Abstract
Elongator is a conserved, multi-protein complex discovered in Saccharomyces cerevisiae, loss of which confers a range of pleiotropic phenotypes. Elongator in higher eukaryotes is required for normal growth and development and a mutation in the largest subunit of human Elongator (Elp1) causes familial dysautonomia, a severe recessive neuropathy. Elongator promotes addition of mcm5 and ncm5 modifications to uridine in the tRNA anticodon ‘wobble’ position in both yeast and higher eukaryotes. Since these modifications are required for the tRNAs to function efficiently, a translation defect caused by hypomodified tRNAs may therefore underlie the variety of phenotypes associated with Elongator dysfunction. The Elp1 carboxy-terminal domain contains a highly conserved arginine/lysine-rich region that resembles a nuclear localization sequence (NLS). Using alanine substitution mutagenesis, we show that this region is essential for Elongator's function in tRNA wobble uridine modification. However, rather than acting to determine the nucleo-cytoplasmic distribution of Elongator, we find that the basic region plays a critical role in a novel interaction between tRNA and the Elp1 carboxy-terminal domain. Thus the conserved basic region in Elp1 may be essential for tRNA wobble uridine modification by acting as tRNA binding motif.
Introduction
Elongator is a six-subunit protein complex (Elp1–6) that was first discovered in Saccharomyces cerevisiae (Otero et al., 1999) but which is present in all eukaryotes. A mutation in the IKBKAP gene encoding the human homologue of Elongator's largest subunit (Elp1) causes Familial Dysautonomia, a severe recessive neurodevelopmental disease (Anderson et al., 2001; Slaugenhaupt et al., 2001), while the human homologues of ELP3 and ELP4 have also been linked to the neurological disorders amyotrophic lateral sclerosis and rolandic epilepsy respectively (Simpson et al., 2009; Strug et al., 2009). In mice deletion of ELP1 is embryonic lethal, suggesting that Elongator function is required during embryogenesis (Chen et al., 2009b) and in other higher eukaryotes, loss of Elongator function is associated with a range of developmental and growth defects (Nelissen et al., 2005; Chen et al., 2009a; Mehlgarten et al., 2010; Singh et al., 2010; DeFraia and Mou, 2011; Walker et al., 2011).
In budding yeast, the genes encoding Elongator are non-essential but when deleted lead to a common set of phenotypes (Otero et al., 1999; Frohloff et al., 2001; Li et al., 2009), one of which is resistance to zymocin, a protein toxin secreted by the yeast Kluyveromyces lactis (Frohloff et al., 2001). Zymocin is a tRNA anticodon nuclease that cleaves tRNAGlu(UUC) and to a lesser extent tRNALys(UUU) and tRNAGln(UUG), three tRNAs that contain 5-methoxycarbonymethyl-2-thiouridine (mcm5s2U) in the anticodon wobble position (Lu et al., 2005). The mcm5s2U modification is essential for substrate recognition by zymocin, which targets tRNAGlu(UUC) for cleavage on the 3′ side of the wobble position (Lu et al., 2005). Elongator mutant strains lack the 5-methyoxycarbonylmethyl (mcm5) and 5-carbamoylmethyl (ncm5) modifications on wobble uridine-containing tRNAs and in the case of the zymocin substrate tRNAs, absence of the mcm5 moiety protects them from recognition and cleavage by zymocin (Huang et al., 2005). The Elongator complex is therefore required for an early step in synthesis of the mcm5 and ncm5 modifications on wobble uridine-containing tRNAs (Huang et al., 2005). In yeast, 11 out of 42 tRNAs carry these Elongator-dependent modifications, which are required for efficient decoding at the wobble position during translation (Huang et al., 2005; Agris, 2008; Johansson et al., 2008). Despite the variety of disparate roles that have been proposed for Elongator (Otero et al., 1999; Wittschieben et al., 1999; Rahl et al., 2005; Creppe et al., 2009), the major role of Elongator in yeast is now thought to be in tRNA wobble uridine modification: increased expression of tRNALys(UUU) and tRNAGln(UUG) is sufficient to rescue the majority of elp mutant phenotypes tested so far (Esberg et al., 2006; Chen et al., 2011), providing strong evidence that these mutant phenotypes are caused by a common translational defect. Like Elongator itself, its requirement for tRNA wobble uridine modification is also conserved in worms, plants and mice (Chen et al., 2009a; Mehlgarten et al., 2010; Lin et al., 2013).
The Elongator holocomplex is currently thought to exist as a dodecamer containing two copies of each of its six subunits. Elp4, Elp5 and Elp6 each contain a common RecA-like NTPase fold and assemble into a heterohexameric ring structure with which two Elp1–3 heterotrimers have been proposed to associate (Glatt et al., 2012; Glatt and Muller, 2013). While it is not yet known whether Elongator acts directly in the catalysis of the tRNA modifications, Elp3 contains two functionally important enzymatic domains that may be involved; an N-terminal Radical S-adenosylmethionine (SAM)-like domain (Chinenov, 2002) and a C-terminal GNAT-type histone acetyltransferase (HAT) like domain (Wittschieben et al., 1999), both of which are essential for tRNA modification (Huang et al., 2005; Chen et al., 2011). The combination of these two functional domains in Elp3 is highly conserved and enzymes containing Radical SAM domains have already been implicated in other RNA modification reactions (Agarwalla et al., 2002; Pierrel et al., 2004; Anton et al., 2008; Yan et al., 2010; Grove et al., 2011). In addition, the Elongator accessory protein Kti12, which is related to O-phosphoseryl-tRNASec kinase (PSTK; Sherrer et al., 2008), is also required for Elongator-dependent tRNA wobble uridine modification (Butler et al., 1994; Frohloff et al., 2001; Fichtner et al., 2002a) although its role is currently unknown.
Elp1 is the largest Elongator subunit and has been thought to act as a core scaffold, since interactions of Elp2 and Elp3 with each other and with components of the Elp4–6 subcomplex are lost when Elp1 is deleted (Fichtner et al., 2002b; Frohloff et al., 2003). Budding yeast Elp1 contains an Arg/Lys-rich basic region in the C-terminal domain that was previously noted as a putative nuclear localization sequence (NLS: Fichtner et al., 2002b, Fichtner et al., 2003). Since tRNA anticodon modifications are generally thought to occur in the cytoplasm (Hopper and Phizicky, 2003; Huang et al., 2005) and it is known that Elp1 is predominantly cytoplasmic in both yeast and human cells (Kim et al., 2002; Pokholok et al., 2002; Huh et al., 2003), it was unclear why the Elongator complex might require a nuclear import sequence. We therefore set out to investigate the importance of the Elp1 basic region in terms of Elongator function and have found that it mediates the binding of tRNA to Elp1, suggesting a critical role for the Elp1 C-terminal domain in the binding of tRNA to the Elongator complex.
Results
A highly conserved basic region in Elp1 is essential for its function
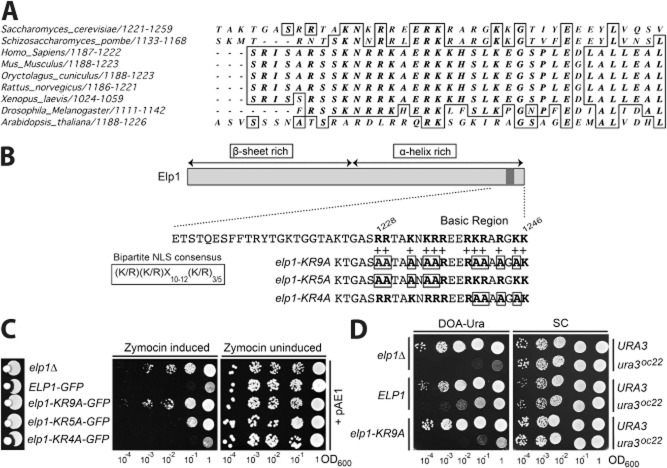
The yeast Elp1 C-terminal domain contains an Arg/Lys-rich basic region that is highly conserved across eukaryotes (Fig. 1A), implying that it could be functionally important. This region had previously been noted to resemble a putative bipartite NLS (Fichtner et al., 2003), which has the classical consensus sequence (K/R)(K/R)X10–12(K/R)3/5 (Lange et al., 2007; Kosugi et al., 2009). To study the functional significance of this conserved basic region a series of mutations were designed throughout its sequence. A mutant allele (elp1–KR9A) was generated in which nine of the basic residues were substituted by alanine, as well as elp1–KR5A and elp1–KR4A alleles containing alanine substitutions in the first and second halves of the basic region respectively (Fig. 1B). All three mutants were incorporated into the genomic ELP1 locus using the ‘delitto perfetto’ approach.
Fig. 1.
A highly conserved basic region in Elp1 is essential for Elongator function.
A. Multiple sequence alignment of the Elp1 subunit of Elongator from various eukaryotic organisms showing a portion of the Elp1 C-terminal domain including the basic region to highlight its conservation. Alignment was carried out using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Sequences used were as follows with accession numbers in brackets; S. cerevisiae (Q06706), S. pombe (O59704), H. sapiens (O95163), M. musculus (Q7TT37), O. cuniculus (Q8WND5), R. norveigus (Q8VHU4), X. laevis (Q2TAQ1), D. melanogaster (Q9VGK7), A. thaliana (Q9FNA4).
