. 1998. On size and area: patterns in mammalian body size extremes across landmasses. Evol. Ecol. 12, 127–139. ( 10.1023/a:1006567227154) [DOI] [Google Scholar]
- 29.Kelt DA, Van Vuren DH. 2001. The ecology and macroecology of mammalian home range area. Am. Nat. 157, 637–645. ( 10.1086/320621) [DOI] [PubMed] [Google Scholar]
- 30.Brandl R, Kristin A, Leisler B. 1994. Dietary niche breadth in a local community of passerine birds: an analysis using phylogenetic contrasts. Oecologia 98, 109–116. ( 10.1007/BF00326096) [DOI] [PubMed] [Google Scholar]
- 31.Layman CA, Winemiller KO, Arrington A, Jepsen DB. 2005. Body size and trophic position in a diverse tropical food web. Ecology 86, 2530–2535. ( 10.1890/04-1098) [DOI] [Google Scholar]
- 32.Costa GC. 2009. Predator size, prey size, and dietary niche breadth relationships in marine predators. Ecology 90, 2014–2019. ( 10.1890/08-1150.1) [DOI] [PubMed] [Google Scholar]
- 33.Olesen JM, Bascompte J, Dupont YL, Jordano P. 2007. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 104, 19 891–19 896. ( 10.1073/pnas.0706375104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fortuna MA, Albaladejo RG, Fernández L, Aparicio A, Bascompte J. 2009. Networks of spatial genetic variation across species. Proc. Natl Acad. Sci. USA 106, 19 045–19 049. ( 10.1073/pnas.0907704106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rezende EL, Albert EM, Fortuna M, Bascompte J. 2009. Compartments in a marine food web associated with phylogeny, body mass, and habitat structure. Ecol. Lett. 12, 779–788. ( 10.1111/j.1461-0248.2009.01327.x) [DOI] [PubMed] [Google Scholar]
- 36.Bellisario B, Cerfolli F, Nascetti G. 2010. Spatial network structure and robustness of detritus-based communities in a patchy environment. Ecol. Res. 25, 813–821. ( 10.1007/s11284-010-0711-5) [DOI] [Google Scholar]
- 37.Carstensen DW, Dalsgaard B, Svenning JCH, Rahbek C, Fjelds J, Sutherland WJ. 2012. Biogeographical modules and island roles: a comparison of Wallacea and the West Indies. J. Biogeogr. 39, 739–749. ( 10.1111/j.1365-2699.2011.02628.x) [DOI] [Google Scholar]
- 38.van de Wolfshaar KE, de Roos AM, Persson L. 2006. Size-dependent interactions inhibit coexistence in intraguild predation systems with life-history omnivory. Am. Nat. 168, 62–75. ( 10.1086/505156) [DOI] [PubMed] [Google Scholar]
- 39.Guimera R, Amaral LAN. 2005. Functional cartography of complex metabolic networks. Nature 433, 895–900. ( 10.1038/nature03288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dupont YL, Olesen JM. 2009. Ecological modules and roles of species in heathland plant–insect flower visitor networks. J. Anim. Ecol. 78, 346–353. ( 10.1111/j.1365-2656.2008.01501.x) [DOI] [PubMed] [Google Scholar]
- 41.Valdovinos FS, Ramos-Jiliberto R, Flores JD, Espinoza C, López G. 2009. Structure and dynamics of pollination networks: the role of alien plants. Oikos 118, 1190–1200. ( 10.1111/j.1600-0706.2009.17364.x) [DOI] [Google Scholar]
- 42.Pinto R, Barría I, Marquet PA. 2006. Geographical distribution of Tillandsia lomas in the Atacama Desert, northern Chile. J. Arid Environ. 65, 543–552. ( 10.1016/j.jaridenv.2005.08.015) [DOI] [Google Scholar]
- 43.Borthagaray AI, Fuentes MA, Marquet PA. 2010. Vegetation pattern formation in a fog-dependent ecosystem. J. Theor. Biol. 265, 18–26. ( 10.1016/j.jtbi.2010.04.020) [DOI] [PubMed] [Google Scholar]
- 44.Ausden M, Drake M. 2006. Invertebrates. In Ecological census techniques (ed. Sutherland WJ.), pp. 214–247. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 45.González AL, Fariña JM, Kay AD, Pinto R, Marquet PA. 2011. Exploring patterns and mechanisms of interspecific and intraspecific variation in body elemental composition of desert consumers. Oikos 120, 1247–1255. ( 10.1111/j.1600-0706.2010.19151.x) [DOI] [Google Scholar]
