Abstract
Phylogeographic endemism, the degree to which the history of recently evolved lineages is spatially restricted, reflects fundamental evolutionary processes such as cryptic divergence, adaptation and biological responses to environmental heterogeneity. Attempts to explain the extraordinary diversity of the tropics, which often includes deep phylogeographic structure, frequently invoke interactions of climate variability across space, time and topography. To evaluate historical versus contemporary drivers of phylogeographic endemism in a tropical system, we analyse the effects of current and past climatic variation on the genetic diversity of 25 vertebrates in the Brazilian Atlantic rainforest. We identify two divergent bioclimatic domains within the forest and high turnover around the Rio Doce. Independent modelling of these domains demonstrates that endemism patterns are subject to different climatic drivers. Past climate dynamics, specifically areas of relative stability, predict phylogeographic endemism in the north. Conversely, contemporary climatic heterogeneity better explains endemism in the south. These results accord with recent speleothem and fossil pollen studies, suggesting that climatic variability through the last 250 kyr impacted the northern and the southern forests differently. Incorporating sub-regional differences in climate dynamics will enhance our ability to understand those processes shaping high phylogeographic and species endemism, in the Neotropics and beyond.
Keywords: phylogeographic endemism, climate, forest refugia, biodiversity prediction, Atlantic Forest
1. Introduction
Spatial measures of phylogenetic diversity [1] and weighted endemism [2] have been combined to map phylogenetic endemism (PE): the degree to which units of phylogenetic diversity are spatially restricted [3]. While these analyses portray the concentration of evolutionary history in geographical space and improve biodiversity studies and conservation strategy, relatively little is known about the biological processes that underscore PE and related metrics. Three main mechanisms contribute to PE: lineage differentiation, lineage maintenance through time and range restriction owing to abiotic or biotic factors [4]. Yet, biome-wide studies that investigate their relative roles are still lacking. These issues are particularly relevant in tropical regions, where deep phylogeographic structuring and concomitant high genetic endemism are prevalent [5,6]. Hypotheses to explain high diversity and endemism in the tropics frequently invoke climatic heterogeneity in space [7] and time [8], often associated with the expectation that tropical species have restricted physiological tolerances [9,10].
We focus on the processes of lineage maintenance and range restriction and ask how environmental shifts in the Late Quaternary (last 120 000 years or 120 kyr) influenced present-day patterns of genetic diversity and endemism in the Atlantic rainforests of Brazil. To understand how the distribution of genetic variation has been impacted by recent history, we develop a new measure of endemism that is based on the tips of the Tree of Life—phylogeographic lineages. Differently from previous usages of higher rank phylogenies in studies of PE and genetic diversity [3,11], the metric is not affected by branch lengths and tree topology at deep phylogenetic levels (e.g. genera or higher), which may reflect processes older than those of interest [12]. This new measure of fine-scale PE (phylogeographic endemism hereafter) describes how much the history of recently evolved lineages is spatially restricted. This depends on the range of the lineage, how much of its history is shared with closely related lineages and how widespread the latter are.
We expect both past and present-day climate to influence lineage range and hence phylogeographic endemism. Former climate dynamics should be a strong predictor of phylogeographic endemism patterns because of their direct and indirect impacts on the three main underlying processes [13,14]. First, environmental barriers may have promoted vicariance and genetic divergence [7,15]. Second, the location of historical climatic refugia (areas of relative habitat stability), particularly over the Late Quaternary, are tied to maintenance and accumulation of species or lineage diversity [16,17]. Lastly, past climates may have played a role in restricting contemporary lineage range by preventing access to areas that are currently suitable for species occurrence. Yet, the influence of contemporary conditions should not be overlooked. Climatic heterogeneity (now and in the past) directly affects the distribution of lineages through species-specific physiological constraints (e.g. low metabolic rates and performance [18]), as well as variation in the availability of energy and water (the productivity–diversity relationship [19]). Indirectly, climates also affect species distributions through suitability to parasites, pathogens, predators or competitors [20].
To test whether climate-based distributions of habitats both now and over the Late Quaternary accurately predict patterns of phylogeographic endemism, we combine multi-species phylogeographic data with spatial modelling of habitats to provide a high-resolution comparison within the hyperdiverse Brazilian Atlantic Forest (AF hereafter). Specifically, we investigate the roles of climate change and forest stability over the last 21 and 120 kyr (potentially impacting lineage maintenance through time), as well as current climatic heterogeneity (measured by net primary productivity (NPP) and a potential driver of lineage range restriction based on contemporary climatic conditions), on phylogeographic endemism. Although fine-scale palaeoecological data demonstrate that climate and forest community composition have been highly dynamic in the AF [21], we use the term ‘forest stability’ as a relative measure of the extent to which a particular grid cell has been continuously occupied by a forest-type environment, irrespective of plant community composition.
2. Material and methods
We used Biodiverse [22] to map phylogeographic endemism across the Atlantic forest based on new and published mitochondrial sequence-level data from 25 vertebrate species or closely related species groups (electronic supplementary material, methods, tables S1 and S2). As we are interested in the distribution of genetic lineage diversity and it is somewhat arbitrary whether lineages are recognized taxonomically (typically depending on whether divergence is morphologically cryptic or diagnosable), we include phylogeographic data both within-species (n = 19) and across very closely related species (n = 06). More than 90% of the data come from amphibians and reptiles, low-dispersal species for which the impacts of environmental change are expected to be the strongest. To calculate phylogeographic endemism, the length of each branch on the genealogy is divided equally across the grid cells in which it or its descendant lineages occur. These adjusted branch lengths are then summed for each cell, as described in [3]. High scores therefore are allocated to regions where a large component of phylogenetic variation is restricted to a small area.
