Abstract
Genetic variants account for more than half of the cases with congenital or prelingual onset hearing loss. Autosomal recessive nonsyndromic hearing loss (ARNSHL) is the most common subgroup. Whole-exome sequencing (WES) has been shown to be effective detecting deafness-causing single-nucleotide variants (SNVs) and insertion/deletions (INDELs). After analyzing the WES data for causative SNVs or INDELs involving previously reported deafness genes in 78 families with ARNSHL, we searched for copy number variants (CNVs) through two different tools in 24 families that remained unresolved. We detected large homozygous deletions in STRC and OTOA in single families. Thus, causative CNVs in known deafness genes explain 2 out of 78 (2.6%) families in our sample set. We conclude that CNVs can be reliably detected through WES and should be the part of pipelines used to clarify genetic basis of hearing loss.
Introduction
Hearing loss is a public health concern affecting 1 to 3 per 1,000 newborns (Kemper and Downs, 2000). More than half of the cases with congenital or prelingual hearing loss are caused by genetic variations and at least 75% of them show autosomal recessive inheritance (Van Camp et al., 1997). Genetics of autosomal recessive nonsyndromic hearing loss (ARNSHL) is extremely heterogeneous involving over 80 loci, with forty nine genes having been identified as containing pathogenic DNA variants thus far (Van Camp and Smith, 2014).
Whole-exome sequencing (WES) has become an essential tool in genomics research and in diagnostic clinical genetics laboratories (Yang et al., 2013). Due to its rapid, low cost, and comprehensive analysis advantages, WES has been successfully used for various genetic disorders, including the genetically heterogeneous ARNSHL (Diaz-Horta et al., 2012). The main applications of WES in most laboratories are identifying single-nucleotide variants (SNVs) and small deletion/insertions (INDELs). While various practical algorithms for the detection of copy number variants (CNVs) have recently been developed (Fromer et al., 2012; Krumm et al., 2012; Tan et al., 2014), their usage in general is not common and their application in deafness has not been reported.
We have performed WES in 78 families with ARNSHL that were negative for mutations in the most common gene, GJB2 (MIM121011). In addition to GATK (Genome Analysis Tool Kit) (McKenna et al., 2010) used for detection of SNVs and INDELs, we included CoNIFER (Copy Number Inference From Exome Reads) (Krumm et al., 2012) and XHMM (eXome-Hidden Markov Model) (Fromer et al., 2012) as part of our genetic analysis pipeline to identify CNVs. Probands in 54 families were found to have pathogenic SNVs or INDELs in one of the previously described ARNSHL genes that explained the phenotype. We present here the detection of causative CNVs in the remaining 24 families and compare the efficiency of different CNV detection algorithms using WES.
Materials and Methods
Our study was approved by the University of Miami Institutional Review Board and Ankara University Medical School Ethics Committee (Turkey). All participants were Turkish and provided written informed consent before enrollment, or in the case of a minor, from parents.
Twenty-four families were included in this study based on the existence of both parental consanguinity and at least two members with nonsyndromic hearing loss. Diagnosis of sensorineural hearing loss was established through standard audiometry in a sound-proofed room according to current clinical standards. Clinical evaluation of all affected individuals by a geneticist and an ENT surgeon included a thorough physical examination and otoscopy. A high-resolution CT scan of the temporal bone was found to be normal at least in one affected person in each family.
DNA was extracted from peripheral leukocytes of each member of the family by using the phenol–chloroform method. WES was performed in 103 affected individuals from 78 unrelated families who were negative for mutations in GJB2. After WES, 24 families remained negative for SNVs or INDELs in 49 genes that are known to cause ARNSHL (the list of the 49 genes is available in the Supplementary Information; Supplementary Data are available online at www.liebertpub.com/gtmb).
In addition to 24 families, two samples known to be heterozygous for a deletion CNV in ESRRB (MIM602167) and TMC1 (MIM606706) were used as positive controls. Deletions in positive control samples were detected by Illumina or Affymetrix arrays and confirmed with quantitative PCR (Duman et al., 2011). CNV analysis was performed in genomic regions comprising the 49 previously reported ARNSHL genes.
The Agilent SureSelect Human All Exon 50 Mb kit was used for in-solution enrichment of coding exons and flanking intronic sequences following the manufacturer's standard protocol. Adapter sequences for the Illumina Hiseq2000 were ligated, and the enriched DNA samples were subjected to standard sample preparation for the Hiseq2000 instrument (Illumina). The Illumina CASAVA v1.8 pipeline was used to create 99 bp sequence reads. BWA (Li and Durbin, 2010) was used to align sequence reads to the human reference genome (NCBI build 37, hg19) and variants (SNVs & small INDELs) were called using the GATK software package (McKenna et al., 2010).
As a part of our pipeline, XHMM v1.0 and CoNIFER v.02.2 were used for CNV analysis in 103 affected individuals from 78 families. Due to differences in the CNV analyzing algorithms, we applied default parameter settings to compare their efficiency with an unbiased evaluation (Fromer et al., 2012; Krumm et al., 2012).
