Abstract
Aim: We have hypothesized a possible relationship between disc degeneration (DD) and VDR FokI/T2C polymorphism. Methods: A case–control study was performed comprising 121 Brazilian patients with confirmed DD by nuclear magnetic resonance and a control group consisting of 131 healthy patients without a history of disc cysts of the lumbar spine. Detection of VDR FokI/T2C polymorphism was performed using restriction fragment length polymorphism–polymerase chain reaction. The chi-square test was used to compare allele and genotype frequencies between groups, and a p-value of <0.05 was considered statistically significant. Results: The results disclosed statistical difference between allele distribution among cases and controls (p=0.025, odds ratio=1.58, confidence interval=1.07–2.32) considering VDR FokI/T2C polymorphism. Conclusion: The results showed a positive association between VDR FokI/T2C polymorphism and DD in Brazilian patients tested.
Introduction
The intervertebral disc is a fibrocartilaginous structure whose main function is to act as a buffer, transmitting compressive loads between vertebral bodies (Buckwalter, 1995; Miller et al., 1988). The intervertebral disc consists of three main structures: the cartilaginous endplates, the central nucleus pulposus, and the annulus fibrosus located at the periphery of the disc. The intervertebral disc loses its hygroscopic properties with aging, leading to a progressive dehydration process, characterizing disc disease. From the intervertebral disc degeneration (DD), the spine begins to show progressive instability of the affected region (Inoue, 1981).
The process of DD is associated with many clinical conditions, including low back pain, which is one of the most common health problems in society, being a major cause of work absenteeism and use of health services. It is estimated that 15–20% of adults have back pain during a single year and 50–80% experience at least one episode of back pain during their lifetime (Rubin, 2007).
The precise etiology of DD is not fully understood. Until recently, it was exclusively attributed to the accumulation of environmental effects, primarily micro or macro, trauma, lifestyle, smoking, atherosclerosis, and the changes that occur in the disc with aging (Zawilla et al., 2013). However, more recent research has demonstrated that the influence of these factors is moderate in DD, reinforcing the notion of genetic involvement in the etiology of the disease (Nunes et al., 2007). Epidemiological studies on families and twins suggest that inheritance is the major determinant of DD (Battie et al., 1995; Matsui et al., 1998; Sambrook et al., 1999). Several disease-associating variations have been found in a number of different genes, suggesting that intervertebral DD is a multigenetic entity (Ala-Kokko, 2002; Kalichman and Hunter, 2008; Zawilla et al., 2013).
To date, several gene loci associated with human DD have been identified (Chan et al., 2006). Variations in the genes involved in inflammation, extracellular matrix components, and protein metabolism have been reported as associating with DD (Kalichman et al., 2008).
Vitamin D is known as a hormone that regulates calcium homeostasis and bone mineralization (Cantorna and Mahon, 2004) and can be found in two forms: ergocalciferol (vitamin D2) and cholecalciferol (vitamin D3). Vitamin D, derived from the diet or the bioactivation of 7-deidrocalciferol, is inert and must be activated to exert the biological functions (Fraser and Kodicek, 1970). The hormonal form of vitamin D (1,25-2-hydroxyvitamin D3) has essential roles in endocrine functions: (1) mineralization process of bone, (2) absorption of calcium from the intestine, (3) control of calcium and phosphorus homeostasis, and (4) regulation of parathyroid hormone (Vilarino et al., 2011), and it has shown antiproliferative and immunosuppressive effects on several cell types, for example, lymphocyte proliferation and immunoglobulin synthesis, besides inhibiting the action of proinflammatory transcription factors and the production of different cytokines, such as IL-2, IL-12, among others (Froicu et al., 2003; Lehman, 2005).