Because Biodiverse [22] requires species presence data for all grid cells to estimate endemism, we used geo-referenced information from each species to generate correlative models of species distribution (electronic supplementary material, methods and dataset S1). Lineage-specific ranges were defined by superimposing the geo-referenced haplotype data on the genealogical trees. The molecular data were represented in Biodiverse [22] as a single, composite phylogeny that includes the within-species genetic information for all species in the study. To focus on recent evolutionary history, we incorporated only those well-supported clades identified within each species (more than 80% maximum-likelihood bootstrap support) and removed all other (higher level) phylogenetic information from our analyses (electronic supplementary material, figure S1). Nuclear data available from 19 target species or species complexes (two widespread, 10 in the south, seven in the north) demonstrate that several (or, in two cases of comprehensive nuclear DNA sampling, all) mtDNA lineages represent independently evolving organismal lineages ([23–27] and R. Damasceno, C. Firkowski, I. Prates, D. Rivera, M. Strangas 2014, unpublished data).
To address whether phylogeographic endemism patterns can be explained by inferred shifts of the forest climate through time, we built correlative models of forest distribution under present-day conditions and projected them onto past climates at 4 kyr intervals extending through a full glacial cycle (120 kyrs), as well as at 1 kyr intervals back to the last glacial maximum (LGM). Because phylogeographic data gathered from AF species with distinct environmental requirements (e.g. lowland versus montane specialists) indicate that preliminary palaeomodels of the forest [15] were inefficient at explaining differential historical responses to climate change across different species pools [23,28], we identified and independently modelled distinct environmental envelopes occupied by the biome. For that, we used simple ordination methods to delimit unique climate spaces within the forest (electronic supplementary material, methods). Because studies of humid air circulation in the AF suggest the existence of two distinct climatological units (a northern forest characterized by rainy winters and a southern forest with rainy summers [29]), we implemented a K-means cluster analysis to our PCA plots by setting the algorithm to identify two distinct climatic spaces. Once the climatic uniqueness of these two forest spaces was verified (see Results), we generated separate correlative models of forest distribution based on randomly extracted points from each of the two climatic regions.
To estimate the impact of historical climate change on forest persistence, we first developed present-day models of the distribution of the entire forest, as well as of each PCA-delimited forest space. As per recent studies [30], we projected these models to snapshot climatic simulations covering the last 120 kyr using the Hadley Centre Climate model (HadCM3 [30,31]), for which climatic reconstructions are available at small (1 or 4 kyr) time intervals, enabling a more continuous and dynamic view of forest shifts through time (electronic supplementary material, methods). Using Viterbi's algorithm [32] to combine predictions of past distributions of the forest across time intervals, we estimated historical, climate-based forest stability per pixel while allowing for 0, 5 and 10 m forest dispersal per year [33]. These maps are henceforth referred to as dynamic refugia maps, and the forest stability values obtained per grid cell were used in our predictive model as proxies for the potential to maintain lineages locally through periods of climate fluctuation.
To evaluate how present-day climatic heterogeneity potentially restricts the distribution of our target lineages and affects patterns of endemism, we applied K-means clustering to a PCA analysis based on values of mean temperature, mean precipitation and annual temperature seasonality from random forest localities. With this, we delimited the largest possible number of distinct, diagnosable climatic spaces within the forest. We then calculated the area of each polygon, or uniquely recognized climatic space, under contemporary conditions and applied this value to all grid cells within each respective polygon as a proxy for contemporary lineage restriction potential.
To assess the relative effects of historical and present-day climate heterogeneity on phylogeographic endemism, we examined correlations between per grid cell values of phylogeographic endemism and scores of forest stability over 120 kyr, forest stability over 21 kyr, the total area of the contemporary climatic space occupied by each cell (potential for lineage restriction in geographical space) and NPP [34] per cell. We used the software Spatial Analyses in Macroecology (SAM [35]) to calculate correlation coefficients, testing for significance after correcting for spatial autocorrelation. Additionally, we used SAM's partial least-squares regression analysis to determine coefficients of determination (r2) and examine the relative explanatory power of historical versus contemporary climatic diversity in predictions of phylogeographic endemism.