- 46.Sokal RR, Rohlf FJ. 2011. Biometry: the principles and practices of statistics in biological research, 4th edn New York, NY: W.H. Freeman and Co. [Google Scholar]
- 47.Newman MEJ. 2006. Modularity and community structure in networks. Proc. Natl Acad. Sci. USA 103, 8577–8582. ( 10.1073/pnas.0601602103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichardt J, Bornholdt S. 2006. Statistical mechanics of community detection. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 74, 016110 ( 10.1103/PhysRevE.74.016110) [DOI] [PubMed] [Google Scholar]
- 49.Newman MEJ, Girvan M. 2004. Finding and evaluating community structure in networks. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 69, 026113 ( 10.1103/physreve.69.026113) [DOI] [PubMed] [Google Scholar]
- 50.Hillborn R, Mangel M. 1997. The ecological detective: confronting models with data. Monographs in population biology. Princeton, NJ: Princeton University Press. [Google Scholar]
- 51.Richards SA. 2005. Testing ecological theory using the information-theoretic approach: examples and cautionary results. Ecology 86, 2805–2814. ( 10.1890/05-0074) [DOI] [Google Scholar]
- 52.Morin PJ. 1999. Community ecology. Oxford, UK: Blackwell Science. [Google Scholar]
- 53.Petchey OL, Hector A, Gaston KJ. 2004. How do different measures of functional diversity perform? Ecology 85, 847–857. ( 10.5167/uzh-61819) [DOI] [Google Scholar]
- 54.Fortuna MA, Popa-Lisseanu AG, Ibáñez C, Bascompte J. 2009. The roosting spatial network of a bird-predator bat. Ecology 90, 934–944. ( 10.1890/08-0174.1) [DOI] [PubMed] [Google Scholar]
- 55.Donatti CI, Guimaraes PR, Galetti M, Pizo MA, Mariquitti FMD, Dirzo R. 2011. Analysis of a hyper-diverse seed dispersal network: modularity and underlying mechanisms. Ecol. Lett. 14, 773–781. ( 10.1111/j.1461-0248.2011.01639.x) [DOI] [PubMed] [Google Scholar]
- 56.White EP, Ernest SKM, Kerkhoff AJ, Enquist BJ. 2007. Relationships between body size and abundance in ecology. Trends Ecol. Evol. 22, 323–330. ( 10.1016/j.tree.2007.03.007) [DOI] [PubMed] [Google Scholar]
- 57.Gaston KJ. 2000. Global patterns in biodiversity. Nature 405, 220–227. ( 10.1038/35012228) [DOI] [PubMed] [Google Scholar]
- 58.Hosmer DW, Lemeshow S. 1989. Applied logistic regression. New York, NY: John Wiley and Sons. [Google Scholar]
- 59.Cox DR, Snell EJ. 1989. Analysis of binary data. Monographs on statistics and applied probability, 2nd edn New York, NY: Chapman and Hall. [Google Scholar]
- 60.Borthagaray AI, Barreneche JM, Abades SR, Arim M. 2014. Modularity along organism dispersal gradients challenges a prevailing view of abrupt transitions in animal landscape perception. Ecography 37, 564–571. ( 10.1111/j.1600-0587.2013.00366.x) [DOI] [Google Scholar]
- 61.Jordano P. 1987. Patterns of mutualistic interactions in pollination and seed dispersal: connectance, dependence asymmetries and coevolution. Am. Nat. 129, 657–677. ( 10.1086/284665) [DOI] [Google Scholar]
- 62.Mello MAR, Marquitti FMD, Guimaraes PR, Jr, Kalko EKV, Jordano P, Martinez de Aguiar MA. 2011. The missing part of seed dispersal networks: structure and robustness of bat–fruit interactions. PLoS ONE 6, e17395 ( 10.1371/journal.pone.0017395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.May RM, Lawton JH, Stork NE. 1995. Assessing extinction rates. Extinction rates. Oxford, UK: Oxford University Press. [Google Scholar]
- 64.Barnosky AD, et al. 2012. Approaching a state shift in Earth's biosphere. Nature 486, 52–58. ( 10.1038/nature11018) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.