B. Schematic of Elp1 highlighting the Arg/Lys rich basic region (basic residues in bold) and alanine substitution mutations (boxed) made within this region. Mutations were made across the entire Elp1 basic region (elp1–KR9A) and within the first and second halves of the region (elp1–KR5A and elp1–KR4A respectively). For comparison the consensus sequence for a bipartite NLS is also shown. The indicated structural predictions were made using PSIPRED (Buchan et al., 2013).
C. Zymocin sensitivity assays of strains containing GFP tagged wild-type ELP1 or elp1 basic region mutants at the ELP1 genomic locus. Wild-type (YRDS253) and elp1Δ (YRDS250) served as controls for zymocin sensitivity and resistance respectively. Left panel, Eclipse assays on YPAD medium measuring growth inhibition adjacent to a colony of a zymocin-secreting K. lactis strain. Centre and right panels, growth of equivalent 10-fold serial dilutions (indicating OD600) of the indicated strains containing pAE1, which expresses the zymocin γ subunit under control of a galactose inducible promoter. Strains were grown on YPARG (centre panel: zymocin induced) and YPAD (right panel: zymocin uninduced).
D. Growth of equivalent 10-fold serial dilutions of the indicated strains containing chromosomal copies of ELP1 or elp1–KR9A together with the SUP4 ochre suppressor and either wild-type URA3 (as a control) or the ura3oc22 ochre allele. SUP4 can read through the premature stop codon in ura3oc22 to allow growth in the absence of uracil on DOA-Ura medium but efficient suppression requires mcm5 modification of the SUP4 tRNA at the wobble uridine position, as shown by comparing growth of the ELP1 and elp1Δ ura3oc22 strains. Growth of the same strains on synthetic complete (SC) medium without uracil selection is shown as a control.
Eclipse assays, in which sensitivity to K. lactis zymocin is scored by formation of a halo of growth inhibition around a colony of K. lactis, are a convenient method of assessing Elongator function because loss of Elongator-dependent tRNA modification confers resistance to this toxin (Huang et al., 2005). Figure 1C (left panel) shows that while strains with wild-type Elongator were sensitive to zymocin as expected, the elp1Δ control, elp1–KR9A and elp1–KR5A mutant strains were all resistant to zymocin, indicating that they are defective in Elongator function. In contrast, the elp1–KR4A mutant showed sensitivity to zymocin (Fig. 1C, left panel). To gain a more quantitative readout of Elongator function in the three mutants, a plasmid (pAE1) expressing the toxic γ subunit of zymocin under control of a galactose inducible promoter (Butler et al., 1994) was introduced into each strain and then intracellular expression of the γ subunit induced by growth on galactose-containing medium. Figure 1C (centre and right panels) shows that the elp1–KR9A mutant was fully resistant to intracellular zymocin expression in comparison with an elp1Δ strain that is completely lacking Elongator function. However, the elp1–KR5A strain showed a lower level of resistance while the elp1–KR4A mutant appeared as sensitive as the wild-type ELP1 control. Taken together, these results indicate that elp1–KR4A, elp1–KR5A and elp1–KR9A show progressive reduction in zymocin sensitivity, with elp1–KR9A having the strongest resistance phenotype, comparable to that of the elp1Δ null strain. Although elp1–KR4A had little effect on its own, it clearly enhanced the defect of elp1–KR5A when the two sets of mutations were combined (elp1–KR9A) and since the elp1–KR9A mutant shows the most severe loss of function, the entire basic region must contribute to Elongator function.
To provide an additional readout of Elongator function, we next employed an assay that monitors readthrough of a ura3 ochre allele (ura3oc22) by the SUP4 ochre suppressor tRNATyr(UUA), which depends on Elongator-dependent modification of its wobble uridine residue for efficient UAA codon readthrough (Huang et al., 2005). Consequently, only strains with functional Elongator can efficiently suppress ura3oc22 and grow well on medium lacking uracil, while strains that have compromised Elongator function and therefore reduced levels of mcm5U tRNA modification show strongly reduced growth on medium lacking uracil (DOA-Ura). The difference in such growth between wild-type ELP1 SUP4 ura3oc22 and elp1Δ SUP4 ura3oc22 strains indicates the extremes of phenotype against which other mutant elp1 alleles can be compared. Figure 1D shows that the elp1–KR9A mutant and elp1Δ control strains conferred comparable defects in SUP4 ochre suppression, confirming that elp1–KR9A is largely lacking in Elongator function. Taken together, these results indicate that the Elp1 basic region is essential for Elongator-dependent tRNA modification function of Elongator.
Elp1 basic region mutations do not alter its nucleo-cytoplasmic distribution
The yeast Elp1 basic region has been previously suggested to resemble a bipartite NLS and when fused to GFP, the Elp1 C-terminal domain (CTD) can direct import of the fusion protein into the nucleus (Fichtner et al., 2003). In spite of this, Elp1 has been reported to be mainly cytoplasmic in both yeast and human cells (Kim et al., 2002; Pokholok et al., 2002; Huh et al., 2003), while other work has shown that addition of a bona fide NLS onto the Elp1 C-terminus restricts Elp1 to the nucleus and causes loss of Elongator function, consistent with a cytoplasmic role for the complex (Rahl et al., 2005). However, the potential role of this region in determining the distribution of Elongator between the nucleus and the cytoplasm has not yet been fully addressed and in order to understand why Elp1's basic region is essential for Elongator function, we therefore wished first to exclude any roles it might have in determining the subcellular distribution of Elongator.
In agreement with earlier work (Fichtner et al., 2003), we found that the Elp1 C-terminal domain (amino acids 1181–1349) was capable of driving nuclear import of GFP and that this was dependent specifically on the basic region. Thus GFP tagged with the wild-type Elp1 C-terminus was concentrated exclusively in the cell nucleus (Fig. 2), while the elp1–KR9A, elp1–KR5A and elp1–KR4A mutants all showed diffuse localization of GFP throughout the cell that was comparable to that of GFP alone, suggesting that when the basic region is mutated the C-terminus can no longer influence GFP localization. The basic region in the Elp1 carboxy-terminal domain therefore has the potential to drive nuclear import.
Fig. 2.
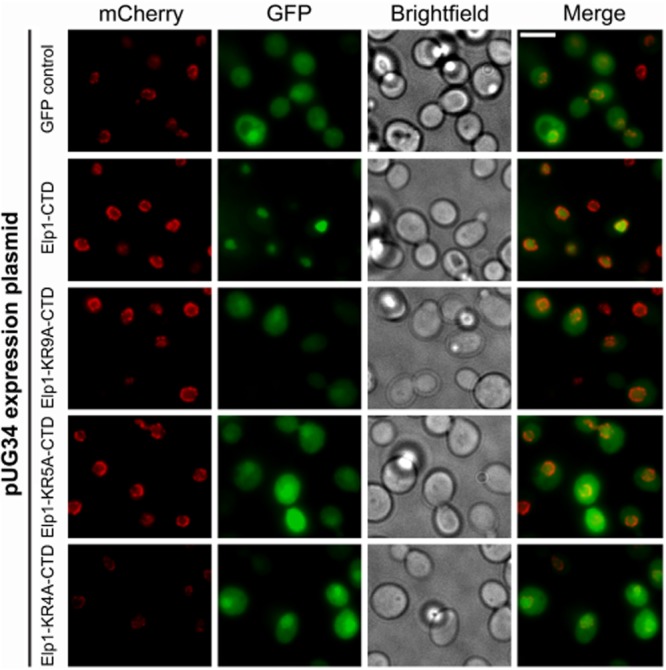
The Elp1 C-terminal domain can drive nuclear import of GFP dependent on the conserved basic region. Representative images of cells expressing Nic96-4mCherry (red) to indicate the nuclear periphery (YRDS84) containing either pUG34 (GFP control) expressing GFP or its derivative plasmids expressing GFP fused at its C-terminus to residues 1181–1349 from wild-type (Elp1–CTD) or the mutant (Elp1–KR4A-CTD, Elp1–KR5A–CTD, Elp1–KR9A–CTD) versions of Elp1 shown in Fig. 1 (green). Cells were grown to log phase in DOA-Met-His medium to induce GFP expression and then fixed for imaging. Both GFP (27 kDa) and the fusion proteins (47 kDa) are expected to be present in both the nucleus and cytoplasm in the absence of active nuclear localization signals as they are small enough to enter the nucleus by diffusion. CTD, C-terminal domain; scale bar: 5 μm.