3. Results
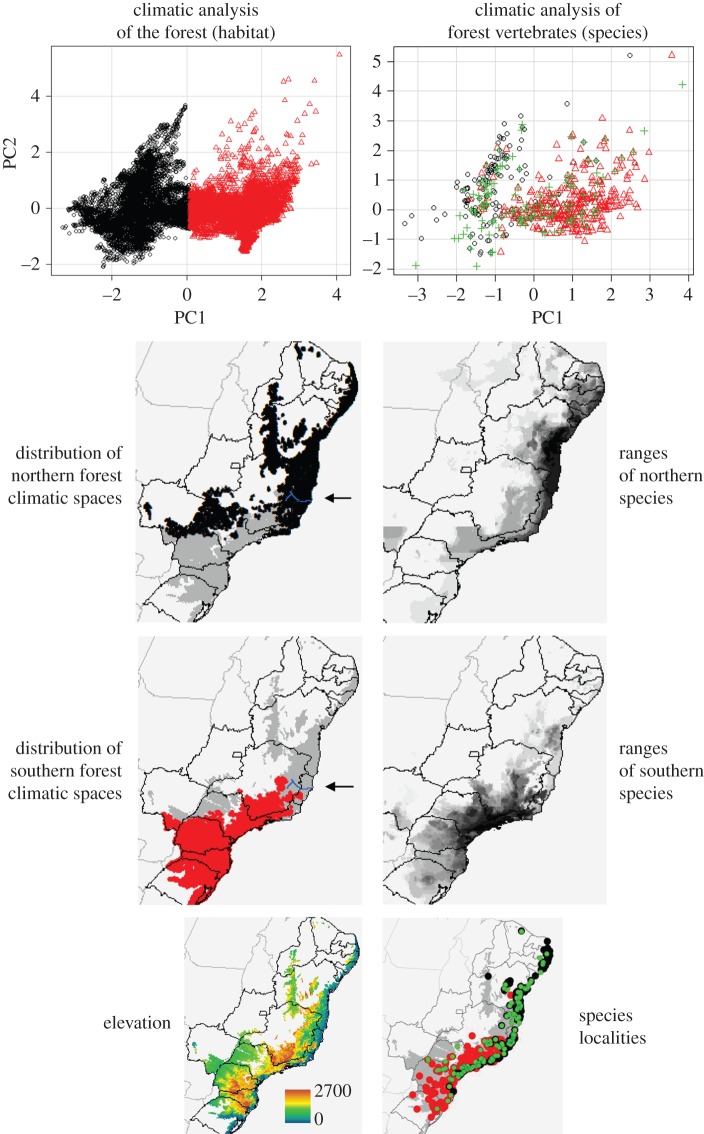
A PCA-based clustering of the AF environmental space demonstrates the distinctiveness of the climatic envelopes occupied by the lowland and mid-elevation forests of the north (above 20°S) relative to the generally cooler and higher elevation forests of southern and southeastern Brazil (figure 1). Climate niche analyses [36] further demonstrate that the environmental spaces occupied by these northern and southern components of the forest are significantly less similar to each other than expected by chance (p < 0.01). This environmental shift begins a few kilometres south of the Rio Doce and is matched by strong biological turnover, as shown by the PCA plots of the climatic spaces occupied by all species included in the study (figure 1, left). Following this PC-based classification, nine species can be categorized as having a mostly northern (lowland, mid-elevation) distribution, whereas 12 have a southern (montane) distribution (four taxa are widespread, figure 1; see the electronic supplementary material, table S1, for classifications). The level of agreement between the geographical limits of these two species assemblages and the northern and southern climatic spaces of the forest is evident (figure 1).
Figure 1.
Left column: PCA-based identification of two climatically distinct spaces within the AF (top graph, black and red points), followed by maps of their respective northern (black) and southern (red) geographical ranges. In both maps, the areas in grey indicate the remaining extent of the forest; the location of the Rio Doce is shown with an arrow. Right column: PCA-based identification of species assemblages along the climatic axes of the AF (top), identifying the environmental spaces occupied by those taxa with northern (black), southern (red) or widespread (green) distributions. The graph is followed by maps of the superimposed ranges of the northern and southern species, respectively. In both maps, darker shades of grey represent pixels with greater species richness, as per superimposed correlative species distribution models. An elevation map is provided at the bottom of the figure (left), followed by a map of point localities of all species (right), shown over a grey mask of the AF. The colour scheme used for the point localities follows that of the PCA analysis (northern species shown in black, southern taxa in red, widespread species in green).
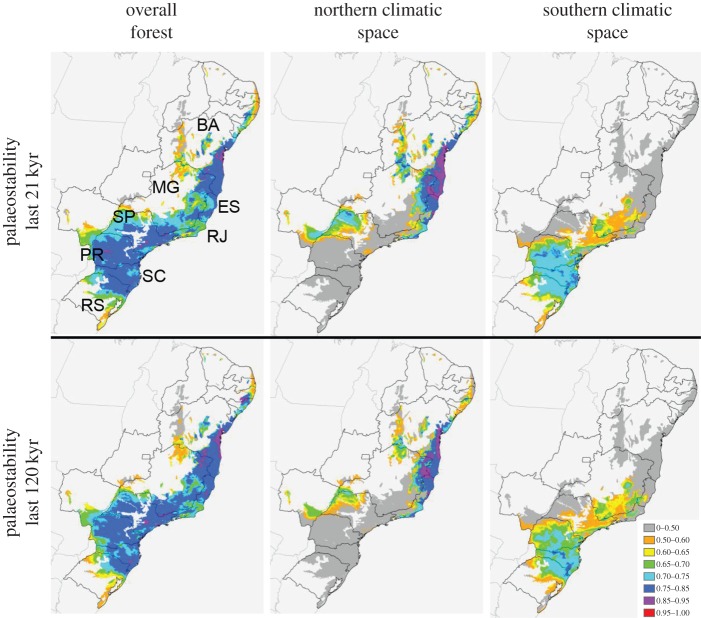
Dynamic refugia maps generated independently for the northern and southern climatic spaces of the forest are also non-overlapping regardless of the time period modelled, as expected given marked environmental turnover (figure 2; electronic supplementary material, figure S2). When forest refugia are estimated under 0 m forest dispersal per year (static forest refugia [33]), the forest stability levels inferred over the last 21 kyr are strikingly similar to those calculated over the last 120 kyr (figure 2). When forest dispersal is allowed at 5 and 10 m per year (dynamic forest refugia [33]), the inferred level of forest stability is considerably larger for calculations going back 120 kyr relative to 21 kyr (electronic supplementary material, figure S2).
Figure 2.
Forest stability models over the last 21 kyr (above) and 120 kyr (below), assuming zero forest dispersal over time (static forest refugia). Legend depicts suitability values, calculated as the sum of negative log probabilities of forest occurrence in each time period. Left panel depicts predictions when the forest is modelled as a whole. Central and right panels portray predictions for the northern and southern climatic spaces of the forest, respectively. State boundaries are included in maps; BA, Bahia; ES, Espírito Santo; MG, Minas Gerais; PR, Paraná; RJ, Rio de Janeiro; RS, Rio Grande do Sul; SC, Santa Catarina; SP, São Paulo. For models that assume forest dispersal over time, refer to the electronic supplementary material, figure S2.
