Abstract
Porphyromonas gingivalis sialidase activity is associated with virulence and initiated by sialic acid (SA) binding to the β-propeller domain (BPD). Sialidase BPD is structurally conserved in various bacterial species and the protein binding interfaces have the tendency to form salt bridges, whereas uncommitted charged residues may affect binding and protein structure. However, it is not clear whether the sialidase BPD of varying strains of the same bacterial species differ, particularly with regards to salt bridge formation. Here, we determined the P. gingivalis ATCC 33277 and W50 sialidase homology models and sialidase activities, while the putative salt bridge residues found in the sialidase BPDs were compared. We established that both ATCC 33277 and W50 have different sialidase homology models and activities, whereas, the BPD (β1–6) is structurally conserved with most salt bridge-forming residues following a common orientation. Moreover, β2D444–β6K338 distance measurement in ATCC 33277 (5.99 Å) and W50 (3.09 Å) differ, while β1K396A substitution alters the β2D444–β6K338 distance measurements in ATCC 33277 (3.09 Å) and W50 (3.01 Å) consequentially affecting each model. P. gingivalis plays a major role in periodontitis induction and its virulence is greatly influenced by the sialidase enzyme wherein the sialidase BPD is highly conserved. Our results suggest that alterations in the salt bridge formation within the BPD interface may affect the P. gingivalis sialidase structure. This would imply that disrupting the salt bridge formation within the P. gingivalis sialidase BPD could serve as a potential therapeutic strategy for the treatment of P. gingivalis-related periodontitis.
Introduction
Two common periodontal diseases affecting the periodontium include: (1) gingivitis described as inflammation of the gingiva whereby the connective tissue attachment to the tooth remains at its original level and is limited to the soft-tissue compartment of the gingival epithelium and connective tissues; and (2) periodontitis which is a destructive nonhomogenous inflammatory disease affecting the gingival tissue and has been attributed to both host susceptibility differences and diversity in pathogenic oral bacteria found in the host (Cekici et al., 2014; Genco, 1996; Pathirana et al., 2008). The main proponent of periodontal diseases are bacteria, and the pathogenesis of periodontal diseases is mediated by the inflammatory response to bacteria in the dental biofilm (Cekici et al., 2014). Among the various pathogenic oral bacteria found in the host, Porphyromonas gingivalis has been implicated as a major etiological agent in the onset and progression of periodontitis (Lamont and Jenkinson, 1998; O' Brien-Simpson et al., 2003). P. gingivalis is a Gram-negative, black pigmented anaerobe implicated in the pathogenesis of severe forms of periodontal disease (Lamont and Jenkinson, 1998; Pathirana et al., 2008) and one of the enzymes that function as a potential virulence factor is sialidase (Li et al., 2011).
Sialidase or neuraminidase is a family of exoglycosidase enzymes hydrolyzing the α-linkage of the terminal sialic acids (SAs) found in various sialoglycoconjugates in diverse organisms (Taylor, 1996). Sialidase-producing microorganisms are either pathogenic or commensal in the mammalian host. Moreover, sialidases can be used either for scavenging SA as a nutrient or to recognize SA exposed on the host cell surface (Li et al., 2011; Vimr et al., 2004). P. gingivalis sialidase can influence biofilm formation, capsule biosynthesis, regulation of gingipain activity, and virulence (Aruni et al., 2011; Li et al., 2011), whereas its abrogation has resulted in reduced virulence (Li et al., 2011), emphasizing the importance of sialidase in influencing P. gingivalis virulence. Among the structural domains found in bacterial sialidases, the β-propeller domain (BPD) is the SA binding interface and functions in cleaving terminal SA from glycoconjugates (Kim et al., 2011; Roggentin et al., 1989).
In general, the BPDs are disc-shaped and composed of twisted β-sheets radially arranged around a central tunnel lined with hydrogen donors and acceptors (Fulop and Jones, 1999). Protein binding interfaces are generally hydrophilic and have the tendency to form salt bridges, while uncommitted charged residues found within the protein binding interfaces may affect binding and overall protein structure (Missimer et al., 2007; Xu et al., 1997). Sialidase BPD is a common feature and conserved among bacterial sialidases (Buschiazzo and Alzari, 2008; Taylor, 1996). However, it is not clear whether the sialidase BPD of varying strains of the same bacterial species differ. In this study, we determined the salt bridge formation within the sialidase BPD of two different P. gingivalis strains known to have varying virulence (Amano et al., 2004).
Materials and Methods
Porphyromonas gingivalis sialidase homology modeling and quality estimation
We used P. gingivalis ATCC 33277 and W50 since both strains differ in terms of invasiveness and adherence (Pathirana et al., 2007, 2008). We utilized the sialidase amino acid sequences of P. gingivalis ATCC 33277 (Genebank accession: YP001929724) and W50 (Genebank accession: ZP14485603) to generate bacterial sialidase homology models using the Phyre 2 server (Kelley and Sternberg, 2009). Briefly, the Phyre server uses a library of known protein structures taken from the SCOP database and augmented with newer depositions in the PDB database. The sequence of each predicted 3D structure is scanned against a non-redundant sequence database, and the top ten highest scoring alignments are then used to construct full 3D models. We used the Jmol applet (Herraez, 2006) to visualize the homology models.