Most of the biological activities of vitamin D are mediated by a high-affinity receptor that acts as a transcription factor activated by ligand—the gene for the vitamin D receptor (VDR), located on chromosome 12 (12q13.11) and a member of the family of steroid receptors that mediates the effects of vitamin D in regulating the transcription of multiple genes. Genetic alterations in the VDR gene lead to significant gene activation defects affecting calcium metabolism, cell proliferation, immune function, and others, which can be explained by changes in protein conformation (Videman et al., 1998). Changes in the sequence of the gene, such as polymorphisms, may occur in the noncoding region of the gene (introns) affecting the level of gene expression and thus protein levels and the coding regions (exons) and lead to changes in the sequence of protein (Valdivielso and Fernandez, 2006; Vilarino et al., 2011).
Based on this observation, a possible relationship between polymorphism FokI of the vitamin D receptor gene and DD has been hypothesized.
Materials and Methods
Patients
A prospective case–control study that included 121 patients in the Outpatient Clinic of the Spinal Surgery at the Hospital Estadual Mário Covas, coordinated by the Department of Diseases of the Locomotor System of the Faculdade de Medicina do ABC, Santo André/SP, Brazil. The sample includes individuals with chronic low back pain associated with degenerative intervertebral disc disease and control subjects.
The inclusion criteria for patients were as follows: (1) patients with chronic low back pain (over 3 months), (2) aged <45 years, and (3) magnetic resonance imaging (MRI) with DD on sagittal T2. The exclusion criteria were as follows: (1) patients who underwent previous surgical treatment, (2) patients with congenital deformities of the spine, and (3) patients who refuse to sign the consent form and donate a blood sample for analysis of genomic DNA.
Considering the control group, the patients were recruited in the Clinical Laboratory of the Faculdade de Medicina do ABC and the inclusion criteria were as follows: (1) aged 20–45 years, (2) without previous surgery, (3) no history of disc hernia treatment, (4) have not been hospitalized for back pain, (5) not taking medication for back pain for more than 7 days, and (6) do not have family members younger than 45 years with disc herniation or clinical treatment for low back pain.
All study subjects answered a clinical and epidemiological questionnaire with data on age, gender, ethnicity, occupation, education, income, weight, height, smoking habits, comorbidities, physical examination, information regarding complaints, painful, and family history. All patients signed the form of free and informed consent approved by the local ethics committee.
The MRI scans of all patients were performed by two experienced radiologists. The DD was classified according to the classification of Pfirrmann et al. (2001), and only patients with Pfirrmann 3, 4, or 5 were included in the case group, once moderate and intense DD is evident.
Molecular analysis
Peripheral blood was collected from each patient and control in an EDTA-containing tube. Genomic DNA was extracted from peripheral blood of all study subjects according to the salting out method (Lahiri and Numberger, 1991).
Genotyping of the VDR polymorphism
The FokI/T2C/rs222857 polymorphism of the VDR gene was studied by restriction fragment length polymorphism–polymerase chain reaction (PCR) according to the protocol of Horst-Sikorska et al. (2007), with modifications. In general, the PCR procedure was carried out in a total volume of 25 μL reaction mixture containing 10X reaction buffer (500 mM KCl, 100 mM Tris–Cl; pH 8.3), 2.5 mM MgCl2, 0.8 mM dNTP, 2.0 U Taq polymerase, and 50 nM of each primer (forward: AGC TGG CCC TGG CAC TGA CTC TGC TCT and reverse: ATG GAA ACA CCT TGC TTC TTC TCC CTC). The cycling profile consisted of denaturation at 95°C for 30 s, 60°C for 60 s for annealing temperature, and extension at 72°C for 30 s, except for the first cycle, when denaturation was extended to 5 min. The PCR product was digested with 5 U of FokI restriction enzyme (New England Biolabs, Ipswich, MA), and the reaction mixture was incubated at 65°C for 15 min. The digestion product was subjected to electrophoresis on a gel containing 2% agarose stained with ethidium bromide and visualized under ultraviolet light.
Using a DNA ladder of 50 base pairs (bp) as a reference, we identified the Fok1 polymorphism genotypes: normal homozygote (TT) presented a unique fragment of 265 bp, heterozygote (TC) presented three fragments of 265, 196, and 69 bp, and mutant homozygote (CC) presented two fragments of 196 and 69 bp (Fig. 1).