Results
The average coverage for each exome in the 24 families was 95%, 83%, and 69% of mappable bases of the Gencode-defined exome, represented with at least 2X, 10X, and 20X reads, respectively. Both tools were first used for the detection of the two previously identified heterozygous CNVs. Both the positive controls with heterozygous ESRRB and TMC1 deletions were detected through XHMM, but not with CoNIFER (Table 1). While CoNIFER failed to report the heterozygous deletions, it did show both deletions in its visualization file (Supplementary Figs. S1 and S2).
Table 1.
Summary of the Copy Number Variant Results
| Family ID | Gene | Deleted region (XHMM) | Deleted region (CoNIFER) |
|---|---|---|---|
| Positive control 1 | ESRRB | chr14:76948183-76967234 | Not available |
| Positive control 2 | TMC1 | chr 9:75420183-75451112 | Not available |
| Family 3 | OTOA | chr16:21610949-21747850 | chr16:21327883-21969920 |
| Family 12 | STRC | chr15:43892635-43903864 | chr15:43850992-43940259 |
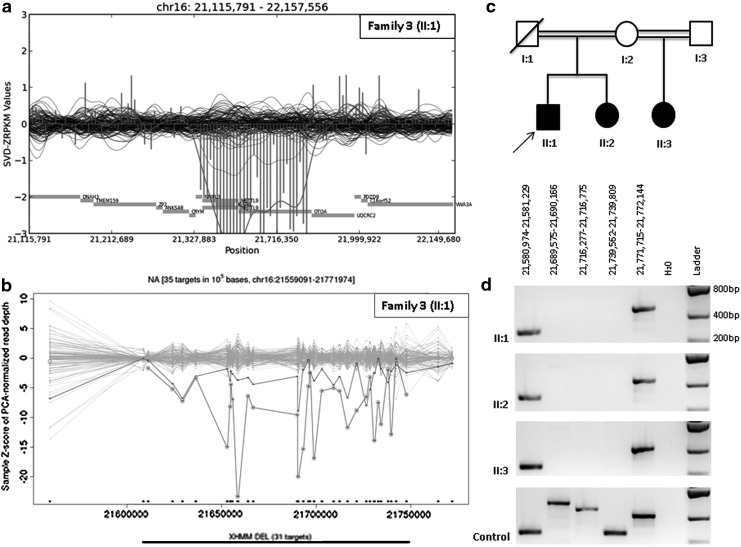
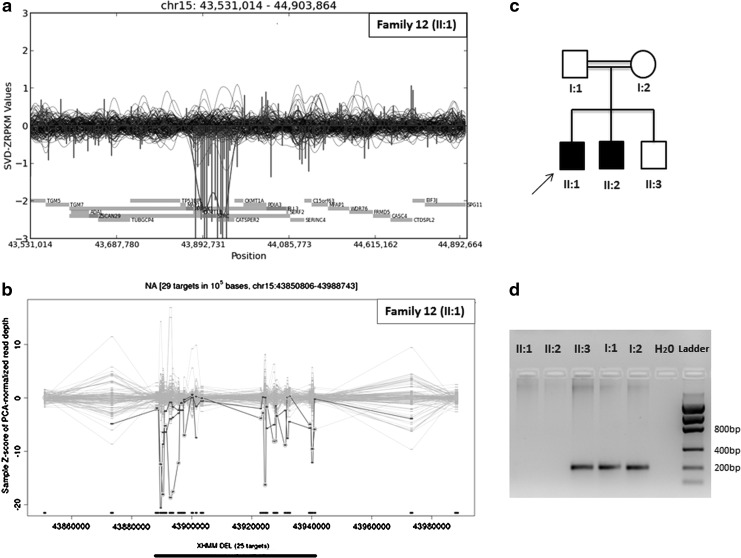
Among the probands of the 24 families, both tools detected two homozygous deletions. A large homozygous deletion, including OTOA (MIM607038), was identified in family 3 with three children having mild to moderate sensorineural hearing loss. This deletion was confirmed with PCR and cosegregated with the phenotype in the entire family (Fig. 1). However, the size of the deleted region was reported to be 642 kb in CoNIFER and only 89kb in XHMM. Further PCR analyses showed that the deleted region was shorter than 190kb (Supplementary Fig. S3). Another homozygous deletion encompassing STRC (MIM606440) was detected in Family 12, where two affected children had mild to severe sensorineural hearing loss. This deletion was also confirmed with PCR and cosegregated with the phenotype in the family (Fig. 2).
FIG. 1.
OTOA gene deletion in family 3. Visualized results of CoNIFER (a) and XHMM (b) showing the homozygous OTOA gene deletion in family 3. OTOA gene deletion in all three affected patients (c) was confirmed with serial PCRs (d).
FIG. 2.
STRC gene deletion in family 12. Visualized results of CoNIFER (a) and XHMM (b) indicating the homozygous STRC gene deletion in family 12. STRC gene deletion in both affected siblings (c) was confirmed with PCR (d).