We validated the quality of the homology models using QMEAN (Qualitative Model Energy Analysis) and QMEAN Z-scores, whereby QMEAN denotes the scoring function based on six terms of normalized potentials related to protein length and QMEAN Z-scores are estimates of the “degree of nativeness” observed in a generated model by describing the probability that a model is of comparable quality to known high-resolution protein structures (Benkert et al., 2011). Homology models with QMEAN scores close to 0.5 and QMEAN Z-scores greater than -5 were considered acceptable for further analyses (Benkert et al., 2011). In addition, P. gingivalis sialidase homology models were superimposed with a known Streptococcus pneumoniae sialidase crystal structure (PDB ID: 4FOQ) using SuperPose (Maiti et al., 2004) to determine sialidase Cα backbone distribution in the BPD. Cα distribution values <1.0 would insinuate high structural similarities.
Sialic acid docking and sialidase activity detection
Molecular docking of SA (PDB ID: 2YA5) to the sialidase BPD were performed using HexServer (Macindoe et al., 2010). We focused on simulations that involve SA docking to the sialidase BPD interface and have the lowest energy requirement. In addition, amino acid residues with a distanc <3.5Å and an angle >90° relative to the docked SA were only considered for this study (Chen and Kurgan, 2009). We visualized the amino acid residues related to SA binding and, likewise, measured distance measurements and angles using the Jmol applet.
P. gingivalis ATCC 33277 and W50 were grown in Gifu anaerobic medium (Nissui, Tokyo, Japan) containing 5 μg mL−1 hemin and 0.5 μg mL−1 menadione under anaerobic conditions (10% CO2, 10% H2, and 80% N2 at 37°C). Growth media was used as negative control. Turbidity in both bacterial cultures was adjusted prior to downstream applications to OD600=1.0. Bacterial supernatants were filtered through sterile 0.22-μm-pore PVDF syringe filters (Millipore). Sialidase activity detection was determined following a previously published work, with modifications (Whiley et al., 1990). Fluorogenic substrate 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (4-MUNANA) (Sigma) dissolved in 50 mM phosphate buffer (pH 7.0) was mixed with the bacterial supernatant and incubated at 37°C for 2 h. Sialidase activity was determined by measuring 4-methylumbelliferone (MU) amounts released to the supernatant using a fluorometer with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. One unit was defined as the enzyme activity to release 1 μmol of 4-MU from 4-MUNANA per minute.
Potential salt bridge residue identification and distance measurement
We designated the six β-sheets found in both P. gingivalis sialidase BPDs as β1–6 following a clockwise orientation. Subsequently, we standardized the residue numbering of amino acid residues found in the sialidase BPD following strain W50 since it is 46 residues less than that of strain ATCC 33277. We identified salt bridge-forming residues (Lys, Arg, Glu, Asp) found in the sialidase BPD; distance measurements between identified positively charged residues (Lys, Arg) and negatively charged residues (Glu, Asp) were measured to establish which residue pair has the propensity to form salt bridges (<4.0 Å) within the sialidase BPD interface (Kumar and Nussinov, 2002). Similarly, we generated altered forms of our original sialidase homology models wherein any salt bridge residue that may influence the potential salt bridge formation within the BPD interface were substituted with Ala. Homology modeling and residue substitution were performed using the Phyre 2 server, while distance measurements and residue identification were determined using the Jmol applet.
Identification of variations in the sialidase homology models
Amino acid empirical distribution of the original and altered sialidase homology models were determined by establishing the Ramachandran plot using RAMPAGE (Lovell et al., 2003). Briefly, the RAMPAGE server utilizes density-dependent smoothing of amino acid residues that would show sharp boundaries at critical edges and would indicate a clear delineation between large empty areas and regions that are allowed but disfavored. Similarly, coarse-grained molecular dynamic simulation (CG-MD) was performed to determine the radius of gyration (Rgyr) of both the original and altered sialidase homology models using MDWeb (Hospital et al., 2012). Briefly, the MDWeb server is a web portal of the original software platform MDMoby, which is based in the Ambertools and VMD packages combined with in-house and publicly available programs. CG-MD simulation conditions were set at 500 ps simulation time with Δt at 0.01 ps. Furthermore, structural differences between the original and altered sialidase homology models were determined through protein superimposition using SuperPose. Root Mean Square Deviation (RMSD) values of the superimposed Cα backbone close to 0 would indicate high structural similarity, whereas, RMSD values above 1 would insinuate high structural difference.