However, when the distribution of wild-type full length Elp1 between the nucleus and cytoplasm was examined using strains with GFP-tagged Elp1 and mCherry-tagged Nic96 (to define the nuclear boundary), localization of wild-type Elp1–GFP was largely cytoplasmic (Fig. 3A) in agreement with previous reports (Pokholok et al., 2002; Huh et al., 2003; Tkach et al., 2012), but with low levels of Elp1–GFP fluorescence in the nucleus. Furthermore, when the localization of the Elp1–KR9A–GFP mutant was also assayed it showed no difference in the nucleo-cytoplasmic distribution of Elp1 in comparison with wild-type Elp1–GFP (Fig. 3A). Thus while the basic region of Elp1 can drive nuclear import in isolation, in the context of the whole polypeptide this does not appear to be functionally relevant and the apparent nucleo-cytoplasmic distribution of Elp1 was unaffected by the basic region mutation. The elp1–KR5A and elp1–KR4A mutants also showed no differences in Elp1 localization (data not shown). The fact that the elp1–KR4A mutant was almost normal in terms of Elongator function in comparison with the defective elp1–KR9A allele (Fig. 1) and yet the C-terminal domain from either mutant protein was equally compromised in its ability to drive nuclear import of GFP (Fig. 2) is consistent with the role of this region being unrelated to potential NLS function. Finally, we confirmed that when the NLS from Cbp80 was inserted into our Elp1–GFP construct that Elp1–NLS–GFP localized exclusively to the nucleus (Fig. 3A) and that this was associated with complete loss of Elongator-dependent tRNA modification as assayed by zymocin resistance (Fig. 3B). In contrast, when four basic amino acids in the Cbp80 NLS sequence were mutated, Elp1 localization reverted to a mainly cytoplasmic distribution and full sensitivity to zymocin was restored (Fig. 3). This extends the previous findings (Rahl et al., 2005) by showing that Elongator's tRNA wobble uridine modification function, as assayed by zymocin sensitivity, is abolished when Elp1 is trapped in the nucleus. This supports a tRNA modification role for Elongator that requires cytoplasmic rather than nuclear localization. Taken together, these data are therefore consistent with the functional importance of the Elp1 basic region being not through acting as a determinant of nucleo-cytoplasmic localization but through playing some other role.
Fig. 3.
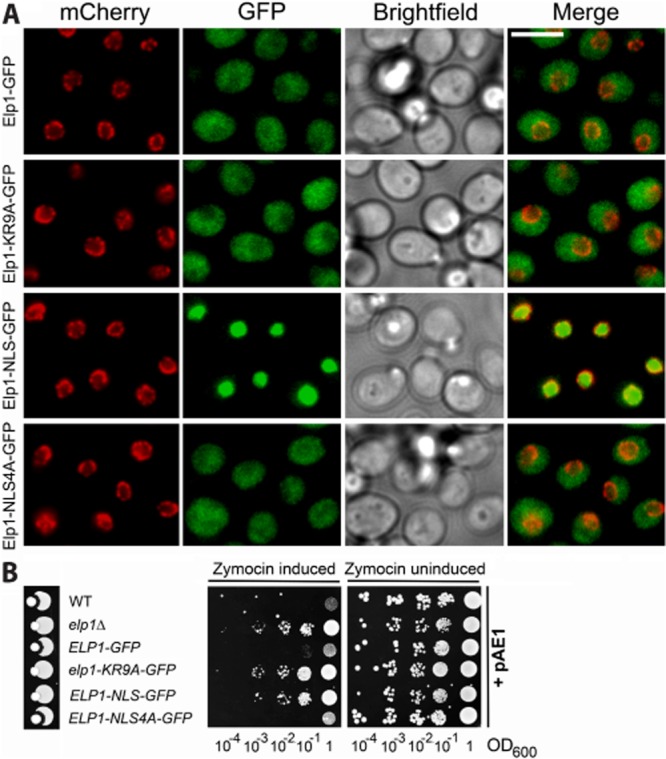
The nucleo-cytoplasmic distribution of Elp1 is unaffected by basic region mutations.
A. Representative images of strains expressing Nic96-4mCherry (red) together with GFP-tagged wild-type Elp1, Elp1–KR9A, Elp1–NLS or Elp1–NLS4A (green). Elp1–NLS contains the NLS from Cbp80 and Elp1–NLS4A contains a mutant version containing 4 alanine substitutions, inserted in each case between the Elp1 and GFP sections of the fusion protein. Cells were grown to log phase in YPAD medium and then fixed for imaging. Scale bar: 5 μm.
B. Zymocin sensitivity assays of the indicated strains as described in the legend to Fig. 2. Wild-type (WT, BY4741) and elp1Δ (YRDS250) served as controls for zymocin sensitivity and resistance respectively.
The Elp1 basic region mutation does not disrupt Elongator complex assembly but leads to subtle differences in subunit association
Recent work has suggested that the dodecameric Elongator holocomplex assembles such that a heterohexameric ring of Elp4–6 bridges two subcomplexes of Elp1–Elp3 (Glatt et al., 2012). Furthermore, the accessory protein Kti12 binds to the preassembled Elongator complex and is also required for its tRNA modification function (Butler et al., 1994; Frohloff et al., 2001; Fichtner et al., 2002a). It was therefore possible that the Elp1 basic region could act as a protein interaction domain that is required for assembly of the holocomplex. To examine Elongator complex assembly in the elp1–KR9A basic region mutant, the interaction of GFP tagged Elp1 and Elp1–KR9A with other HA tagged Elongator complex members was tested. Representative members of the two subcomplexes of Elongator, Elp3 and Elp5 respectively, as well as Kti12, were HA tagged in strains expressing GFP tagged Elp1 or Elp1–KR9A. As a control for the specificity of these protein–protein interactions, Elp3, Elp5 and Kti12 were also HA tagged in a strain expressing Arg4–GFP, a protein with no known connection to Elongator but reported to be present at similar levels.
GFP–Trap immunoprecipitations of Elp1–GFP and Elp1–KR9A–GFP were assayed for association with HA tagged Kti12, Elp3 and Elp5 and these were each present as co-precipitants under physiological salt conditions (Fig. 4A). This confirmed both that the Elp1–3 sub complex was intact and that interactions with Elp5 and Kti12 are not abolished by the basic region mutation, and so the Elp1 basic region is not required for the overall gross assembly of the Elongator complex. However, when these interactions were analysed carefully by densitometry, Elp1–KR9A–GFP was consistently found associated with ∼ 30% less Elp5-3HA and ∼ 40% less Kti12-3HA than the wild-type Elp1–GFP protein (Fig. 4B). These minor interaction defects are highly consistent and statistically significant. In contrast, Elp3 levels and the interaction between Elp1–KR9A–GFP and Elp3-3HA appeared largely unchanged (Fig. 4B), consistent with earlier work in which Elp3 was found to become very unstable in strains where it could not interact with Elp1 (Petrakis et al., 2004). The small defects in Kti12 and Elp5 interactions with the Elp1–KR9A mutant may reflect the fact that Elongator is no longer functional. For example, if the complex needs to shuttle between multiple forms and is stalled due to the basic region mutations, this may lead to reduced complex stability and subsequently affect the interaction with other subunits.
Fig. 4.
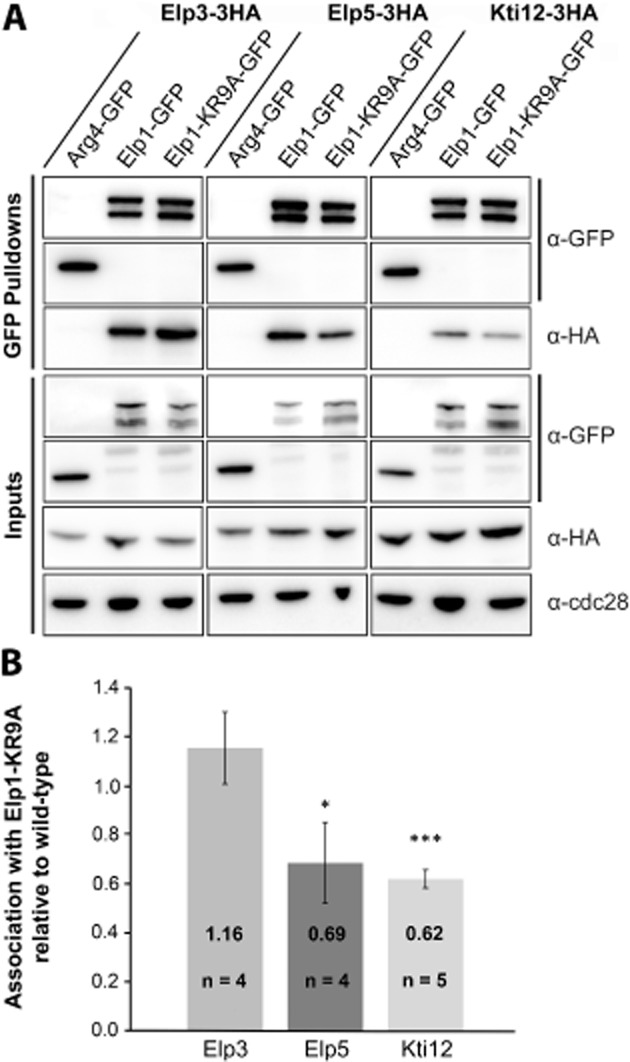
The elp1–KR9A mutation does not disrupt Elongator complex assembly but causes subtle differences in association with Elp5 and Kti12.
A. Western blot analysis of inputs and GFP–Trap immunoprecipitates of Elp1–GFP and Elp1–KR9A–GFP from strains expressing HA-tagged Elp3, Elp5 or Kti12. All samples were analysed with anti–GFP and anti-HA. Input samples were also analysed using anti-Cdc28 as a loading control. Arg4–GFP was confirmed to have similar pulldown efficiency to Elp1–GFP and was used as a control to demonstrate that interaction between Elp1 and the various HA tagged complex members was specific. All pulldown samples were analysed from the same blot. It should be noted that Elp1 is always observed by Western blot analysis as both a full-length and a faster migrating form (truncated at its N-terminus) that may be a degradation product or serve an as yet unknown function (Fichtner et al., 2003).