Discussion
In this study, we aimed to detect pathogenic CNVs through WES in a sample set of families with ARNSHL. We identified deafness-causing deletions involving two previously reported ARNSHL genes in two out of 24 families (8.3%). Overall, causative CNVs were present in 2.6% of the 78 studied families. Causative GJB2 mutations are present in 19% of multiplex and consanguineous Turkish families with ARNSHL (Tekin and Arici, 2007). Thus, the frequency of causative CNVs in this population is 2.1% (95% CI: 0% to 4.9%). This result is similar to that of a recent study that reported pathogenic CNVs in 14 of 636 cases with nonsyndromic sensorineural hearing loss through SNP arrays (Tsai et al., 2013).
Homozygous deletions of STRC were initially reported in 2001 (Verpy et al., 2001). More recent studies report STRC deletions with varying sizes in approximately 2.5% of cases with sensorineural hearing loss in the American population (Francey et al., 2012). The deletion of OTOA has been previously reported in a consanguineous Palestinian family with 1% carrier frequency in that population (Shahin et al., 2010). Boundaries of the deleted region for OTOA (MIM607038) have not been clearly defined. The size of the deleted region detected by XHMM in family 3 is similar to those previously published (Supplementary Fig. S3).
Both CoNIFER and XHMM are based on a read depth approach to detect CNVs in WES data. While XHMM utilizes the principal component analysis followed by the Hidden Markov model to identify CNVs, CoNIFER uses a singular value decomposition technique to correct systematic biases and identifies a CNV call if the corrected signal reaches a predefined threshold at no less than three subsequent exons (Fromer et al., 2012; Krumm et al., 2012). Breakpoint detection is an advantage of the XHMM (Fromer et al., 2012; Tan et al., 2014). According to our data, XHMM showed more reliable results for the size of the OTOA deletion. We also noted that CoNIFER CNV analysis missed two heterozygous deletions used as positive controls. Recent CNV comparison studies in WES showed that by using the previously described heterozygosity check method (Zhu et al., 2012), CoNIFER detects less (40%) heterozygous false-positive deletions (for regions >1 kb) compared to XHMM (64%) (Tan et al., 2014), suggesting that it might be missing some true positives to increase specificity. Conservative predefined thresholds in default settings of the CoNIFER might be the reason for missing heterozygous deletions in positive controls in our data set.
Identifying causative genetic changes in hearing loss provides substantial information for the etiological diagnosis and genetic counseling. Our study shows that CNVs are an important cause of deafness and should be a part of the multigene panel or WES pipelines performed for sensorineural hearing loss.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant R01DC009645 to M.T.
Author Disclosure Statement
Authors declare that there are no conflicts of interest to report.
References
- Diaz-Horta O, Duman D, Foster J 2nd, et al. (2012) Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One 7:e50628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman D, Sirmaci A, Cengiz FB, et al. (2011) Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in Turkey. Genet Test Mol Biomarkers 15:29–33 [DOI] [PubMed] [Google Scholar]
- Francey LJ, Conlin LK, Kadesch HE, et al. (2012) Genome-wide SNP genotyping identifies the Stereocilin (STRC) gene as a major contributor to pediatric bilateral sensorineural hearing impairment. Am J Med Genet A 158A:298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, et al. (2012) Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet 91:597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper AR, Downs SMA. (2000) A cost-effectiveness analysis of newborn hearing screening strategies. Arch Pediatr Adolesc Med 154:484–488 [DOI] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, et al. (2012) Copy number variation detection and genotyping from exome sequence data. Genome Res 22:1525–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, et al. (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Rayyan AA, et al. (2010) Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet 18:407–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R, Wang Y, Kleinstein SE, et al. (2014) An evaluation of copy number variation detection tools from whole-exome sequencing data. Hum Mutat 35:899–907 [DOI] [PubMed] [Google Scholar]
- Tekin M, Arici ZS. (2007) Genetic epidemiological studies of congenital/prelingual deafness in Turkey: population structure and mating type are major determinants of mutation identification. Am J Med Genet A 143A:1583–1591 [DOI] [PubMed] [Google Scholar]
- Tsai EA, Berman MA, Conlin LK, et al. (2013) PECONPI: a novel software for uncovering pathogenic copy number variations in non-syndromic sensorineural hearing loss and other genetically heterogeneous disorders. Am J Med Genet A 161:2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp G, Smith RJH. Hereditary Hearing Loss Homepage. Available at http://hereditaryhearingloss.org (accessed April24, 2014)
- Van Camp G, Willems PJ, Smith RJ. (1997) Nonsyndromic hearing impairment: unparalleled heterogeneity. Am J Hum Genet 60:758–764 [PMC free article] [PubMed] [Google Scholar]
- Verpy E, Masmoudi S, Zwaenepoel I, et al. (2001) Mutations in a new gene encoding a protein of the hair bundle cause non-syndromic deafness at the DFNB16 locus. Nat Genet 29:345–349 [DOI] [PubMed] [Google Scholar]
- Yang Y, Muzny DM, Reid JG, et al. (2013) Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369:1502–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Need AC, Han Y, et al. (2012) Using ERDS to infer copy-number variants in high-coverage genomes. Am J Hum Genet 91:408–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.