Akin to the Carnaval–Moritz model [37], climatic suitability for forest occurrence over time is high in the northern forests, across Bahia, northeastern Minas Gerais and Espírito Santo. Moreover, the inferred stability of forest distributions is higher in the north than the south when the two forest climatic spaces are modelled independently (figure 2). However, both the overall forest model and the southern climate space model suggest higher levels of forest stability in the south relative to previous hypotheses [37] (figure 2). Particularly notable are the regions of high relative stability of forest distributions in the highlands of the states of São Paulo, Paraná and Santa Catarina. Dynamic refugia maps, allowing for forest dispersal across time periods, also support this (electronic supplementary material, figure S2).
Applying K-means clustering to a PCA analysis of the forest climatic space, we found the upper limit of K that allows for the recognition of the largest number of climatically discrete spaces to be 14 (electronic supplementary material, figure S3). We therefore used the geographical area of each uniquely recognized climatic space as a conservative proxy for present-day potential for lineage restriction in space.
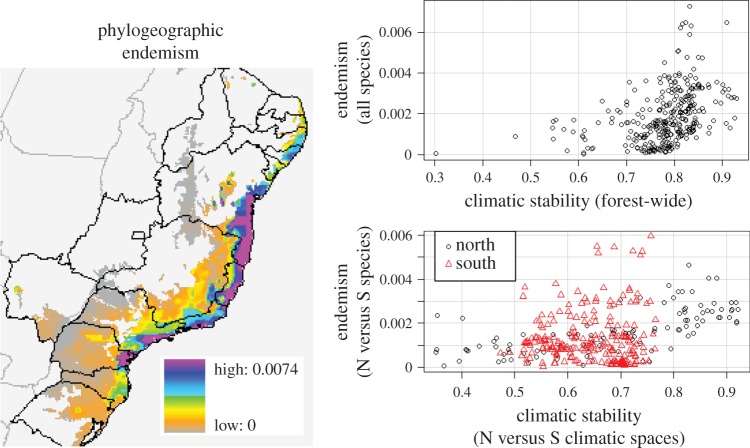
Across the 25 species examined, we identified 109 major phylogeographic lineages from mtDNA analyses (electronic supplementary material, methods and figure S1). Phylogeographic endemism for these taxa is unarguably high across the forest. Relatively smaller values were estimated for the northernmost and southernmost edges of the biome, yet samples from these localities are available only from a relatively small number of species (figures 1 and 3). Across the entire biome, higher levels of phylogeographic endemism are observed in areas of higher forest stability over the last 21 kyr (r [overall PE, 21 kyr stability] = 0.265, corrected p = 0.033; table 1). The same is detected if stability values span the last 120 kyr (r [overall PE, 120 kyr stability] = 0.387, corrected p = 0.007; table 1 and figure 3). More importantly for model verification purposes, no region with high phylogeographic endemism shows low forest stability values. High phylogeographic endemism is more frequently observed in grid cells located within climatic spaces that are currently small-ranged (r [overall PE, area] = −0.243, p = 0.033; table 1), attesting to the potential role of present-day climatic heterogeneity in restricting lineage ranges. Phylogeographic endemism is also positively correlated with NPP (r [overall PE, NPP] = 0.415, p = 0.001; table 1).
Figure 3.
Phylogeographic endemism map based on available data from all AF species included in this study (left), and the relationship between phylogeographic endemism and forest stability over the last 120 kyr (right). The top graph describes how overall phylogeographic endemism relates to forest stability when the AF is modelled as a whole. The bottom graph portrays phylogeographic endemism of the northern species (black circles) and of the southern species (red triangles) in relation to forest stability values of the northern and southern climatic spaces, respectively. Stability values represent the sum of negative log probabilities of forest occurrence in each time period. Similar results are observed when values are estimated over the last 21 kyr (see the electronic supplementary material, figure S4).
Table 1.
Correlation analyses between phylogeographic endemism and potential contemporary and historical determinants. (Present-day variables include the geographical extent (area) of unique climatic spaces and NPP. Historical determinants are climate-based forest stability values over the last 21 and 120 kyr. Correlations are examined across the entire forest (top rows), as well as the northern (middle rows) and the southern (bottom rows) climatic spaces, in isolation. Significant variables (p < 0.05) are italicized.)
| phylogeographic endemism | variables | r | p |
|---|---|---|---|
| overall | area | −0.243 | 0.033 |
| NPP | 0.415 | 0.001 | |
| 120 kyr stability | 0.387 | 0.007 | |
| 21 kyr stability | 0.265 | 0.033 | |
| north | area | 0.097 | 0.595 |
| NPP | 0.076 | 0.670 | |
| 120 kyr stability | 0.677 | 0.019 | |
| 21 kyr stability | 0.642 | 0.04 | |
| south | area | −0.397 | 0.01 |
| NPP | 0.467 | 0.001 | |
| 120 kyr stability | 0.069 | 0.733 | |
| 21 kyr stability | −0.001 | 0.995 |
However, the strengths of the correlations differ significantly between the north and the south. Whereas historical climate dynamics (forest stability) predict phylogeographic endemism in the north, contemporary climatic heterogeneity is a stronger predictor in the south. More specifically, phylogeographic endemism of the northern taxa is correlated with forest stability over the last 120 and 21 kyr (r [N PE, 120 kyr stability] = 0.677, p = 0.019; r [N PE, 21 kyr stability] = 0.642, p = 0.04; table 1 and figure 3), but is not correlated with either the geographical extent of current climatic spaces (r[N PE, area] = 0.097, p = 0.595; table 1) or NPP (r [N PE, NPP] = 0.076, p = 0.670). By contrast, phylogeographic endemism of the southern taxa is not correlated with spatial patterns of southern forest stability (r [S PE, 120 kyr stability] = 0.069, p = 0.733; r [S PE, 21 kyr stability] = −0.001, p = 0.995; table 1 and figure 3), yet it is significantly negatively correlated with the geographical extent of current climatic spaces (r [S PE, area] = −0.397, p = 0.01; table 1) and positively correlated with NPP (r [S PE, NPP) = 0.467, p = 0.001; table 1). These correlations hold when values are estimated through a dynamic refugia approach, allowing for forest dispersal over time, for both the overall forest and southern climatic spaces (electronic supplementary material, methods, table S3 and figure S4). The correlation between historical forest stability and phylogeographic endemism in the north nonetheless weakens as forest dispersal is included in the analysis, becoming marginally significant over the 21 kyr period and non-significant over the last 120 kyr period (electronic supplementary material, table S3 and figure S4).