B. Quantification of co-immunoprecipitation efficiency. Immunoprecipitation of HA-tagged proteins was quantified by densitometry of the HA tag signals and normalized using the Elp1–GFP signals across the indicated number of replicates (n), setting the value for the wild-type strain in each case to 1.0. Error bars represent the standard deviation of the mean and the significance of the differences was analysed using a one way ANOVA, with ‘*’ indicating a P-value of < 0.05 (significant) and ‘***’ indicating a P-value of < 0.001 (very highly significant). Any small differences in Elp3 association were not statistically significant when analysed in this manner.
Since it was recently shown that Elongator forms a dodecamer containing two copies each of its six subunits (Glatt et al., 2012), it was possible that Elp1–KR9A could be defective in assembly into this higher order complex. To test this, YCp–ELP1–6HA and YCp–elp1–KR9A–6HA expression plasmids were transformed into ELP1–GFP and elp1–KR9A–GFP strains to generate cells in which two differentially tagged copies of Elp1 were present. The ELP1–GFP strain remained sensitive to zymocin following introduction of YCp–ELP1–6HA, confirming that the increased ELP1 copy number did not cause significant loss of function (data not shown). Wild-type Elp1–GFP and Elp1–KR9A–GFP were immunoprecipitated using GFP–Trap and assayed for co-precipitation of the 6HA-tagged versions of Elp1 (Fig. 5A). GFP-tagged wild-type Elp1 co-precipitated Elp1–6HA as previously reported (Glatt et al., 2012) and could also associate to a similar extent with Elp1–KR9A–6HA. In the reciprocal experiment, GFP tagged Elp1–KR9A also co-precipitated both Elp1–6HA and Elp1–KR9A–6HA, again with no obvious defect in the interaction between any of the different combinations. Thus there is no apparent defect in Elp1 self-association within the Elongator complex due to the Elp1 basic region mutation.
Fig. 5.
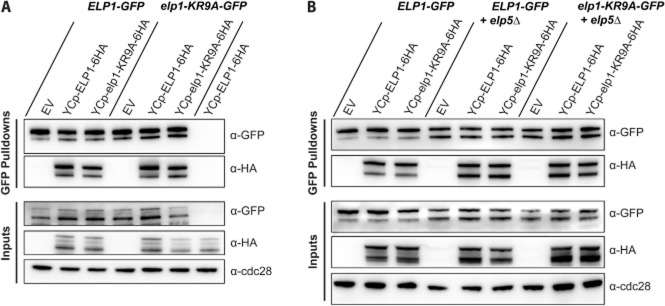
Self-association of Elp1 is unaffected by the elp1–KR9A mutation and is independent of the assembly of the Elp4–6 hexamer.
A. Western blot analysis of inputs and GFP–Trap immunoprecipitates of Elp1–GFP and Elp1–KR9A–GFP from strains containing empty vector (EV), YCplac111–ELP1–6HA or YCplac111–elp1–KR9A–6HA expressing HA-tagged wild-type or mutant Elp1 respectively. All samples were analysed by Western blotting with anti–GFP and anti-HA. Inputs were also analysed using anti-Cdc28 as a loading control.
B. Western blot analysis of inputs and GFP–Trap immunoprecipitates of Elp1–GFP and Elp1–KR9A–GFP from strains containing empty vector (EV), YCplac111–ELP1–6HA or YCplac111–elp1–KR9A–6HA expression plasmids either with or without deletion of the ELP5 gene (elp5Δ).
Since there was no obvious deficiency in Elp1–KR9A self-association despite its reduced interaction with Elp5, it seemed possible that Elp1 might associate independently of interaction between Elp1–3 subcomplexes and the Elp4–6 hexamer. To test this, the above immunoprecipitations were repeated in the context of an elp5 deletion, which would prevent assembly of the Elp4–6 hexamer (Glatt et al., 2012). Fig. 5B shows that neither wild-type Elp1–GFP nor Elp1–KR9A–GFP showed any defect in self-association in the absence of Elp5, suggesting that this interaction occurs independently and does not require bridging of Elp1–3 subcomplexes by an Elp4–6 hexamer. Taken together, mutation of the Elp1 basic region does not disrupt the overall assembly of Elongator, although it leads to small reductions in the association of Elp1 with Elp5 and Kti12 that may be due to loss of Elongator function.
The basic region in Elp1 is required for the binding of tRNA to the Elp1 C-terminal domain
The conserved basic region in Elp1 is essential for tRNA wobble uridine modification but despite being capable of functioning as an NLS, it does not appear to influence the distribution of the Elp1 between the nucleus and cytoplasm and is not required for overall assembly of the Elongator complex. In keeping with Elongator's role in tRNA wobble uridine modification, it was recently shown by electrophoretic mobility shift assay (EMSA) that the Elp4–6 hexameric subcomplex of Elongator directly binds tRNAGlu(UUC), consistent with a direct role for Elongator in tRNA binding and subsequent modification (Glatt et al., 2012). Since the basic region in Elp1 was clearly important for Elongator function we hypothesized that it could be involved in binding tRNA molecules to the Elp1–3 subcomplex for wobble uridine modification, given that the Elp3 subunit within this subcomplex is the most likely candidate for carrying out the modification reaction. Elp1 was therefore analysed for potential RNA binding sites using the binding prediction program BindN+, which predicts RNA binding regions from primary sequence based on the properties of RNA binding residues from known structures (Wang and Brown, 2006). This analysis highlighted the conserved carboxy-terminal basic region in both yeast and human Elp1 as being likely to bind RNA (Fig. 6A), consistent with the notion that it may be required for tRNA binding. Such a role might therefore explain the profound consequences of the elp1–KR9A allele on Elongator function.
Fig. 6.
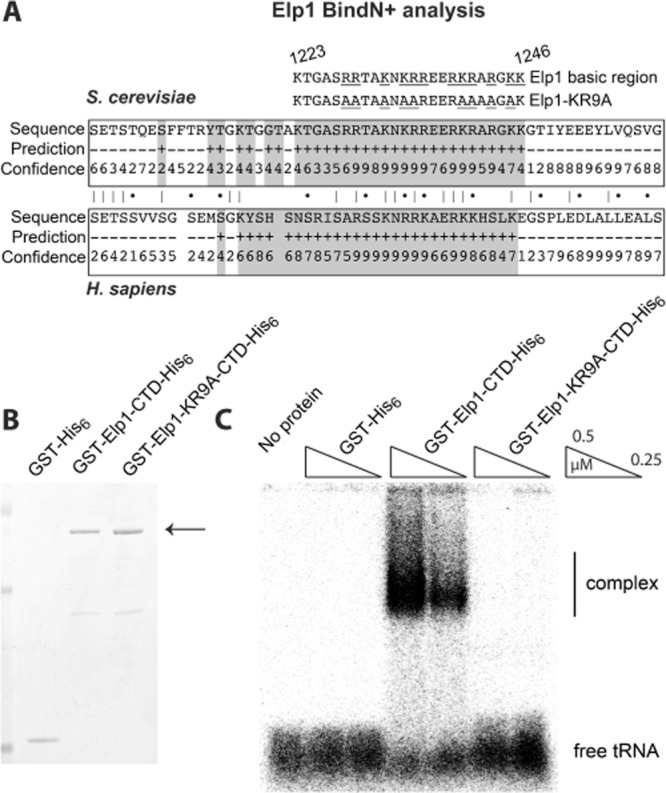
The Elp1 basic region is predicted to bind RNA and is required for formation of a complex with yeast tRNA.
A. Analysis for predicted RNA binding sites within the S. cerevisiae and H. sapiens Elp1 protein sequence using BindN+ (http://bioinfo.ggc.org/bindn+/) showing the high confidence prediction of the basic region as a putative RNA binding region (confidence is scored on a scale of 1–9 with 9 as highest confidence). The locations of the Elp1 basic region and the elp1–KR9A alanine substitution mutations are shown for comparison. Overall conservation between the human and yeast sequences is also indicated (|, identity; •, similar residues).
B. Recombinant Elp1 and Elp1–KR9A C-terminal domains (CTDs). Elp1 CTDs with N-terminal GST and C-terminal His6 tags were expressed and purified from E. coli along with GST-His6 as a control. The corresponding proteins were analysed on a 4–12% polyacrylamide gel and stained with instant blue. The GST–Elp1–CTD-His6 fusion proteins are indicated by an arrow.
C. Electrophoretic mobility shift assay (EMSA) using ∼ 1 nM 32P-labelled yeast tRNA and the indicated concentrations of recombinant GST, GST–Elp1 and GST–Elp1–KR9A fusion proteins shown in (B). Binding reactions were separated on a 1.5% native agarose gel and analysed by autoradiography. A complex was formed between tRNA and the wild-type Elp1 CTD but not with the Elp1–KR9A CTD.