Results
Sialidase BPD found in the ATCC 33277 and W50 sialidase homology models are structurally consistent
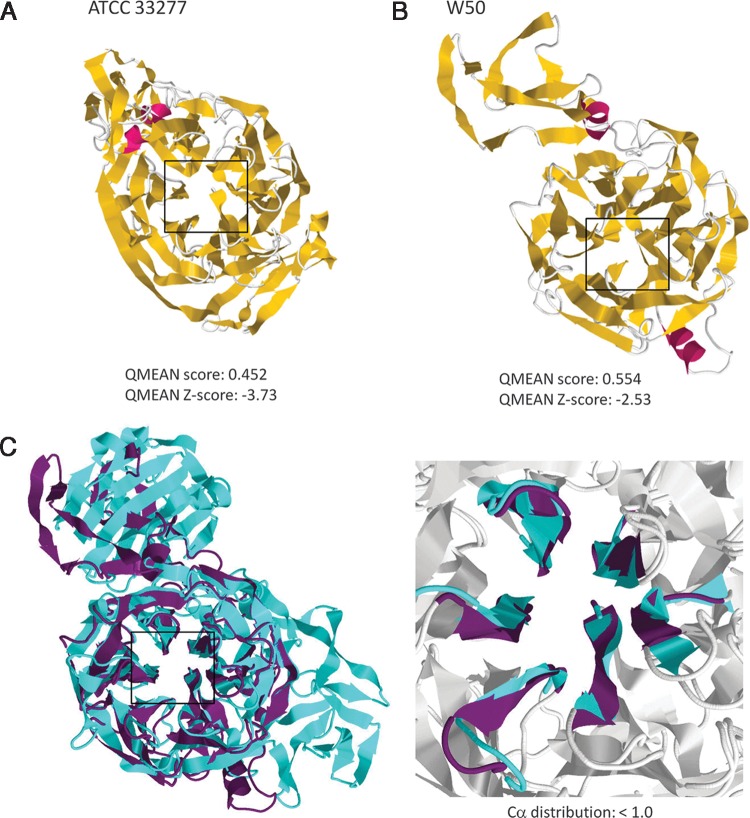
To generate and validate the ATCC 33277 and W50 sialidase structures, homology modeling and model quality estimation were performed. As seen in Figure 1A and 1B (upper panels), visual comparison of both P. gingivalis sialidase homology models show that they are structurally different; however, the location of the sialidase BPD (boxed region) is consistent. Similarly, based on pre-define values, we found that all models have acceptable QMEAN and QMEAN Z-scores (Fig. 1A and 1B, lower panels).
FIG. 1.
P. gingivalis ATCC 33277 and W50 sialidase homology modeling and model quality estimation. P. gingivalis strains (A) ATCC 33277 and (B) W50. Upper panel: Sialidase homology models. α-helix (pink), β-sheets (yellow), and coils (white) are shown. β-Propeller domain is indicated by a box. Lower panel: Model quality estimation. QMEAN and QMEAN Z-scores are shown. (C) Representative P. gingivalis sialidase homology model and S. pneumoniae sialidase crystal structure superimposition. Left panel: Ribbon structure with the sialidase domain region is indicated by a box. Right panel: Structural similarities of the sialidase domain are shown. P. gingivalis W50 (purple) and S. pneumoniae (blue) are indicated.
To confirm the structural accuracy of the sialidase BPD in the generated sialidase homology models, we compared our P. gingivalis sialidase homology models with the S. pneumoniae sialidase crystal structure. We found that superimposition of the BPD in the P. gingivalis sialidase homology models and S. pneumoniae crystal structure has high structural similarities (Fig. 1C). This would imply that the P. gingivalis sialidase BPD based on homology modeling and the S. pneumoniae sialidase BPD based on crystallization are structurally accurate.
ATCC 33277 and W50 sialidase activities differ regardless of having a common sialidase BPD
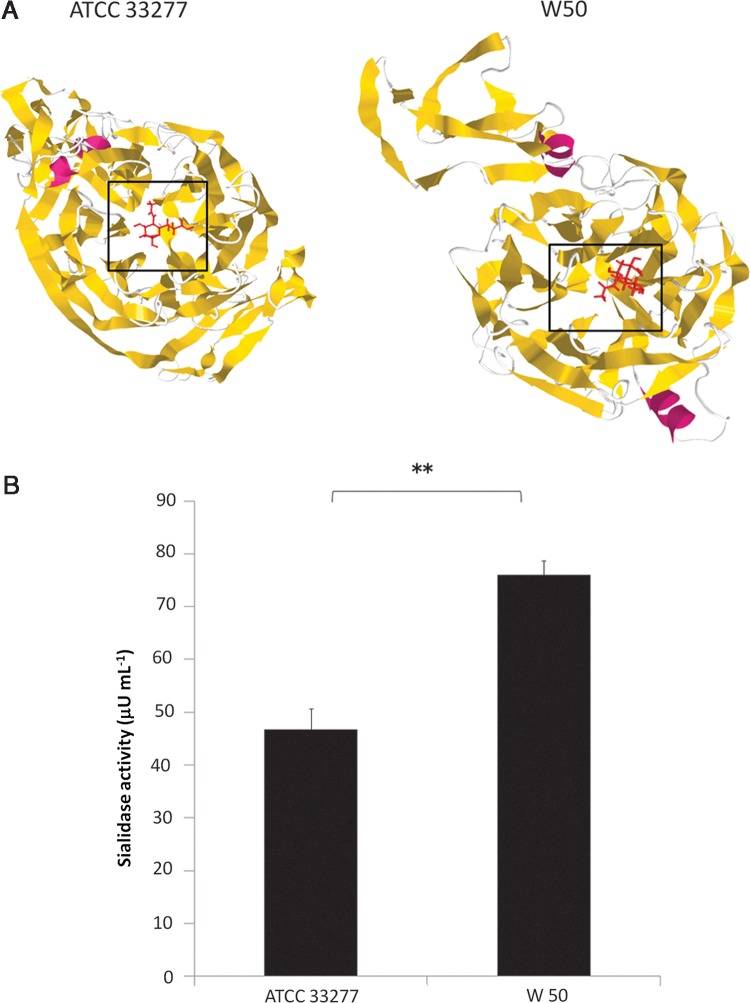
To further distinguish the ATCC 33277 and W50 sialidases, SA docking and sialidase activity assay were performed. We established that SA docks to both ATCC 33277 and W50 sialidase domains (Fig. 2A, boxed region). In addition, as seen in Figure 2B, we found that sialidase activities differ between the ATCC 33277 and W50 strains wherein W50 (75.95 μU mL−1) has a higher sialidase activity as compared to ATCC 33277 (46.74 μU mL−1). Moreover, no sialidase activity was detected in the bacterial growth media (data not shown), validating the accuracy of the sialidase activity assay used.