Partial least-squares regression analyses based on overall phylogeographic endemism and stability values of the forest modelled as a single entity suggest that both present-day and historical climates, together, best explain phylogeographic endemism (r² [overall PE, 120 kyr stability + area + NPP] = 0.303, Akaike information criterion (AIC) weight = 0.706; electronic supplementary material, table S4). Independent analyses of northern-only and southern-only climatic spaces of the forest, and the respective northern and southern species, also confirm the previously observed correlation trends and indicate that distinct processes impact the distribution of phylogeographic endemism in each region. In the north, historical climate change better predicts local phylogeographic endemism patterns (r² [N PE, N 120 kyr stability] = 0.458; AIC weight 0.459; electronic supplementary material, table S4). Conversely, in the south, current climatic conditions (range restriction and NPP) explain endemism much more effectively than former climates (r² [S PE, area + NPP] = 0.264, AIC weight 0.736; electronic supplementary material, table S4). Major predictors of phylogeographic endemism also differ between the north and south when values are estimated through a dynamic refugia approach that allows for forest dispersal over time (electronic supplementary material, table S5).
4. Discussion
Identifying how current and historical climate variation has shaped diversity within Neotropical (and other hyperdiverse) biomes remains a challenge. Through an ordination-based analysis of the AF climate, we identified two broadly different climatic regimes—one mostly distributed in the north, the other in the southern areas of the biome. The transition between these climatic spaces happens roughly around the Rio Doce—a known biogeographic divide [38,39]. This major bioclimatic transition also matches patterns of species turnover between the northern and the southern portions of the AF based on locality data for 25 vertebrate species (figure 1), published observations of precipitation seasonality discontinuities [29], as well as faunal changes revealed by panbiogeographical analyses [40]. This demonstrates that the geographical patterns of environmental shifts along the AF are intimately linked to the current and former distribution of its fauna and flora, and may suggest a climate-physiological basis, rather than river-associated vicariance [41], for the observed patterns of diversity and endemism.
By independently modelling these two bioclimatic domains within the AF, we are able to generate predictions of climate-based forest refugia and accumulation of unique lineages that match phylogeographical patterns across distinct species pools. Climatic refugia in the lowland and mid-elevation forests detected in this study are predicted to have been larger in the north relative to the south, in agreement with the Carnaval–Moritz [37] model and the historical scenarios of demographic change inferred from phylogeographic data for widely ranged species [17,42]. Yet, these new models of dynamic refugia of the southern forests, which have a strong montane and sub-tropical component, match the recently accumulated phylogeographic data for seemingly cold-associated taxa [23,28]. This supports the notion that microrefugia [43] played an important role in maintaining diversity over time in the highlands and higher latitudes of the AF. The models also highlight the existence of forest refugia in poorly sampled regions of southern Brazil (e.g. in the Serra Geral, in Paraná and Santa Catarina). We predict that targeted surveys will uncover yet undescribed species and cryptic lineages in those areas, similarly to the findings of recent inventories in the forests of Bahia [44].
The results unequivocally show that the southern and northern forests in Brazil, as well as the endemism patterns of its assemblages, are subject to different current and palaeoclimatic drivers. Because improvement in biodiversity prediction is here observed through the introduction of the smallest possible amount of complexity (i.e. splitting the forest into just two bioclimatic domains), the recognition of additional environmentally distinct regions may further increase the explanatory power of habitat and refugia models, particularly in biomes more complex than the one we target. The incorporation of spatially explicit models of lineage differentiation, guided by well-documented topographic changes of the Brazilian relief and validated with time-calibrated genealogies, shall further advance prediction of phylogeographic endemism in the biome.
Correlation analyses and regression models demonstrate that spatial patterns of phylogeographic endemism are closely linked to both historical climate dynamics (through relative forest stability) and current climatic drivers of lineage ranges and productivity (table 1; electronic supplementary material, tables S4 and S5). The results show that forest persistence over time is a necessary condition, although not sufficient, for high phylogeographic endemism. Nowhere was high phylogeographic endemism observed in a region of low forest stability as per the new predictive models. Yet, the analyses demonstrate that the contemporary (interglacial) conditions impact the distribution of evolutionary history in the southern taxa more strongly.
Whether historical or current climatic drivers of lineage ranges will more strongly determine endemism patterns in this biome or others depends, we suggest, on a combination of two factors: the general environmental tolerances of the focal biological assemblages and the current phase of the climate cycle. In the AF, these aspects may explain why the balance between contemporary and past drivers of phylogeographic endemism differs between the north and the south. Cold-associated (largely southern) taxa are now in the range-contracted part of their climate cycle, and thus strongly constrained by the interglacial environment that we now experience. Yet for the low- and mid-altitude taxa, several of which are widespread or present in the northern forests and whose ranges are fairly expanded given today's environment, historical climate conditions (especially the LGM) are more effective at explaining phylogeographic endemism because they better reflect past (contracted) ranges of the biota. Comparative physiological data from lineages pertaining to these distinct species pools will shed light on the mechanisms underlying the strong correlative patterns we detect and the biotic processes here proposed.