To address a potential role in tRNA binding, the C-terminal domains (CTD) of Elp1 and Elp1–KR9A were expressed and purified from E. coli using N-terminal GST and C-terminal His6 tags (Fig. 6B). As a control, GST-His6 was also purified in parallel. An electrophoretic mobility shift assay (EMSA) was carried out using these fusion proteins and [32P]-tRNA, resolving the tRNA-protein binding reactions by agarose gel electrophoresis. Figure 6C clearly shows that the wild-type Elp1 fusion protein was able to shift the radiolabelled tRNA to a much slower migrating form, indicating formation of a tRNA-protein complex that was absent both in the GST control and in the Elp1–KR9A binding reactions (Fig. 6C). The Elp1–CTD can therefore bind tRNA, but this ability is lost when the basic region is mutated in Elp1–KR9A.
To test the specificity of the interaction between the Elp1–CTD and tRNA, competition binding experiments were carried out (Fig. 7A). While addition of unlabelled yeast tRNA at a 100-fold excess efficiently competed formation of the [32P]-tRNA–Elp1–CTD complex, addition of a similar excess of either poly(U) or ssDNA left complex formation between the tRNA probe and Elp1–CTD essentially unaffected. The Elp1 C-terminal domain therefore preferentially binds tRNA over poly(U) RNA or ssDNA. We also compared the ability of tRNA purified from either wild-type or elp1Δ mutant yeast (in which wobble uridine-containing tRNAs lack the Elongator-dependent mcm5 and ncm5 modifications) as competitors of [32P]-tRNA binding to the Elp1 CTD. However, no differences were found (data not shown), suggesting that there are unlikely to be very large differences in affinity resulting from whether or not the tRNA carries Elongator-dependent modifications.
Fig. 7.
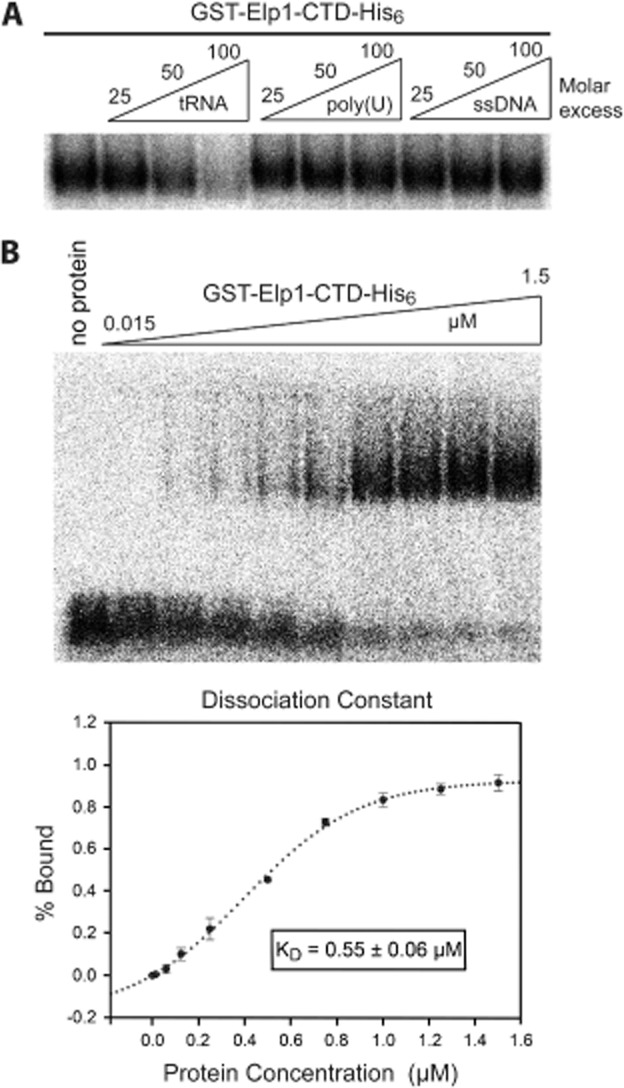
Binding of the Elp1 C-terminal domain to tRNA occurs with a dissociation constant in the low micromolar range and is not competed by poly(U) or ssDNA.
A. Electrophoretic mobility shift assay (EMSA) using ∼ 1 nM 32P-labelled yeast tRNA, 0.4 μM recombinant Elp1 C-terminal domain and the indicated molar excess of unlabelled yeast tRNA, poly(U) or ssDNA.
B. EMSA using ∼ 0.1 nM 32P-labelled yeast tRNA and the indicated concentrations of recombinant Elp1 C-terminal domain. Bound and free tRNA bands were quantified from three replicate experiments and used to estimate a dissociation constant (KD) of 0.55 μM for the interaction by non-linear curve fitting (f = a*xb/[cb + xb]). The Hill coefficient (b) for the fitted curve was 1.76 ± 0.19. All binding reactions were separated on a 1.5% native agarose gel and analysed by autoradiography.
To estimate the dissociation constant (KD) of the Elp1–tRNA complex, binding reactions were set up with [32P]-tRNA and increasing concentrations of wild-type Elp1–CTD, ranging from 0.015 to 1.5 μM (Fig. 7B). The % bound values were quantified, plotted against the protein concentration and fitted to a non-linear sigmoidal curve. A KD value of 0.55 μM was estimated from determining the protein concentration that gave ∼ 50% binding and this value is similar to that which was reported for the interaction between tRNAGlu(UUC) and recombinant Elp4–6 hexamers (Glatt et al., 2012).
These data therefore demonstrate a novel role for the C-terminal domain of Elp1 in tRNA binding and show that Elp1–KR9A, in which the basic region of Elp1 is mutated, has lost the ability to form this complex. We therefore propose that this basic region in Elp1 functions as a conserved tRNA binding motif that is important for Elongator-dependent wobble uridine modification.
Discussion
A conserved basic region in Elp1 is essential for Elongator function
The Elp1 subunit of Elongator contains a conserved lysine/arginine-rich basic region in its carboxy-terminal domain and we have shown that mutation of this region leads to zymocin resistance and loss of Elongator-dependent SUP4 ochre suppressor function, phenotypes indicative of a defect in Elongator-dependent tRNA wobble uridine modification. Although the basic region can function as an NLS in isolation, in comparison with full length wild-type Elp1–GFP we found no difference in the distribution of the Elp1–KR9A mutant protein between the nucleus and cytoplasm. Thus in the context of the full-length Elp1 protein that can be assembled into Elongator the Elp1 basic region does not function as an NLS, leaving open the question of why this region is essential for Elongator-dependent tRNA modification. Our finding that the basic region mediates binding of tRNA to Elp1 that is resistant to competition by non-specific polynucleotides and that is dependent on the residues altered in the elp1–KR9A allele provides a likely answer to this question. We therefore propose that the conserved Elp1 basic region is involved in the interaction of tRNA with the Elongator complex.
The Elp1 basic region constitutes a novel tRNA binding domain
Although a direct role for Elongator in tRNA modification remains to be demonstrated, it seems likely that this is the primary role of the complex at least in yeast (Huang et al., 2005; Esberg et al., 2006). Our finding that the Elp1 C-terminal domain can bind tRNA with high nanomolar affinity, together with the previous demonstration of direct tRNA binding by the Elp4–6 hexamer (Glatt et al., 2012), lends credence to the idea that Elongator is directly involved in the tRNA modification reaction. Our calculated KD of ∼ 0.55 μM for interaction between tRNA and the Elp1 C-terminal domain and tRNA complex indicates an interaction of similar strength to that observed between tRNA and the Elp4–6 hexamer (KD of 1.2–1.4 μM; Glatt et al., 2012). Interestingly, the Hill coefficient value calculated from our binding curves (1.76) is indicative of positive cooperativity, suggesting either that the Elp1–C-terminal region may interact with tRNA as a dimer or that each Elp1 molecule can bind > 1 tRNA, although further work would be needed to confirm this. Given an estimated 3.3 × 106 tRNA molecules per yeast cell (Waldron and Lacroute, 1975) and a mean cell volume of 37 fl (Tyson et al., 1979) we can estimate that the cellular total tRNA concentration is of the order of ∼ 150 μM, which would imply that binding in the cell would be saturated, even if only the 11 out of 42 tRNA species that require Elongator-dependent tRNA modification (Johansson et al., 2008) are bound.
However, it seems likely that there is little binding selectivity by either the Elp1 C-terminal domain or the Elp4–6 hexamer for tRNAs containing modifiable wobble uridines. Thus the tRNA–Elp4–6 complex was still formed regardless of the identity of the base at the wobble position of the tRNA probe used (Glatt et al., 2012), while in our study it is clear that a large proportion of the tRNA probe, which was prepared using total yeast tRNA, is capable of binding to Elp1. Furthermore, we found no difference in the ability of tRNA isolated from wild-type or Elongator-deficient elp1Δ mutant yeast cells (in which wobble uridines would be unmodified) to compete for binding with the [32P]-tRNA probe (data not shown). Thus if Elongator does directly modify wobble uridine-containing tRNAs it appears likely that substrate specificity is not determined at the level of tRNA binding to the complex. This could be considered analogous to how ribosomes select aminoacyl-tRNAs during protein synthesis: any charged tRNA can enter the ribosomal A-site, but only one that has a cognate anticodon–codon interaction has its binding stabilized so that a new peptide bond can form. Future work will need to identify whether tRNA interaction with Elp1 or Elongator as a whole shows any specificity towards substrate tRNAs containing a uridine in the wobble position, why it now seems likely that both subcomplexes of Elongator can bind tRNA, and whether extra stability and specificity of tRNA binding could be imparted within the context of the holo–Elongator complex.