FIG. 1.
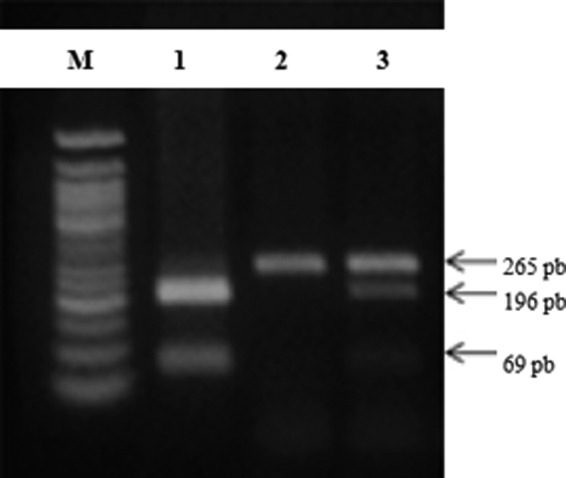
The figure shows a 2% agarose gel representative of the VDR Fok1 polymorphism, where (M) is the DNA ladder 50pb, (1) is a mutated subject (CC), (2) is a wild homozygote (TT), and (3) is a heterozygous subject (CT).
Statistical analyses
Statistical analyses were carried out using SPSS for Windows 11.0 (SPSS, Inc., Chicago, IL). The chi-square test was used to compare allele and genotype frequencies between groups and to estimate the Hardy–Weinberg equilibrium. Genetic Power Calculator (Purcell et al., 2003) was used to estimate the statistical power of the results concerning Fok1 polymorphism data and showed at least 89% of power to detect the genetic effects regarding the association with DD for the allele frequencies and sample size in the present study. The odds ratio (OR) and range with 95% confidence interval (CI) were calculated for the presence of the reference genotype using a logistic regression model. All p-values were two-tailed, and 95% CIs were calculated. A p-value of <0.05 was considered statistically significant.
Results
Among the 121 subjects studied with DD, 46.3% (56/121) were male (mean age 46.0±5.4 years) and 53.7% (65/121) were female (mean age 45.2±5.9 years). Considering the control group, 26.7% (31/131) were male (mean age 33.8±8.2 years) and 76.3% (100/131) were female (mean age 33.9±8.1 years).
The genotype and allele distributions of the Fok1 polymorphisms of the VDR gene in DD patients and controls are shown in Table 1.
Table 1.
The Genotype and Allelic Distributions of the FokI Polymorphism of the VDR Gene in Intervertebral Disc Degeneration Patients and Controls
| Genotypes | Alleles | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP VDR | Population studied | n | n (%) | n (%) | n (%) | n (%) | n (%) | p | OR (95% CI) | HWE |
| TT | TC | CC | T | C | ||||||
| FokI | DD patients | 121 | 54 (44.6) | 50 (41.4) | 17 (14.0) | 158 (65.3) | 84 (34.7) | 0.025 | 1.58 (1.07–2.32) | 0.624 |
| rs2228570 | Controls | 131 | 75 (57.2) | 46 (35.2) | 10 (7.6) | 196 (74.8) | 66 (25.2) | 0.737 | ||
CI, confidence interval; HWE, Hardy–Weinberg equilibrium; IDD, intervertebral disc degeneration; OR, odds ratio; SNP, single-nucleotide polymorphism.
Considering the genotype distribution in the DD patients, 44.6% (54/121) presented normal homozygote genotype (TT), 41.3% (50/121) presented a heterozygote genotype (TC), and 14.1% (17/121) presented a mutant homozygote genotype (CC). In the control group, the genotypes TT, TC, and CC were found in 57.2% (75/121), 35.2% (46/131), and 7.6% (10/131) of the subjects.
Regarding the allele frequencies, the wild allele T was found in 65.3% of the DD patients and in 74.8% of the control group; and the mutant allele C were observed in 34.7% of the DD patients and in 25.2% of the control group (p=0.025; OR=1.58, CI=1.07–2.32).