These results are well integrated with recently gathered speleothem and pollen fossil data, suggesting that orbital- and millennial-scale climatic variability through the last 250 kyr impacted the northern and the southern regions of the AF differently: while precipitation patterns in the northern forests have been in phase with those of Eastern Amazonia, those in the south matched that of Western Amazonia and the Andean forests [45]. As suggested by the palaeoclimatic evidence, these two large systems have been acting as a dipole: climatic conditions in the northern AF and Eastern Amazonia were similarly drier in the LGM relative to today's climate, and generally wetter in the Mid-Holocene, whereas the opposite occurred in the southern AF, Western Amazonia and the Andes [46]. These differential mechanistic links affecting the southern and northern regions of the forest are consistent with emerging biological data that indicate distinct histories of interconnection between northern and southern AF species with other South American forests [47]. More broadly, they reinforce our conclusion that the incorporation of sub-regional differences in contemporary and former climates is relevant to and improves overall prediction of endemism and historical demography across taxa with radically different environmental correlates. We expect that more nuanced climatic analyses of other large complex biomes (e.g. sub-Saharan Africa [48], Amazonia [49] and Neotropical savannahs [50]) will help advance biodiversity prediction and conservation in tropical hotspots worldwide.
Supplementary Material
Acknowledgements
M. Teixeira Jr., R. Recoder, F. Dal Vechio, A. Camacho, J. Cassimiro, M. Sena and R. Amaro assisted with the collection of vouchers and tissues. S. Baroni, M. Miceno, S. Geurgas, L. Zeidler, M. Cheu and M. Lyra provided laboratory assistance or contributed with small sequence datasets. A subset of the tissues of Vitreorana species were obtained from the Celio F. B. Haddad Herpetological Collection.
Data accessibility
DNA sequences: GenBank accession nos. KM204320–KM204375. Climate data and correlative model output data: Dryad doi: (doi:10.5061/dryad.8kc1v).
Funding statement
This work profited from interactions enabled by the NESCent Montane Biodiversity Working Group and used data generated through National Science Foundation awards to A.C. and C.M. (DEB-817035, DEB-1035184, DEB-1120487) and FAPESP grants to M.R. (2003/10335-8 and 2011/50146-6). R.D. was supported by CAPES-Fullbright (BEX-2740/06-0) FAPESP 213/22477-3. This work is partially co-funded by FAPESP (BIOTA, 2013/50297-0), NSF (DEB 1343578) and NASA, through the Dimensions of Biodiversity Program.
References
- 1.Faith DP. 1992. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10. ( 10.1016/0006-3207(92)91201-3) [DOI] [Google Scholar]
- 2.Williams PH, Humphries CJ. 1994. Biodiversity, taxonomic relatedness, and endemism in conservation. In Systematics and conservation evaluation (eds Forey PL, Humphries CJ, Vane-Wright RI.), pp. 269–287. Oxford, UK: Oxford University Press. [Google Scholar]
- 3.Rosauer DF, Laffan SW, Crisp MD, Donnellan SC, Cook LG. 2009. Phylogenetic endemism: a new approach to identifying geographical concentrations of evolutionary history. Mol. Ecol. 18, 4061–4072. ( 10.1111/j.1365-294X.2009.04311.x) [DOI] [PubMed] [Google Scholar]
- 4.Pressey RL, Johnson IR, Wilson PD. 1994. Shades of irreplaceability: towards a measure of the contribution of sites to a reservation goal. Biodivers. Conserv. 3, 242–262. ( 10.1007/BF00055941) [DOI] [Google Scholar]
- 5.Weir JT, Schluter D. 2007. The latitudinal gradient in recent speciation and extinction rates of birds and mammals. Science 315, 1574–1576. ( 10.1126/science.1135590) [DOI] [PubMed] [Google Scholar]
- 6.Moritz C, Patton JL, Scheider CJ, Smith TB. 2000. Diversification of rainforest faunas: an integrated molecular approach. Annu. Rev. Ecol. Syst. 31, 533–563. ( 10.1146/annurev.ecolsys.31.1.533) [DOI] [Google Scholar]
- 7.Janzen DH. 1967. Why mountain passes are higher in the tropics. Am. Nat. 101, 233–249. ( 10.1086/282487) [DOI] [Google Scholar]
- 8.Haffer J. 1969. Speciation in Amazonian forest birds. Science 165, 131–137. ( 10.1126/science.165.3889.131) [DOI] [PubMed] [Google Scholar]
- 9.Addo-Bediako A, Chown SL, Gaston KJ. 2000. Thermal tolerance, climatic variability and latitude. Proc. R. Soc. Lond. B 267, 739–745. ( 10.1098/rspb.2000.1065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunday JM, Bates AE, Dulvy NK. 2010. Global analysis of thermal tolerance and latitude in ectotherms. Proc. R. Soc. B 278, 1823–1830. ( 10.1098/rspb.2010.1295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vieites DR, Wollenberg KC, Andreone F, Köhler J, Glaw F, Vences M. 2009. Vast underestimation of Madagascar's biodiversity evidenced by an integrative amphibian inventory. Proc. Natl Acad. Sci. USA 106, 8267–8272. ( 10.1073/pnas.0810821106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosauer DF, Ferrier S, Williams KJ, Manion G, Keogh JS, Laffan SW. 2013. Phylogenetic generalised dissimilarity modelling: a new approach to analysing and predicting spatial turnover in the phylogenetic composition of communities. Ecography 36, 1–12. ( 10.1111/j.1600-0587.2012.07785.x) [DOI] [Google Scholar]