FIG. 2.
P. gingivalis ATCC 33277 and W50 have different sialidase activity measurements. (A) Proposed sialic acid-docking in P. gingivalis strains (left panel) ATCC 33277 and (right panel) W50. α-Helix (pink), β-sheets (yellow), coils (white), and sialic acid (red) are shown. β-Propeller domain is indicated by a box. (B) Sialidase activity assay. Results shown are mean±SD with n=3. Statistical analyses were performed using Student's t test (**represents p<0.01).
β1K396 residue influences the β2D444–β6K338 distance measurement and affects the sialidase homology model structure
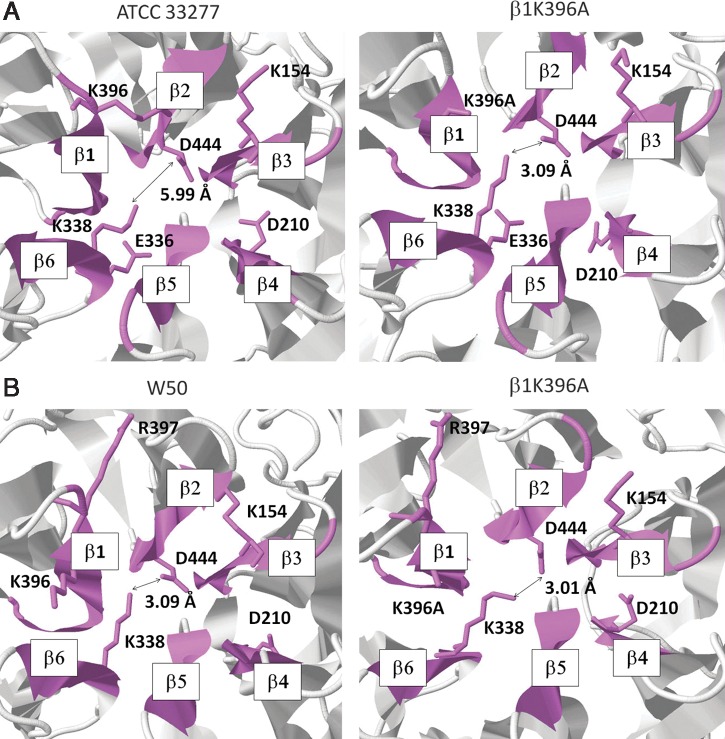
To establish the amino acid residues that have the tendency to form salt bridges, we identified salt bridge-forming residues found in both ATCC 33277 and W50 sialidase BPDs and indicated the distance measurements between potential salt bridge residues. As shown in Figure 3A and 3B (left panels), we observed that most salt bridge-forming residues have a common distribution pattern and, moreover, distance measurements between potential salt bridge residues (β2D444–β6K338) found in strains ATCC 33277 (5.99 Å) and W50 (3.09 Å) vary, possibly ascribable to a difference in β6K338 orientation. Interestingly, we noticed that the neighboring β1K396 residue is uncommitted and is positioned in close proximity to β6K338 that we suspect may affect β6K338 orientation.
FIG. 3.
β1K396 residue influences the P. gingivalis sialidase β-propeller domain β2D444–β6K338 distance measurement. Sialidase β-propeller domains of (A) ATCC 33277 and (B) W50. (Left panel) original ATCC 33277 and W50 (β1K396) and (Right panel) altered ATCC 33277 and W50 (β1K396A) sialidase β-propeller domains. Potential salt bridge-forming residues found in the sialidase β-propeller domain are in wireframe orientation. Six β-sheets found in the sialidase β-propeller domain are labeled as β1–6 following a clockwise orientation. Salt bridge distance measurement is indicated in angstrom (Å).