Effects of elp1–KR9A on Elongator's protein–protein interactions
Since none of the Elongator protein–protein interactions tested were lost as a result of the elp1–KR9A mutation, including the self-association of Elp1, it is unlikely that the basic region of Elp1 constitutes a domain essential for complex assembly. However, we observed subtle reductions in the association of Elp1–KR9A with both Elp5 and Kti12 that may occur because the complex is non-functional. For example, if Elongator cycles between different forms in order to bind, modify and release tRNA molecules then a mutation that disrupts tRNA binding could stall the complex in a particular conformation in which certain protein–protein interactions are destabilized. The Elp4–6 hexamer can be easily dissociated from Elp1–3 (Winkler et al., 2001), and rather than forming a static structure might undergo cycles of conformational changes, as seen with other hexameric RecA-like NTPase complexes (Lyubimov et al., 2011). Assuming this is the case, reduced association of Elp5 with Elp1–KR9A may occur due to an arrest in the mechanism of action of Elongator that weakens the interaction between the two subcomplexes. The Kti12 accessory protein associates with the assembled holocomplex (Butler et al., 1994; Fichtner et al., 2002a) and also contains a highly conserved NTP binding domain (Fichtner et al., 2002a), suggesting it could cycle between different nucleotide bound forms during the process of tRNA modification. Since truncation of the Elp1 carboxy-terminus including the basic region (deletion of residues 1229–1349) still allowed the same level of Kti12 association as with full-length Elp1 (Fichtner et al., 2002b), it seems unlikely that the basic region is a major determinant of Kti12 interaction, consistent with the reduced association that we see being a secondary consequence of the elp1–KR9A mutation. A better understanding of the relationship between subunit interactions, Kti12 association and the mechanism of tRNA binding is required to understand the significance of these observations. Another exciting possibility is that tRNA binding and/or modification could be regulated by Elp1 phosphorylation, which is known to be correlated with Elongator function (Mehlgarten et al., 2009).
In summary, our work has shown that the Elp1 basic region mediates tRNA binding to Elp1 and it therefore forms a highly conserved tRNA binding motif in Elongator. This is the first evidence that Elp1, the largest subunit of Elongator, is involved in the interaction with tRNA and supports a direct role for the complex in tRNA modification.
Experimental procedures
Plasmids
Cloning was carried out using standard methods (Sambrook and Russell, 2001). pAE1, in which the zymocin γ subunit can be expressed from the GAL1 promoter, is based on the YCp50 (URA3) plasmid pBM150 (Johnston and Davis, 1984) and has been described previously (Butler et al., 1991). Plasmids containing elp1 mutations were generated by DNA2.0, Inc. from YCplac111–ELP1–6HA by replacing the relevant part of the gene with a synthesized elp1 mutant sequence. YCplac111–ELP1–6HA was kindly provided by R. Schaffrath (University of Kassel) and consists of YCplac111 (Gietz and Akio, 1988) containing the ELP1 open reading frame with 632 bp of upstream sequence, fused to a 6HA tag and followed by the K. lactis IPP1 transcription terminator (D. Jablonowski and R. Schaffrath, unpublished).
For conditional expression of GFP fused to the Elp1 C-terminal domain (amino acids 1181–1349), the relevant region from YCplac111–ELP1–6HA or its mutant derivatives were amplified by PCR and cloned into the GFP tagging vector pUG34 (GenBank accession AF298784; a kind gift from Johannes Hegemann), thereby allowing expression of the GFP–Elp1 fusion proteins from the MET25 promoter.
To generate templates for C-terminal tagging of Elp1 with an NLS sequence, a 462 bp DNA sequence (‘gBlock’; Integrated DNA technologies) was used to replace the plasmid sequence between endogenous PacI and BsrGI sites in pFA6a–GFP(S65T)–HIS3MX6 (Longtine et al., 1998) in order to include upstream of the GFP gene a sequence encoding the first 30 amino acids of yeast Cbp80 that contains a nuclear localization sequence (Izaurralde et al., 1995), generating pFA6a-NLS–GFP(S65T)–HIS3MX6. A mutant version of this construct [pFA6a-NLS4A–GFP(S65T)–HIS3MX6] containing four alanine substitution mutations to disrupt NLS function was similarly generated. For recombinant expression of Elp1, the region encoding the C-terminal half (amino acids 730–1349) was amplified by PCR so as to include a C-terminal His6 tag and cloned into pGEX4T-1 (GE Healthcare; GenBank accession U13853).
Yeast strains and methods
Yeast strains used in this study are summarized in Table 1. Basic yeast methods, growth media, and routine recombinant DNA methodology were performed as previously described (Amberg et al., 2005). Haploid budding yeast strains were cultured in rich yeast-peptone-adenine-dextrose (YPAD) medium or the appropriate selective dropout media (DOA) at 30°C. Yeast transformations were carried out using the Lithium acetate/PEG method (Gietz and Woods, 2002). Eclipse assays were carried out as described (Kishida et al., 1996) using Kluyveromyces lactis (NCYC1368) and photographed after 1–2 days.
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| Kluyveromyces lactis | |
| NCYC1368 | prototroph pGLK2+ pGLK1+a |
| Saccharomyces cerevisiae | |
| BY4741 | MATa his3Δ1 ura3Δ0 leu2Δ0 met15Δ0b |
| SBY128 | BY4741 his3Δ1::pSB1(URA3, SUP4, NatRMX) |
| SBY132 | BY4741 his3Δ1::pSB3 (ura3oc22, SUP4, NatRMX) |
| YRDS1 | BY4741 NIC96-4mcherry::NatRMX |
| YRDS59 | YRDS1 ELP1-L-GFP-HIS3MXc |
| YRDS84 | BY4741 NIC96-4mcherry::NatRMX lys2Δ0 |
| YRDS228 | BY4741 elp1::CORE |
| YRDS229 | YRDS59 elp1::CORE-L-GFP-HIS3MX |
| YRDS250 | BY4741 elp1Δ::HIS3MX |
| YRDS253 | YRDS229 ELP1-L-GFP-HIS3MXd |
| YRDS255 | YRDS229 elp1-KR9A-L-GFP-HIS3MXd |
| YRDS259 | SBY128 elp1Δ::HIS3MX |
| YRDS261 | SBY132 elp1Δ::HIS3MX |
| YRDS270 | YRDS228 ELP1 his3Δ1::pSB1(URA3, SUP4, NatRMX)d |
| YRDS272 | YRDS228 ELP1 his3Δ1::pSB3 (ura3oc22, SUP4, NatRMX)d |
| YRDS278 | YRDS228 elp1-KR9A his3Δ1::pSB1(URA3, SUP4, NatRMX)d |
| YRDS280 | YRDS228 elp1-KR9A his3Δ1::pSB3 (ura3oc22, SUP4, NatRMX)d |
| YRDS317 | YRDS229 elp1-KR5A-L-GFP-HIS3MXd |
| YRDS319 | YRDS229 elp1-KR4A-L-GFP-HIS3MXb |
| YRDS439 | YRDS1 ARG4-L-GFP-HIS3MX |
| YRDS461 | YRDS253 KTI12-L-3HA-KanMX |
| YRDS463 | YRDS253 ELP3-L-3HA-KanMX |
| YRDS467 | YRDS255 KTI12-L-3HA-KanMX |
| YRDS474 | YRDS253 ELP5-L-3HA-KanMX |
| YRDS482 | YRDS255 ELP3-L-3HA-KanMX |
| YRDS487 | YRDS255 ELP5-L-3HA-KanMX |
| YRDS526 | YRDS439 KTI12-L-3HA-KanMX |
| YRDS528 | YRDS439 ELP5-L-3HA-KanMX |
| YRDS537 | YRDS439 ELP3-L-3HA-KanMX |
| YRDS539 | YRDS1 ELP1-L-NLS4A-L-GFP-HIS3MX |
| YRDS544 | YRDS1 ELP1-L-NLS-L-GFP-HIS3MX |
National Collection of Yeast Cultures (http://www.ncyc.co.uk).
Brachmann et al. (1998).
‘L’ indicates inclusion of a linker region (see Experimental procedures).
Created by ‘delitto perfetto’ method of integration (see Experimental procedures).
Gene knockout and C-terminal tagging cassettes for transformation were generated as described previously (Longtine et al., 1998) unless otherwise indicated. In several cases, the forward primer was designed to incorporate a linker (encoding the amino acid sequence –GSAGSAAGSG–) between the end of the gene-specific sequence and the tag. To visualize the nuclear periphery in yeast cells, the nuclear pore complex component Nic96 was tagged with 4mCherry as outlined above, using pT909 as the template (a kind gift from Tomoyuki Tanaka, University of Dundee). Correct integration of gene knockouts or C-terminal tagging cassettes were confirmed by colony PCR and if appropriate by fluorescence microscopy and Western blotting.
The ‘delitto perfetto’ method was used to introduce site-directed mutations into the genomic copy of ELP1 (Storici and Resnick, 2006), using YCplac111–ELP1–6HA and its mutant derivatives as a template to generate the DNA fragments required to replace marker genes inserted at the point where the mutation was to be made. Correct integration of the mutant fragment was confirmed by DNA sequencing of a PCR product encompassing the sequences replaced by the mutant fragment.