- 13.Dynesius M, Jansson R. 2000. Evolutionary consequences of changes in species’ geographical distributions driven by Milankovitch climate oscillations. Proc. Natl Acad. Sci. USA 97, 9115–9120. ( 10.1073/pnas.97.16.9115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ricklefs RE. 2006. Evolutionary diversification and the origin of the diversity environment relationship. Ecology 87, S3–S13. ( 10.1890/0012-9658(2006)87[3:EDATOO]2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 15.Hewitt GM. 1996. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol. J. Linn. Soc. 58, 247–276. ( 10.1111/j.1095-8312.1996.tb01434.x) [DOI] [Google Scholar]
- 16.Araújo MB, Nogués-Bravo D, Diniz-Filho JAF, Haywood AM, Valdes PJ, Rahbek C. 2008. Quaternary climate changes explain diversity among reptiles and amphibians . Ecography 31, 8–15. ( 10.1111/j.2007.0906-7590.05318.x) [DOI] [Google Scholar]
- 17.Carnaval AC, Moritz C, Hickerson M, Haddad C, Rodrigues M. 2009. Stability predicts diversity in the Brazilian Atlantic Forest hotspot. Science 323, 785–789. ( 10.1126/science.1166955) [DOI] [PubMed] [Google Scholar]
- 18.Navas CA. 2002. Herpetological diversity along Andean elevational gradients: links with physiological ecology and evolutionary physiology. Comp. Biochem. Physiol. A. Mol. Integr. Physiol. 133, 469–485. ( 10.1016/S1095-6433(02)00207-6) [DOI] [PubMed] [Google Scholar]
- 19.Hawkins BA, et al. 2003. Energy, water, and broad-scale geographic patterns of species richness. Ecology 84, 3105–3117. ( 10.1890/03-8006) [DOI] [Google Scholar]
- 20.Ricklefs RE. 2010. Evolutionary diversification, coevolution between populations and their antagonists, and the filling of niche space. Proc. Natl Acad. Sci. USA 107, 1265–1272. ( 10.1073/pnas.0913626107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montade V, Ledru MP, Burte J, Martins ESPR, Verola CF, Costa IR, Magalhães e Silva FH. 2014. Stability of a Neotropical microrefugium during climatic instability. J. Biogeogr. 41, 1215–1226. ( 10.1111/jbi.12283) [DOI] [Google Scholar]
- 22.Laffan SW, Lubarsky E, Rosauer DF. 2010. Biodiverse, a tool for the spatial analysis of biological and related diversity. Ecography 33, 643–647. ( 10.1111/j.1600-0587.2010.06237.x) [DOI] [Google Scholar]
- 23.Amaro RC, Yonenaga-Yassuda Y, Rodrigues MT, Carnaval AC. 2012. Demographic processes in the Montane Atlantic rainforest: molecular and cytogenetic evidence from the endemic frog Proceratophrys boiei. Mol. Phylogenet. Evol. 62, 880–888. ( 10.1016/j.ympev.2011.11.004) [DOI] [PubMed] [Google Scholar]
- 24.Brunes TO, Sequeira F, Haddad CF, Alexandrino J. 2010. Gene and species trees of a Neotropical group of treefrogs: genetic diversification in the Brazilian Atlantic Forest and the origin of a polyploid species. Mol. Phylogenet. Evol. 57, 1120–1133. ( 10.1016/j.ympev.2010.08.026) [DOI] [PubMed] [Google Scholar]
- 25.Cabanne GS, d'Horta FM, Sar EH, Santos FR, Miyaki CY. 2008. Nuclear and mitochondrial phylogeography of the Atlantic forest endemic Xiphorhynchus fuscus (Aves: Dendrocolaptidae): biogeography and systematics implications. Mol. Phylogenet. Evol. 49, 760–773. ( 10.1016/j.ympev.2008.09.013) [DOI] [PubMed] [Google Scholar]
- 26.Carnaval ACOQ, Bates JM. 2007. Amphibian DNA shows marked genetic structure and tracks Pleistocene climate change in northeastern Brazil. Evolution 61, 2942–2957. ( 10.1111/j.1558-5646.2007.00241.x) [DOI] [PubMed] [Google Scholar]
- 27.Thomé MTC, Zamudio KR, Giovanelli JG, Haddad CF, Baldissera FA, Alexandrino J. 2010. Phylogeography of endemic toads and post-Pliocene persistence of the Brazilian Atlantic Forest. Mol. Phylogenet. Evol. 55, 1018–1031. ( 10.1016/j.ympev.2010.02.003) [DOI] [PubMed] [Google Scholar]
- 28.Batalha-Filho H, Cabanne GS, Miyaki CY. 2012. Phylogeography of an Atlantic forest passerine reveals demographic stability through the last glacial maximum. Mol. Phylogenet. Evol. 65, 892–902. ( 10.1016/j.ympev.2012.08.010) [DOI] [PubMed] [Google Scholar]
- 29.Grimm AM. 2003. The El Nino impact on the summer monsoon in Brazil: regional processes versus remote influences. J. Clim. 16, 263–280. () [DOI] [Google Scholar]
- 30.Fuchs J, Parra JL, Goodman SM, Raherilalao MJ, Vanderwal J, Bowie RC. 2013. Extending ecological niche models to the past 120 000 years corroborates the lack of strong phylogeographic structure in the crested drongo (Dicrurus forficatus forficatus) on Madagascar. Biol. J. Linn. Soc. Lond. 108, 658–676. ( 10.1111/j.1095-8312.2012.02022.x) [DOI] [Google Scholar]