To confirm the β1K396 structural influence on the β2D444–β6K338 distance measurements, we generated altered ATCC 33277 and W50 (β1K396A) sialidase homology models and determined the β2D444–β6K338 distance measurement. We found that β1K396A substitution changed ATCC 33277 (3.09 Å) and W50 (3.01 Å) β2D444–β6K338 distance measurements. To elucidate the structural effects of altering the β2D444–β6K338 distance measurement, empirical distribution, folding activity, and structural differences of both the original (β1K396) and altered (β1K396A) sialidase homology models were determined through Ramachandran plot analyses, CG-MD simulation, and homology model superimposition, respectively.
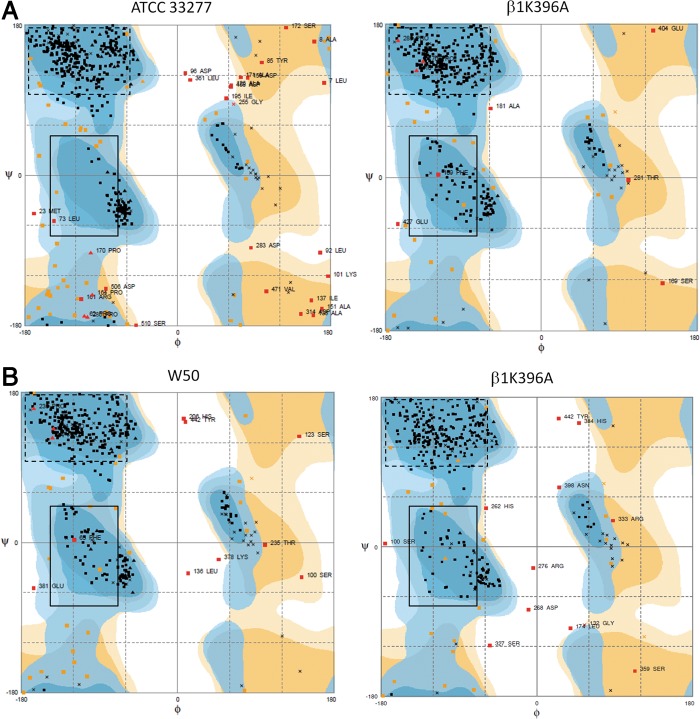
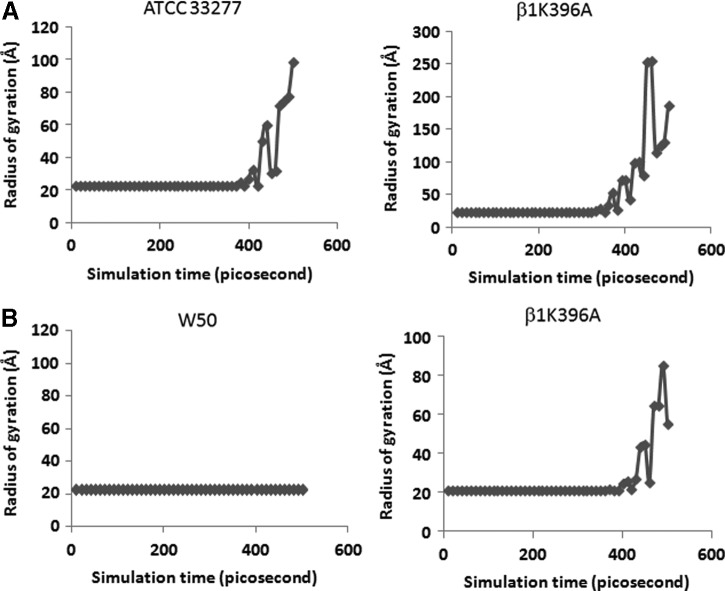
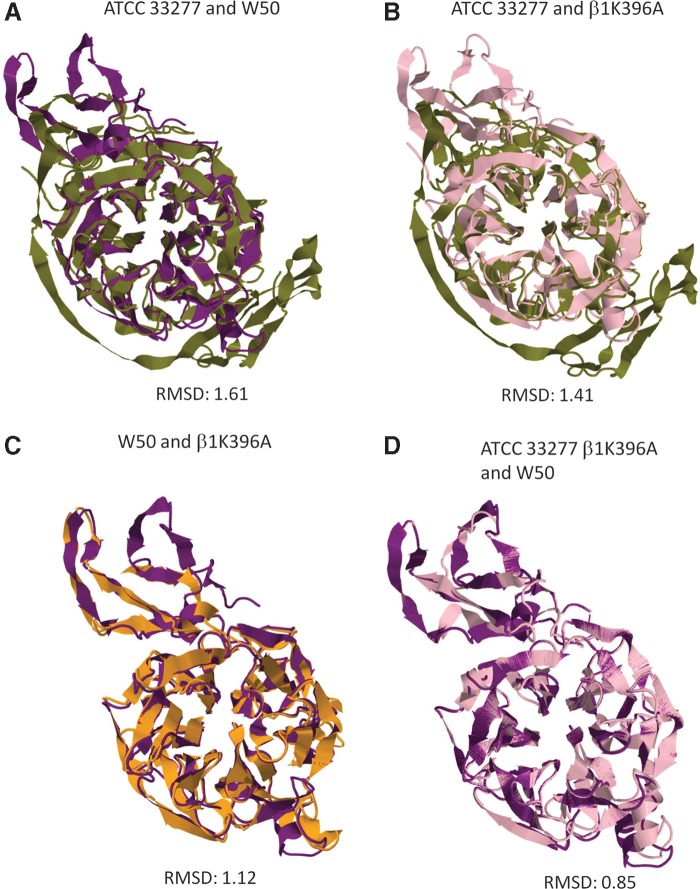
Our results showed that there were major differences in amino acid empirical distribution between the original (Fig. 4A, boxed left panels) and altered (Fig. 4A, boxed right panels) ATCC 33277 sialidase homology models, while minor differences were observed between the original (Fig. 4B, boxed left panels) and altered (Fig. 4B, boxed right panels) W50 sialidase homology models. Moreover, we found that the Rgyr between the original (Fig. 5A and 5B, upper and lowers left panels) and altered (Fig. 5A and 5B, upper and lower right panels) sialidase homology models differed, insinuating a change in folding activity. Furthermore, structural comparison between the original ATCC 33277 and W50 sialidase homology models (Fig. 6A) and cross comparison between the original and altered ATCC 33277 and W50 sialidase homology models (Fig. 6B and 6C) showed low structural similarity (RMSD: 1.61, 1.41, and 1.12, respectively). Surprisingly, visual inspection of the homology models generated suggests that the altered ATCC 33277 and the original W50 sialidase homology models are structurally similar. To corroborate the possible structural similarity between the two homology models, we superimposed both sialidase homology models (Fig. 6D) and found that both have high structural similarity (RMSD: 0.85).
FIG. 4.
β1K396A residue alters the amino acid empirical distribution of P. gingivalis ATCC 33277 and W50 sialidase. Ramachandran plots of (A) ATCC 33277 and (B) W50. (Left panel) original (β1K396) and (right panel) altered (β1K396A) ATCC 33277 and W50 sialidase homology models. Altered amino acid empirical distribution of α-helices and β-sheets are indicated in solid and dashed lines, respectively.
FIG. 5.
Sialidase homology model folding is affected by β1K396A substitution. Radius of gyration obtained from coarse-grained molecular dynamics simulation of (A) ATCC 33277 and (B) W50. (Left panel) original (β1K396) and (right panel) altered (β1K396A) ATCC 33277 and W50 sialidase homology models.