Statistical analyses showed that the genotype distribution in DD and the control group for FokI polymorphism were in the Hardy–Weinberg equilibrium.
Discussion
The publication of the human genome sequence led to a great increase in the biomarker study field. The search for genetic associations with various diseases, including cancer, infertility, and other complex diseases, had great momentum to identify prognostic or preventive markers. Polymorphisms may serve as genetic markers, are responsible for human diversity, and can directly influence the risk factors associated with common diseases. Thus, the polymorphisms are key elements in the research and practice of human genetics (Kalichman and Hunter, 2008; Bag et al., 2012).
Nowadays, polymorphisms are the basis for the attempt to provide personalized medicine based on genomics, based on the fact that if an individual carries polymorphic variants that increase or decrease the risk for common diseases in adulthood (such as coronary heart disease, cancer, diabetes, endometriosis, or DD) probably presents more complications after the surgery, or influence the effectiveness or safety of specific medications (Bag et al., 2012).
Polymorphism refers to a variant of a particular gene sequence found in more than 1% of the general population. Single-nucleotide polymorphisms (SNPs) generally have two alleles corresponding to two different bases that occupy a particular position in the genome (locus). There are over 3 million SNPs documented and are found in virtually all genes, but only a minority results in changes in amino acids and lead to alteration of protein conformation. When this polymorphism results in a change of amino acid encoding a protein with altered function, these characteristics make them excellent markers to generate genetic maps such as those needed to evaluate the potential contribution of a given gene for a complex disorder. SNPs have been identified in genes responsible for metabolism, cell proliferation, transport, inflammatory response, immune response, and DNA repair that may be related to the development and progression of a disease and also in response to a specific treatment or preventive disease development (Kalichman and Hunter, 2008). There is not much information regarding how the polymorphisms affect vitamin D receptor transcription. The VDR FokI polymorphism in exon 2 leads to an alternative transcription initiation site, resulting in a VDR protein with the addition of three amino acids (Mory et al., 2009; Vilarino et al., 2011).
In this study, we hypothesized a possible relationship between FokI polymorphism of VDR gene and intervertebral DD. We found a significant difference in the frequencies of the polymorphism studied between DD patients and controls.
Previously, a series of studies have been conducted to evaluate the associations between FokI polymorphism of VDR gene and the risk of intervertebral DD but produced conflicting results. In 2003, Noponen-Hietala et al. (2003) studied FokI polymorphism in 29 Finnish probands and 56 controls and a difference in the genotype and allele frequencies was not found. Eskola et al. (2010) evaluated 352 Danish children with early DD, and no statistical difference was found. Eser et al. (2010) investigated 300 young Turkish individuals regarding DD and herniation. An association was found in the patients having VDR gene TT (“T” wild allele and “t” mutant allele of TaqI polymorphism of VDR gene), Tt, FF (“F” wild allele and “f” mutant allele of FokI polymorphism of VDR gene), and Ff genotypes with the protrusion type of disc herniation, whereas the patients having tt and ff genotypes were associated with extrusion/sequestration types of the disease. In addition, an association was observed between TT and FF genotypes of the VDR gene and mild forms of DD and also among tt, ff, and Ff genotypes and severe forms of the disease. Kelempisioti et al. (2011) investigated 538 young adults belonging to the 1986 Northern Finland Birth Cohort. The results disclosed that no association was found between DD and the FokI polymorphism.
The discrepant findings in the literature suggest genetic heterogeneity within the VDR gene in different diseases and populations, possibly due to divergent evolutionary lineages resulting in separate clusters of distinct geography (Vogel et al., 2002). Consequently, the structure of linkage disequilibrium differs markedly across genomic regions and populations, and the extent of linkage disequilibrium is highly dependent on the population in which it is measured (Neale and Sham, 2004). Thus, the same allele may have different patterns of association with markers in different populations (Goldstein, 2001). Other aspects should be considered is the sample selection.