- 31.Singarayer JS, Valdes PJ. 2010. High-latitude climate sensitivity to ice-sheet forcing over the last 120 kyr. Q. Sci. Rev. 29, 43–55. ( 10.1016/j.quascirev.2009.10.011) [DOI] [Google Scholar]
- 32.Viterbi AJ. 1967. Error bounds for convolutional codes and an asymptotically optimum decoding algorithm. IEEE Trans. Inf. Theor. 13, 260–269. ( 10.1109/TIT.1967.1054010) [DOI] [Google Scholar]
- 33.Graham CH, VanDerWal J, Phillips S, Williams SE, Moritz C. 2010. Dynamic refugia and species persistence: tracking spatial shifts in habitat through time. Ecography 33, 1062–1069. ( 10.1111/j.1600-0587.2010.06430.x) [DOI] [Google Scholar]
- 34.Zhao M, Running SW. 2010. Drought-induced reduction in global terrestrial net primary production from 2000 through 2009. Science 329, 940–943. ( 10.1126/science.1192666) [DOI] [PubMed] [Google Scholar]
- 35.Rangel T, Diniz-Filho JAF, Bini LM. 2010. SAM: a comprehensive application for spatial analysis in macroecology. Ecography 33, 46–50. ( 10.1111/j.1600-0587.2009.06299.x) [DOI] [Google Scholar]
- 36.Warren DL, Glor RE, Turelli M. 2010. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33, 607–611. ( 10.1111/j.1600-0587.2009.06041.x) [DOI] [Google Scholar]
- 37.Carnaval ACOQ, Moritz CM. 2008. Historical climate change predicts current biodiversity patterns in the Brazilian Atlantic rainforest. J. Biogeogr. 35, 1187–1201. ( 10.1111/j.1365-2699.2007.01870.x) [DOI] [Google Scholar]
- 38.Bates JM, Hackett SJ, Cracraft J. 1998. Area-relationships in the Neotropical lowlands: a hypothesis based on raw distributions of passerine birds. J. Biogeogr. 25, 783–793. ( 10.1046/j.1365-2699.1998.2540783.x) [DOI] [Google Scholar]
- 39.Costa LP, Leite YLR, Fonseca GAB, Fonseca MT. 2000. Biogeography of South American forest mammals: endemism and diversity in the Atlantic forest. Biotropica 32, 872–881. ( 10.1111/j.1744-7429.2000.tb00625.x) [DOI] [Google Scholar]
- 40.Silva SM, Moraes-Barros N, Ribas CC, Ferrand N, Morgante JS. 2012. Divide to conquer: a complex pattern of biodiversity depicted by vertebrate components in the Brazilian Atlantic Forest. Biol. J. Linn. Soc. Lond. 107, 39–55. ( 10.1111/j.1095-8312.2012.01919.x) [DOI] [Google Scholar]
- 41.Pellegrino K, Rodrigues MT, Waite AN, Morando M, Yassuda YY, Sites JW. 2005. Phylogeography and species limits in the Gymnodactylus darwinii complex (Gekkonidae, Squamata): genetic structure coincides with river systems in the Brazilian Atlantic forest. Biol. J. Linn. Soc. Lond. 85, 13–26. ( 10.1111/j.1095-8312.2005.00472.x) [DOI] [Google Scholar]
- 42.Martins FM. 2011. Historical biogeography of the Brazilian Atlantic forest and the Carnaval–Moritz model of Pleistocene refugia: what do phylogeographical studies tell us? Biol. J. Linn. Soc. Lond. 104, 499–509. ( 10.1111/j.1095-8312.2011.01745.x) [DOI] [Google Scholar]
- 43.Rull V. 2009. Microrefugia . J. Biogeogr. 36, 481–484. ( 10.1111/j.1365-2699.2008.02023.x) [DOI] [Google Scholar]
- 44.Teixeira M, Jr, Dal Vechio F, Recoder RS, Carnaval AC, Strangas S, Damasceno RP, Sena MA, Rodrigues MT. 2012. Two new species of marsupial tree-frogs genus Gastrotheca Fitzinger, 1843 (Anura, Hemiphractidae) from the Brazilian Atlantic Forest. Zootaxa 3437, 1–23. [Google Scholar]
- 45.Cheng H, et al. 2013. Climate change patterns in Amazonia and biodiversity. Nat. Commun. 4, 1411 ( 10.1038/ncomms2415) [DOI] [PubMed] [Google Scholar]
- 46.Cruz FW, et al. 2009. Orbitally driven east–west antiphasing of South American precipitation. Nat. Geosci. 2, 210–214. ( 10.1038/ngeo444) [DOI] [Google Scholar]
- 47.Batalha-Filho H, Fjeldsa J, Fabre P-H, Miyaki CY. 2012. Connections between the Atlantic and the Amazonian forest avifaunas represent distinct historical events. J. Ornithol. 154, 41–50. ( 10.1007/s10336-012-0866-7) [DOI] [Google Scholar]
- 48.Levinsky I, Araújo MB, Nogués-Bravo D, Haywood AM, Valdes PJ, Rahbek C. 2013. Climate envelope models suggest spatio-temporal co-occurrence of refugia of African birds and mammals. Glob. Ecol. Biogeogr. 22, 351–363. ( 10.1111/geb.12045) [DOI] [Google Scholar]
- 49.Ribas CC, Aleixo A, Nogueira ACR, Miyaki CY, Cracraft J. 2012. A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proc. R. Soc. B 1729, 681–689. ( 10.1098/rspb.2011.1120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werneck FP, Gamble T, Colli GR, Rodrigues MT, Sites JW., Jr 2012. Deep diversification and long-term persistence in the South American ‘Dry Diagonal’: integrating continent-wide phylogeography and distribution modeling of geckos. Evolution 66, 3014–3034. ( 10.1111/j.1558-5646.2012.01682.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
DNA sequences: GenBank accession nos. KM204320–KM204375. Climate data and correlative model output data: Dryad doi: (doi:10.5061/dryad.8kc1v).