FIG. 6.
β1K396A residue alters overall P. gingivalis ATCC 33277 and W50 sialidase homology models. Superimposition of the (A) original ATCC 33277 and W50 (β1K396); (B) original ATCC 33277 (β1K396) and altered ATCC 33277 (β1K396A); (C) original W50 (β1K396) and altered W50 (β1K396A); and (D) altered ATCC 33277 (β1K396A) and original W50 (β1K396) sialidase homology models. RMSD scores below 1 are considered structurally similar, whereas scores above 1 are considered structurally different. Original ATCC 33277 (green), original W50 (purple), altered ATCC 33277 (pink), and altered W50 (orange) sialidase homology models are indicated.
Discussion
Bacterial sialidases can catalyze the hydrolysis of terminal sialic acids with α(2,3)-, α(2,6)-, or α(2,8)-linkages found in a diverse range of substrates either for nutrition or as a virulence factor (Varki and Varki, 2007; Vimr and Lichtensteiger, 2002; Vimr et al., 2004). Throughout this study, we established that both ATCC 33277 and W50 sialidases vary in structure and activity. Moreover, the sialidase homology models are structurally different except for the BPD which is a common feature among bacterial sialidases (Buschiazzo and Alzari, 2008; Taylor, 1996). This insinuates that P. gingivalis strains have varying sialidase homology models and activities regardless of having a common BPD. We propose that salt bridge formation within the sialidase BPD interface differs since binding interfaces are composed of salt bridges (Missimer et al., 2007; Xu et al., 1997).
We identified potential salt bridge-forming residues within the sialidase BPD interface at topologically identical positions that we suspect could be involved in protein folding and stability, enzyme activation, and biomolecular recognition (Breuker et al., 2011; Buschiazzo and Alzari, 2008; Meot-Ner Mautner, 2012; Wilks et al., 1988). In addition, we established differences in both ATCC 33277 and W50 β2D444–β6K338 distance measurements, whereby strain ATCC 33277 does not favor salt bridge formation while strain W50 favors salt bridge formation. Salt bridges have <4.0 Å distance measurement between residue pairs and are composed of two noncovalent interactions (hydrogen bonding and electrostatic interactions) with the most common salt bridges occurring between the anionic carboxylate of either Asp or Glu and the cationic ammonium from Lys or the guanidinium of Arg (Kumar and Nussinov, 2002). This would insinuate that the sialidase BPD interface in strains ATCC 33277 and W50 differ in salt bridge formation consistent with our earlier proposal. Consequently, we suspect that among the potential salt bridge-forming residues, uncommitted residues may affect salt bridge formation ascribable to amino acid charge.