Monitoring Elongator function through efficiency of SUP4 ochre suppression
To monitor the efficiency of suppression by the Elongator-dependent SUP4 ochre suppressor tRNATyr(UUA) we constructed a plasmid carrying SUP4 and a ura3 ochre allele (ura3oc22) that could be used to introduce both the tRNA and a suppressible allele into yeast cells. The ura3oc22 mutation was generated by site-directed mutagenesis using the QuikChange® Multi Site-Directed Mutagenesis Kit (Stratagene), the URA3 gene from YDpU (Berben et al., 1991) as template and a synthetic oligonucleotide carrying the desired ochre point mutation (5′-CATGTCGAAAGCTACATATAAGTAACGTGCTGCTACTCATCC-3′, mutational change is underlined). The mutated gene was moved as an 1126 bp NheI–BamHI fragment into the BamHI–XbaI interval of pRS303 (Sikorski and Hieter, 1989), following which a 270 bp fragment from pTC3 (Shaw and Olson, 1984) carrying SUP4 was inserted into the unique BamHI site to generate pRS303-URA3-SUP4. However, when selection for growth in the absence of uracil was applied to cells containing this plasmid, little difference in Ura phenotype was seen in cells lacking Elongator function despite loss of SUP4 efficiency in such cells (Huang et al., 2005), probably because selection for growth without uracil can drive up plasmid copy number. A 1225 bp EcoRI–NdeI fragment from pRS303-URA3-SUP4 was therefore cloned between the same sites within the YIplac211 backbone (Gietz and Akio, 1988), followed by insertion of a 1245 bp EcoRI–SalI fragment carrying the NatRMX marker from pAG25 (Goldstein and McCusker, 1999) between the EcoRI and SalI sites. Finally, a synthetic DNA segment (Integrated DNA Technologies) was inserted between the unique HindIII and SalI sites of this new construct to generate pSB3. After linearizing pSB3 with MscI, the entire construct is flanked by HIS3 sequences that allow targeting of a single copy to the wild-type HIS3 locus or its his3-11,15 and his3Δ1 derivatives under ClonNat selection (see Fig. S1 in Supporting Information). pSB1 (in which the URA3 gene lacks the ochre mutation) was generated similarly. In contrast to wild-type cells, Elongator-deficient cells containing integrated pSB3 showed strongly reduced ability to grow in the absence of added uracil, whereas no significant difference was observed using pSB1. Integration of pSB3 therefore confers the ability to read out the efficiency of SUP4 ochre suppression by monitoring growth in the absence of uracil.
Fluorescence microscopy
For fixed cell imaging, overnight cultures were subcultured to log-phase (OD600 ∼ 1) at 30°C in the appropriate medium. Cells of each strain equivalent of 4.5 OD600 units were fixed by adding formaldehyde to 3.7% (v/v) for 30 min with gentle agitation. Cells were washed three times with 1× PBS and pipetted onto 0.2% (w/v) Concanavalin A (Sigma) treated coverslips. After immobilizing for 3–5 min, coverslips were placed on microscope slides with a small spot of antifade solution [0.5% (w/v) p-phenylenediamine, 20 mM Tris-HCl (pH 8.8), 90% (v/v) glycerol] and sealed with nail varnish. Cells were visualized on a Delta Vision RT microscope (Applied Precision) connected to a CCD camera (CoolsnapHQ / ICX285) using a 100× lens (Olympus 1X-71). 2D images (512 × 512) were captured with Bin 2 × 2 mode (0.132 μm pixel size) using an ET filter set (Chroma, 89006) to visualize GFP and mCherry signals. Flat field calibration was carried out before imaging for the GFP channel using a fluorescent calibration slide (488–519 nm, Applied Precision) and applied during image acquisition.
Immunoprecipitation and Western blot analysis
Strains were grown to log phase (OD600 ∼ 1.0) and typically 50 OD600 units of cells were harvested by centrifugation, snap frozen using liquid nitrogen and then lysed by shaking vigorously at 4°C with 0.4 mm glass beads in cold Lysis Buffer [50 mM HEPES-KOH (pH 7.5), 2 mM MgCl2, 150 mM NaCl, 0.1% (v/v) Triton X-100, 50 mM NaF, 50 mM Na-βglycerophosphate] supplemented with Complete EDTA free Protease inhibitor (Roche), PhosSTOP (Roche), 1 mM AEBSF and 2 mM Na3VO4. All further steps were carried out at 4°C. Lysates were cleared by centrifugation at 14 000 rpm for 10 min and normalized using Bradford Coomassie reagent (Pierce). Protein extracts (∼ 0.5 ml) were pre-cleared with 25 μl Sepharose-4B beads (Sigma) for 30 min and then added to 50 μl Sepharose-4B beads conjugated to GFP–Trap (Chromotek) for 2 h with end-over rotation. GFP–Trap beads were washed three times with Lysis Buffer and proteins were eluted from the beads twice by adding 50 μl 2× Laemmli Sample Buffer and incubating at 75°C for 15 min, pooling the two eluates.
Protein samples prepared as above were analysed by Western blotting using anti–GFP (Roche, 11814460001), anti-HA (Santa-Cruz, sc-7392) and anti-Cdc28 (Santa-Cruz, sc-6709). Densitometry of bands was carried out using Aida Image Analyzer v.3.27 and Statistical analyses were carried out using SigmaPlot 12.0 (Systat Software, Inc.).
Recombinant Elp1 expression and purification
The C-terminal domains of Elp1 and Elp1–KR9A including a C-terminal His6 tag were cloned into pGEX4T-1 and expressed in E. coli BL21 (DE3) (Stratagene). A plasmid encoding GST-His6 (a kind gift from Tom Owen-Hughes) was used to express GST-His6 for use as a control. Cultures were grown in Autoinduction Medium (Studier, 2005) at 30°C with shaking for 4–5 h and then the temperature was reduced to 25°C overnight. All further steps were carried out at 4°C. The cells were lysed in cold Lysis Buffer [20 mM Tris-HCl (pH 7.5), 300 mM NaCl] supplemented with Complete EDTA free Protease inhibitors (Roche) and cleared by centrifugation at 20 000 g for 30 min. Fusion proteins were purified using HisPur Cobalt resin (Thermo Scientific) and the eluate supplemented with 10 mM DTT before further purification with Glutathione-Sepharose Fast Flow 4 resin (GE Healthcare). Proteins were eluted from the glutathione resin using GST elution buffer [50 mM Tris-HCl (pH 8.0), 50 mM reduced glutathione] and diafiltered into Protein Binding Buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM MgCl2].
EMSA assays
Yeast tRNA (R9001, Sigma) was treated with Fast alkaline phosphatase (Fermentas) to remove the 5′ phosphate group before radiolabelling with [γ-32P]-ATP (Specific activity 6000 Ci mmol−1, Perkin Elmer) using T4 polynucleotide kinase (Fermentas). Unincorporated nucleotides were removed using a G-25 microspin column (GE healthcare). For EMSAs using radiolabelled tRNA, each binding reaction consisted of approximately 1 nM radiolabelled tRNA, mixed with the appropriate concentration of protein in Binding Buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 8% (v/v) glycerol and 10 U Ribolock (Thermo scientific) RNase inhibitor] and allowed to equilibrate for 45 min at room temperature. Binding reactions were separated on 1.5% agarose gels in ultrapure 0.5× TBE Buffer (Life Technologies) at 100–120 V, cooling the gel apparatus in an ice bucket during electrophoresis. Gels were fixed and then dried at 50°C for 90 min on a vacuum gel drier (Bio-Rad) before exposure to Phosphoimager film. Bands were visualized by scanning using a fluorescent image analyser (FLA-5100 Fujifilm). Competitor EMSA experiments were performed similarly except that competitor was added to the binding reaction before the radiolabelled tRNA. Competitors used were ssDNA (5′-GCTGAAAGACTTGGTCTAAGC-3′) and poly(U) RNA (chain length 20). For estimation of dissociation constant from EMSA experiments (see Ryder et al., 2008), the conditions were as described except that approximately 0.1 nM [32P]-tRNA was used per binding reaction. Analysis of bands by densitometry was carried out using Aida Image Analyzer v.3.2.
To prepare yeast tRNA, total RNA was extracted from yeast using Tripure reagent (Roche) according to manufacturer's instructions and large RNAs (greater than 200 bp) were precipitated for 3 h at −20°C with 0.33 volume of 8 M LiCl. The small RNAs in the supernatant were precipitated overnight at −20°C with 0.1 volume of 3 M sodium acetate, 2.5 volumes of 100% ethanol, 20 μg glycogen and a final concentration of 10 mM MgCl2. The pellet was washed once in 75% ethanol before being air dried and dissolved in water. Small RNA preps were run on a 10% TBE-Urea gel (Novex) and the tRNA band was extracted and purified using a small-RNA PAGE recovery kit (Zymo Research).