The first meta-analysis performed by Xu et al. (2012) included five studies that evaluated FokI polymorphism and DD (Noponen-Hietala et al., 2003; Chen et al., 2007; Eser et al., 2010; Eskola et al., 2010; Kelempisioti et al., 2011). No significant association was found considering the polymorphism and the development of DD. However, the authors point to the fact that since potential confounders could not be ruled out completely, further studies are needed to confirm these results.
In conclusion, our results suggest that, in the Brazilian population studied, intervertebral DD risk is associated with FokI polymorphism of the VDR gene. However, further studies on much larger samples are needed to evaluate whether or not this association is real.
Acknowledgments
The authors wish to thank FAPESP (Fundação de Amparo a Pesquisa do Estado de São Paulo) for grants No. 2013/00902-4 and for granting student Patricia Leme De Marchi a student scholarship (No. 2012/21886-4).
Author Disclosure Statement
The authors disclosed no conflicts of interest.
References
- Ala-Kokko L. (2002) Genetic risk factors for lumbar disc disease. Ann Med 34:42–47 [DOI] [PubMed] [Google Scholar]
- Bag A, Jyala NS, Bag N. (2012) Indian studies on genetic polymorphisms and cancer risk. Indian J Cancer 49:144–162 [DOI] [PubMed] [Google Scholar]
- Battie MC, Videman T, Gibbons LE, et al. (1995) Determinants of lumbar disc degeneration. A study relating lifetime exposures and magnetic resonance imaging findings in identical twins. Spine 20:2601–2612 [PubMed] [Google Scholar]
- Buckwalter JA. (1995) Aging and degeneration of the human intervertebral disc. Spine 20:1307–1314 [DOI] [PubMed] [Google Scholar]
- Cantorna MT, Mahon BD. (2004) Mounting evidence for vitamin D as an environmental factor affecting autoimmune disease prevalence. Exp Biol Med (Maywood) 229:1136–1142 [DOI] [PubMed] [Google Scholar]
- Chan D, Song Y, Sham P, Cheung KM. (2006) Genetics of disc degeneration. Eur Spine J 15Suppl 3:S317–S325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WJ, Ye W, Ding Y, et al. (2007) Association of vitamin D receptor gene TruI and FokI polymorphisms with lumbar degenerative disc disease in Han nationality. Orthop J China 15:373–375 [Google Scholar]
- Eser B, Cora T, Eser O, et al. (2010) Association of the polymorphisms of vitamin D receptor and aggrecan genes with degenerative disc disease. Genet Test Mol Biomarkers 14:313–317 [DOI] [PubMed] [Google Scholar]
- Eskola PJ, Kjaer P, Daavittila IM, et al. (2010) Genetic risk factors of disc degeneration among 12–14-year-old Danish children: a population study. Int J Mol Epidemiol Genet 1:158. [PMC free article] [PubMed] [Google Scholar]
- Fraser DR, Kodicek E. (1970) Unique biosynthesis by kidney of a biological active vitamin D metabolite. Nature 228:764–766 [DOI] [PubMed] [Google Scholar]
- Froicu M, Weaver V, Wynn TA, et al. (2003) A crucial role for the vitamin D receptor in experimental inflammatory bowel diseases. Mol Endocrinol 17:2386–2392 [DOI] [PubMed] [Google Scholar]
- Goldstein DB. (2001) Islands of linkage disequilibrium. Nat Genet 29:109–111 [DOI] [PubMed] [Google Scholar]
- Horst-Sikorska W, Kalak R, Wawrzyniak A, et al. (2007) Association analysis of the polymorphisms of the VDR gene with bone mineral density and the occurrence of fractures. J Bone Miner Metab 25:310–319 [DOI] [PubMed] [Google Scholar]
- Inoue H. (1981) Three-dimensional architecture of lumbar intervertebral discs. Spine 6:139–146 [DOI] [PubMed] [Google Scholar]
- Kalichman L, Hunter DJ. (2008) The genetics of intervertebral disc degeneration. Associated genes. Joint Bone Spine 75:388–396 [DOI] [PubMed] [Google Scholar]