Among the potential salt bridge-forming residues we identified, we found that residue β1K396 in both strains ATCC 33277 and W50 has the potential to influence the β2D444–β6K338 distance measurement. Similarly, we showed that β1K396 substitution affected the sialidase amino acid empirical distribution, folding activity, and overall structure. Lys under physiological conditions is a positively charged basic amino acid mostly exposed to the protein surface (Kumar et al., 2000) and is known to form ionic interactions, hydrogen bonds, and interact with water (Barlow and Thornton, 1983; Strickler et al., 2006). This highlights the significance of salt bridge formation within the conserved sialidase BPD and its putative effects on the sialidase protein structure.
We propose that β1K396 may exert an influence on residue β6K338 and affect the β2D444–β6K338 distance measurement, consequentially changing the sialidase amino acid empirical distribution, folding activity, and overall structure. Admittedly, additional study is needed to further prove this proposal. In addition, it is not clear whether structural changes associated to β1K396 substitution could influence P. gingivalis sialidase activity. However, it would be interesting to establish this in a future study.
P. gingivalis plays a major role in periodontitis induction (O' Brien-Simpson et al., 2003) and its virulence is greatly influenced by the sialidase enzyme (Aruni et al., 2011; Li et al., 2011) wherein the sialidase BPD is highly conserved among various bacterial species (Buschiazzo and Alzari, 2008). Our results suggest that alterations in the salt bridge formation within the BPD interface may affect the P. gingivalis sialidase structure. This would imply that disrupting the salt bridge formation within the P. gingivalis sialidase BPD could serve as a potential therapeutic strategy for the treatment of P. gingivalis-related periodontitis.
It is worth mentioning that both the altered ATCC 33277 and original W50 sialidase homology models have high structural similarity possibly attributable to having the same β2D444–β6K338 distance measurements (3.09 Å). In addition, this may suggest that both strains ATCC 33277 and W50 shared a common ancestry (Igboin et al., 2009). Protein evolution is influenced by several factors that include, among others, protein structure and function (Pal et al., 2006; Wilson et al., 1977). We hypothesize that the W50 sialidase evolved from the ATCC 33277 sialidase. It would be interesting to verify this hypothesis in a future study.
Conclusion
In summary, we established that ATCC 33277 and W50 sialidases differ in structure and activity. Moreover, we found that the sialidase BPD in the generated P. gingivalis sialidase homology models and crystal structure have high structural similarities consistent with other bacterial sialidase proteins. In addition, we identified residue β1K396 to have the propensity to influence the salt bridge β2D444–β6K338 distance measurement altering the amino acid empirical distribution, folding activity, and overall structure highlighting a potential therapeutic strategy against P. gingivalis-related periodontitis.
Acknowledgments
This study was supported by the following grants: Grants-in-Aid for Young Scientist (B) 25862088 and 26861580, Nihon University President's Grant for Multidisciplinary Research, Uemura Fund and Dental Research Center-Nihon University School of Dentistry (Tokyo), “Strategic Research Base Development” Program for Private Universities subsidized by the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) 2010 (S1001024).
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Amano A, Nakagawa I, Okahashi N, and Hamada N. (2004). Variations of Porphyromonas gingivalis fimbriae in relation to microbial pathogenesis. J Periodontal Res 39, 136–142 [DOI] [PubMed] [Google Scholar]
- Aruni W, Vanterpool E, Osbourne D, Roy F, Muthiah A, Dou Y, and Fletcher HM. (2011). Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect Immun 79, 2779–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DJ, and Thornton JM. (1983). Ion-pairs in proteins. J Mol Biol 168, 867–885 [DOI] [PubMed] [Google Scholar]
- Benkert P, Biasini M, and Schwede T. (2011). Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27, 343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuker K, Bruschweiler S, and Tollinger M. (2011). Electrostatic stabilization of a native protein structure in the gas phase. Angew Chem Int Ed Engl 50, 873–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschiazzo A, and Alzari PM. (2008). Structural insights into sialic acid enzymology. Curr Opin Chem Biol 12, 565–572 [DOI] [PubMed] [Google Scholar]
- Cekici A, Kantarci A, Hasturk H, and Van Dyke TE. (2014). Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000 64, 57–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, and Kurgan L. (2009). Investigation of atomic level patterns in protein–small ligand interactions. PLoS One 4, e4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop V, and Jones DT. (1999). Beta propellers: Structural rigidity and functional diversity. Curr Opin Struct Biol 9, 715–721 [DOI] [PubMed] [Google Scholar]