Acknowledgments
Thanks are due to Raffael Schaffrath, Johannes Hegemann, Tom Owen-Hughes and Tomoyuki Tanaka for providing plasmids used in this work and to Sam Swift and Alan Prescott for help and advice with microscopy. We are grateful to the Leonardo Da Vinci programme for funding a research internship for Susanne Bandau. Rachael Di Santo was funded by a Wellcome Trust 4-year PhD Studentship (089690/Z/09/Z) and we also gratefully acknowledge additional support from the Wellcome Trust (083524/Z/07/Z) and from the College of Life Sciences, University of Dundee for this work.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
References
- Agarwalla S, Kealey JT, Santi DV, Stroud RM. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J Biol Chem. 2002;277:8835–8840. doi: 10.1074/jbc.M111825200. [DOI] [PubMed] [Google Scholar]
- Agris PF. Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications. EMBO Rep. 2008;9:629–635. doi: 10.1038/embor.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg DC, Burke D, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Anderson SL, Coli R, Daly IW, Kichula EA, Rork MJ, Volpi SA, et al. Familial dysautonomia is caused by mutations of the IKAP gene. Am J Hum Genet. 2001;68:753–758. doi: 10.1086/318808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton BP, Saleh L, Benner JS, Raleigh EA, Kasif S, Roberts RJ. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:1826–1831. doi: 10.1073/pnas.0708608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle PA, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler AR, Porter M, Stark MJR. Intracellular expression of Kluyveromyces lactis toxin gamma subunit mimics treatment with exogenous toxin and distinguishes two classes of toxin-resistant mutant. Yeast. 1991;7:617–625. doi: 10.1002/yea.320070610. [DOI] [PubMed] [Google Scholar]
- Butler AR, White JH, Folawiyo Y, Edlin A, Gardiner D, Stark MJR. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol Cell Biol. 1994;14:6306–6316. doi: 10.1128/mcb.14.9.6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Tuck S, Bystrom AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet. 2009a;5:e1000561. doi: 10.1371/journal.pgen.1000561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Huang B, Eliasson M, Ryden P, Bystrom AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet. 2011;7:e1002258. doi: 10.1371/journal.pgen.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YT, Hims MM, Shetty RS, Mull J, Liu L, Leyne M, Slaugenhaupt SA. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol Cell Biol. 2009b;29:736–744. doi: 10.1128/MCB.01313-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinenov Y. A second catalytic domain in the Elp3 histone acetyltransferases: a candidate for histone demethylase activity? Trends Biochem Sci. 2002;27:115–117. doi: 10.1016/s0968-0004(02)02058-3. [DOI] [PubMed] [Google Scholar]
- Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, et al. Elongator controls the migration and differentiation of cortical neurons through acetylation of α-tubulin. Cell. 2009;136:551–564. doi: 10.1016/j.cell.2008.11.043. [DOI] [PubMed] [Google Scholar]
- DeFraia C, Mou Z. The role of the Elongator complex in plants. Plant Signal Behav. 2011;6:19–22. doi: 10.4161/psb.6.1.14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esberg A, Huang B, Johansson MJ, Bystrom AS. Elevated levels of two tRNA species bypass the requirement for Elongator complex in transcription and exocytosis. Mol Cell. 2006;24:139–148. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Frohloff F, Burkner K, Larsen M, Breunig KD, Schaffrath R. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol. 2002a;43:783–791. doi: 10.1046/j.1365-2958.2002.02794.x. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Frohloff F, Jablonowski D, Stark MJR, Schaffrath R. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol Microbiol. 2002b;45:817–826. doi: 10.1046/j.1365-2958.2002.03055.x. [DOI] [PubMed] [Google Scholar]
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJR, Schaffrath R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol. 2003;49:1297–1307. doi: 10.1046/j.1365-2958.2003.03632.x. [DOI] [PubMed] [Google Scholar]
- Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J. 2001;20:1993–2003. doi: 10.1093/emboj/20.8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohloff F, Jablonowski D, Fichtner L, Schaffrath R. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (γ-toxin target (TOT)) complex. J Biol Chem. 2003;278:956–961. doi: 10.1074/jbc.M210060200. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Akio S. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Glatt S, Muller CW. Structural insights into Elongator function. Curr Opin Struct Biol. 2013;23:235–242. doi: 10.1016/j.sbi.2013.02.009. [DOI] [PubMed] [Google Scholar]
- Glatt S, Letoquart J, Faux C, Taylor NM, Seraphin B, Muller CW. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat Struct Mol Biol. 2012;19:314–320. doi: 10.1038/nsmb.2234. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grove TL, Benner JS, Radle MI, Ahlum JH, Landgraf BJ, Krebs C, Booker SJ. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Devel. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Huang B, Johansson MJ, Bystrom AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA. 2005;11:424–436. doi: 10.1261/rna.7247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Izaurralde E, McGuigan C, Mattaj IW. Nuclear localization of a cap-binding protein complex. Cold Spring Harb Symp Quant Biol. 1995;60:669–675. doi: 10.1101/sqb.1995.060.01.072. [DOI] [PubMed] [Google Scholar]
- Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28:3301–3312. doi: 10.1128/MCB.01542-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M, Davis RW. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lane WS, Reinberg D. Human Elongator facilitates RNA polymerase II transcription through chromatin. Proc Natl Acad Sci USA. 2002;99:1241–1246. doi: 10.1073/pnas.251672198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishida M, Tokunaga M, Katayose Y, Yajima H, Kawamura-Watabe A, Hishinuma F. Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci Biotechnol Biochem. 1996;60:798–801. doi: 10.1271/bbb.60.798. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin α. J Biol Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin α. J Biol Chem. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. The Elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet. 2009;5:e1000684. doi: 10.1371/journal.pgen.1000684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FJ, Shen L, Jang CW, Falnes PO, Zhang Y. Ikbkap/Elp1 deficiency causes male infertility by disrupting meiotic progression. PLoS Genet. 2013;9:e1003516. doi: 10.1371/journal.pgen.1003516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–248. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJR, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol. 2009;73:869–881. doi: 10.1111/j.1365-2958.2009.06811.x. [DOI] [PubMed] [Google Scholar]
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, et al. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol. 2010;76:1082–1094. doi: 10.1111/j.1365-2958.2010.07163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Fleury D, Bruno L, Robles P, De Veylder L, Traas J, et al. The elongata mutants identify a functional Elongator complex in plants with a role in cell proliferation during organ growth. Proc Natl Acad Sci USA. 2005;102:7754–7759. doi: 10.1073/pnas.0502600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, et al. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- Petrakis TG, Wittschieben BO, Svejstrup JQ. Molecular architecture, structure-function relationship, and importance of the Elp3 subunit for the RNA binding of holo-elongator. J Biol Chem. 2004;279:32087–32092. doi: 10.1074/jbc.M403361200. [DOI] [PubMed] [Google Scholar]
- Pierrel F, Douki T, Fontecave M, Atta M. MiaB protein is a bifunctional radical-S-adenosylmethionine enzyme involved in thiolation and methylation of tRNA. J Biol Chem. 2004;279:47555–47563. doi: 10.1074/jbc.M408562200. [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell. 2002;9:799–809. doi: 10.1016/s1097-2765(02)00502-6. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell. 2005;17:841–853. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Recht MI, Williamson JR. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shaw KJ, Olson MV. Effects of altered 5′-flanking sequences on the in vivo expression of a Saccharomyces cerevisiae tRNATyr gene. Mol Cell Biol. 1984;4:657–665. doi: 10.1128/mcb.4.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrer RL, O'Donoghue P, Soll D. Characterization and evolutionary history of an archaeal kinase involved in selenocysteinyl-tRNA formation. Nucleic Acids Res. 2008;36:1247–1259. doi: 10.1093/nar/gkm1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson CL, Lemmens R, Miskiewicz K, Broom WJ, Hansen VK, van Vught PW, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Human Mol Genet. 2009;18:472–481. doi: 10.1093/hmg/ddn375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Lorbeck MT, Zervos A, Zimmerman J, Elefant F. The histone acetyltransferase Elp3 plays in active role in the control of synaptic bouton expansion and sleep in Drosophila. J Neurochem. 2010;115:493–504. doi: 10.1111/j.1471-4159.2010.06892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaugenhaupt SA, Blumenfeld A, Gill SP, Leyne M, Mull J, Cuajungco MP, et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am J Hum Genet. 2001;68:598–605. doi: 10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Resnick MA. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 2006;409:329–345. doi: 10.1016/S0076-6879(05)09019-1. [DOI] [PubMed] [Google Scholar]
- Strug LJ, Clarke T, Chiang T, Chien M, Baskurt Z, Li W, et al. Centrotemporal sharp wave EEG trait in rolandic epilepsy maps to Elongator Protein Complex 4 (ELP4) Eur J Hum Genet. 2009;17:1171–1181. doi: 10.1038/ejhg.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson CB, Lord PG, Wheals AE. Dependency of size of Saccharomyces cerevisiae cells on growth rate. J Bacteriol. 1979;138:92–98. doi: 10.1128/jb.138.1.92-98.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J, Kwon SY, Badenhorst P, East P, McNeill H, Svejstrup JQ. Role of elongator subunit Elp3 in Drosophila melanogaster larval development and immunity. Genetics. 2011;187:1067–1075. doi: 10.1534/genetics.110.123893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown SJ. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:W243–W248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P, Svejstrup JQ. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem. 2001;276:32743–32749. doi: 10.1074/jbc.M105303200. [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, et al. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- Yan F, LaMarre JM, Rohrich R, Wiesner J, Jomaa H, Mankin AS, Fujimori DG. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J Am Chem Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web-site.