- Kelempisioti A, Eskola PJ, Okuloff A, et al. (2011) Genetic susceptibility of intervertebral disc degeneration among young Finnish adults. BMC Med Genet 12:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahiri DK, Nurnberger JI., Jr (1991) A rapid non-enzymatic method for the 10. preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res 19:5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman B. (2005) The vitamin D3 pathway in human skin and its role for regulation of biological processes. Photochem Photobiol 81:1246–1251 [DOI] [PubMed] [Google Scholar]
- Matsui H, Kanamori M, Ishihara H, et al. (1998) Familial predisposition for lumbar degenerative disc disease. A case-control study. Spine 23:1029–1034 [DOI] [PubMed] [Google Scholar]
- Miller JA, Schmatz C, Schultz AB. (1988) Lumbar disc degeneration: correlation with age, sex, and spine level in 600 autopsy specimens. Spine 13:173–178 [PubMed] [Google Scholar]
- Mory DB, Rocco ER, Miranda WL, et al. (2009) Prevalence of vitamin D receptor gene polymorphisms FokI and BsmI in Brazilian individuals with type 1 diabetes and their relation to beta-cell autoimmunity and to remaining beta-cell function. Hum Immunol 70:447–451 [DOI] [PubMed] [Google Scholar]
- Neale BM, Sham PC. (2004) The future of association studies: gene-based analysis and replication. Am J Hum Genet 75:353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noponen-Hietala N, Kyllonen E, Mannikko M, et al. (2003) Sequence variations in the collagen IX and XI genes are associated with degenerative lumbar spinal stenosis. Ann Rheum Dis 62:1208–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes FTB, Conforti-Froes NDT, Negrelli WF, Souza DRS. (2007) Genetic and environmental factors involved on intervertebral disc degeneration. Acta Ortop Bras 15:9–13 [Google Scholar]
- Pfirrmann CW, Metzdorf A, Zanetti M, et al. (2001) Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 26:1873–1878 [DOI] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC. (2003) Genetic power calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics 19:149–150 [DOI] [PubMed] [Google Scholar]
- Rubin DI. (2007) Epidemiology and risk factors for spine pain. Neurol Clin 25:353–371 [DOI] [PubMed] [Google Scholar]
- Sambrook PN, MacGregor AJ, Spector TD. (1999) Genetic influences on cervical and lumbar disc degeneration: a magnetic resonance imaging study in twins. Arthritis Rheum 42:366–372 [DOI] [PubMed] [Google Scholar]
- Valdivielso JM, Fernandez E. (2006) Vitamin D receptor polymorphisms and diseases. Clin Chim Acta 371:1–12 [DOI] [PubMed] [Google Scholar]
- Videman T, Leppavuori J, Kaprio J, et al. (1998) Intragenic polymorphisms of the vitamin D receptor gene associated with intervertebral disc degeneration. Spine 23:2477. [DOI] [PubMed] [Google Scholar]
- Vilarino FL, Bianco B, Christofolini DM, et al. (2011) Analysis of VDR gene polymorphism Fok1 in infertile women with endometriosis. Rev Bras Ginecol Obstet 33:65–69 [PubMed] [Google Scholar]
- Vogel A, Strassburg CP, Manns MP. (2002) Genetic association of vitamin D receptor polymorphisms with primary biliary cirrhosis and autoimmune hepatitis. Hepatology 35:126–131 [DOI] [PubMed] [Google Scholar]
- Xu G, Mei Q, Zhou D, et al. (2012) Vitamin D receptor gene and aggrecan gene polymorphisms and the risk of intervertebral disc degeneration—a meta-analysis. PLoS One 7:e50243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawilla NH, Darweesh H, Mansour N, et al. (2014) Matrix metalloproteinase-3, vitamin D receptor gene polymorphisms, and occupational risk factors in lumbar disc degeneration. J Occup Rehabil 24:370–381 [DOI] [PubMed] [Google Scholar]