- Genco RJ. (1996). Current view of risk factors for periodontal diseases. J Periodontol 67, 1041–1049 [DOI] [PubMed] [Google Scholar]
- Herraez A. (2006). Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ 34, 255–261 [DOI] [PubMed] [Google Scholar]
- Hospital A, Andrio P, Fenollosa C, Cicin-Sain D, Orozco M, and Gelpi JL. (2012). MDWeb and MDMoby: An integrated web-based platform for molecular dynamics simulations. Bioinformatics 28, 1278–1279 [DOI] [PubMed] [Google Scholar]
- Igboin CO, Griffen AL, and Leys EJ. (2009). Porphyromonas gingivalis strain diversity. J Clin Microbiol 47, 3073–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, and Sternberg MJ. (2009). Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc 4, 363–371 [DOI] [PubMed] [Google Scholar]
- Kim S, Oh DB, Kang HA, and Kwon O. (2011). Features and applications of bacterial sialidases. Appl Microbiol Biotechnol 91, 1–15 [DOI] [PubMed] [Google Scholar]
- Kumar S, and Nussinov R. (2002). Close-range electrostatic interactions in proteins. Chembiochem 3, 604–617 [DOI] [PubMed] [Google Scholar]
- Kumar S, Tsai CJ, and Nussinov R. (2000). Factors enhancing protein thermostability. Protein Eng 13, 179–191 [DOI] [PubMed] [Google Scholar]
- Lamont RJ, and Jenkinson HF. (1998). Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev 62, 1244–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Kurniyati Hu B, Bian J, Sun J, Zhang W, Liu J, and Pan Y. (2011). Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis. Infect Immun 80, 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell SC, Davis IW, Arendall WB 3rd, et al. (2003). Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- MacIndoe G, Mavridis L, Venkatraman V, Devignes MD, and Ritchie DW. (2010). HexServer: An FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 38, W445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti R, Van Domselaar GH, Zhang H, and Wishart DS. (2004). SuperPose: A simple server for sophisticated structural superposition. Nucleic Acids Res 32, W590–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meot-Ner Mautner M. (2012). Update 1 of: Strong ionic hydrogen bonds. Chem Rev 112, PR22–103 [DOI] [PubMed] [Google Scholar]
- Missimer JH, Steinmetz MO, Baron R, et al. (2007). Configurational entropy elucidates the role of salt-bridge networks in protein thermostability. Protein Sci 16, 1349–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O' Brien-Simpson NM, Veith PD, Dashper SG, and Reynolds EC. (2003). Porphyromonas gingivalis gingipains: The molecular teeth of a microbial vampire. Curr Protein Pept Sci 4, 409–426 [DOI] [PubMed] [Google Scholar]
- Pal C, Papp B, and Lercher MJ. (2006). An integrated view of protein evolution. Nat Rev Genet 7, 337–348 [DOI] [PubMed] [Google Scholar]
- Pathirana RD, O'Brien-Simpson NM, Visvanathan K, Hamilton JA, and Reynolds EC. (2007). Flow cytometric analysis of adherence of Porphyromonas gingivalis to oral epithelial cells. Infect Immun 75, 2484–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathirana RD, O'Brien-Simpson NM, Visvanathan K, Hamilton JA, and Reynolds EC. (2008). The role of the RgpA-Kgp proteinase-adhesin complexes in the adherence of Porphyromonas gingivalis to fibroblasts. Microbiology 154, 2904–2911 [DOI] [PubMed] [Google Scholar]
- Roggentin P, Rothe B, Kaper JB, Galen J, Lawrisuk L, Vimr ER, and Schauer R. (1989). Conserved sequences in bacterial and viral sialidases. Glycoconj J 6, 349–353 [DOI] [PubMed] [Google Scholar]
- Strickler SS, Gribenko AV, Keiffer TR, Tomlinson J, Reihle T, Loladze VV, and Makhatadze GI. (2006). Protein stability and surface electrostatics: A charged relationship. Biochemistry 45, 2761–2766 [DOI] [PubMed] [Google Scholar]
- Taylor G. (1996). Sialidases: Structures, biological significance and therapeutic potential. Curr Opin Struct Biol 6, 830–837 [DOI] [PubMed] [Google Scholar]
- Varki NM, and Varki A. (2007). Diversity in cell surface sialic acid presentations: Implications for biology and disease. Lab Invest 87, 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E, and Lichtensteiger C. (2002). To sialylate, or not to sialylate: That is the question. Trends Microbiol 10, 254–257 [DOI] [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, and Steenbergen SM. (2004). Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev 68, 132–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley RA, Fraser H, Hardie JM, and Beighton D. (1990). Phenotypic differentiation of Streptococcus intermedius, Streptococcus constellatus, and Streptococcus anginosus strains within the “Streptococcus milleri group”. J Clin Microbiol 28, 1497–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks HM, Hart KW, Feeney R, et al. (1988). A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science 242, 1541–1544 [DOI] [PubMed] [Google Scholar]
- Wilson AC, Carlson SS, and White TJ. (1977). Biochemical evolution. Annu Rev Biochem 46, 573–639 [DOI] [PubMed] [Google Scholar]
- Xu D, Tsai CJ, and Nussinov R. (1997). Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng 10, 999–1012 [DOI] [PubMed] [Google Scholar]