Abstract
We synthesized unmodified Fe3O4 nanoparticles (NPs) with particles size from 10 nm to 100 nm. We cultured NRK-52E cell lines (rat, kidney) and treated with Fe3O4 NPs to investigate and evaluate the cytotoxicity of NPs for NRK-52E cells. Through global proteomics analysis using dimethyl labeling techniques and liquid phase chromatography coupled with a tandem mass spectrometer (LC-MS/MS), we characterized 435 proteins including the programmed cell death related proteins, ras-related proteins, glutathione related proteins, and the chaperone proteins such as heat shock proteins, serpin H1, protein disulfide-isomerase A4, endoplasmin, and endoplasmic reticulum resident proteins. From the statistical data of identified proteins, we believed that NPs treatment causes cell death and promotes expression of ras-related proteins. In order to avoid apoptosis, NRK-52E cell lines induce a series of protective effects such as glutathione related proteins to reduce reactive oxygen species (ROS), and chaperone proteins to recycle damaged proteins. We suggested that, in the indigenous cellular environment, Fe3O4 NPs treatment induced an antagonistic effect for cell lines to go to which avoids apoptosis.
1. Introduction
In general, nanoparticles (NPs) are a group of particles with diameter size between one nanometer to one hundred nanometers. NPs have properties such as a high specific surface area and have a high potential to be a good catalytic agent, and therefore NPs have been widely used in fields such as antibacterial [1], antimicrobial [2, 3], drug delivery [4], and global use in research [5, 6]. However, having high catalytic potential, NPs were modified with polysaccharide or chitosan to avoid cytotoxicity [2, 3]. Based on the response of toxicity, NPs such as those that use silver and titanium dioxide were evaluated for cytotoxicity, DNA damage, and reactive oxygen species (ROS) [7–9]. Especially in AgNPs, numerous studies showed that in cellular responses, 100 nm AgNPs induced serine/threonine protein kinase (PAK), phosphatase 2A, and mitogen-activated protein kinase (MAPK) pathways, and 20 nm AgNPs induced ROS, SUMOylation, and protein carbonylation [10]. Moreover, published papers increasingly show that proteomics analysis and LC-MS/MS were utilized to demonstrate the effects of NPs, such as quantitative proteomics to evaluate AgNPs exert cellular responses [10], AgNPs treatment directly involved in ROS, metal detoxification according to genomics and proteomics results [11], and the toxic effects and behavior in Caenorhabditis elegans [12].
Previous studies reported that in vitro experiments and AgNPs exposure directly induced ROS, cell death, apoptosis, and inflammation [13, 14]. In animal model studies, female rats had AgNPs accumulation in kidney regions, especially in the glomerulus [15]. A variety of reports showed evidence that AgNPs treatment has cytotoxicity in proteomics and genomics; however no evaluated studies of other NPs are described with Fe3O4 or gold nanoparticles.
MS-based quantitative proteomics has been developed at a marvelous rate in the past two decades for biomarker discovery and drug screening. In addition, proteomics provides numerous proteins expression profiles using quantitative techniques to estimate and established the relationships using bioinformatics software. Current quantitative comparisons between specimens both with drug treatment and without treatment or normal and abnormal tissues are beneficial to identify the proteins with upregulation or downregulation and to set up biologic mechanisms and pathways [16, 17]. However in complicated tissue specimens, abundant proteins interfere with the detection of rare proteins. Multiple dimensional separation systems were used to fractionate peptides into different fractions through liquid phase chromatography (LC) to decrease the samples' complication and to increase the amount of protein identification [16, 18].
In this work, we synthesized bare Fe3O4 NPs and characterized the size of NPs by transmission electron microscope (TEM). After treatment with unmodified Fe3O4 NPs, we next utilized dimethyl labeling quantitative reagents to label the tryptic peptides of NRK-52E cell lines with treated and untreated Fe3O4 NPs [19]. In global proteomics research coupled with LC-MS/MS, we demonstrated 435 identified proteins in NRK-52E cell lines by Mascot and simultaneously quantitated 311 proteins through the use of Mascot Distiller bioinformatics software. We classified proteins into chaperone proteins, cell death related and apoptotic proteins, ras-related proteins, and glutathione related proteins. Using STRING for protein-protein interaction [20], we demonstrated the relationships between Fe3O4 NPs and multifarious proteins.
2. Materials and Methods
2.1. Chemical Reagents
Sodium acetate, ferric chloride (FeCl3), ferrous chloride (FeCl2), sodium cyanoborohydride (NaBCNH3), and trifluoroacetic acid (TFA) were obtained from Sigma-Aldrich (St. Louis, MO, USA). Acetonitrile (MeCN) was bought from Merck (Seelze, Germany). Ammonium hydrogen carbonate (NH4HCO3), hydrochloric acid, sodium hydroxide, sodium dodecyl sulphate (SDS), and ethanol were purchased from J. T. Baker (Phillipsburg, NJ, USA). Formaldehyde-D2 solution (20% solution in D2O) was purchased from Isotec Corp. (Miamisburg, OH, USA), while formaldehyde-H2 solution (36.5%–38% in H2O), potassium chloride (KCl), sodium chloride (NaCl), sodium dihydrogen phosphate (NaH2PO4), potassium dihydrogen phosphate (KH2PO4), dimethyl sulfoxide (DMSO), formic acid (FA, 98%–100%), and iodoacetamide (IAM) were purchased from Sigma (St. Louis, MO, USA). Trypsin was purchased from Promega (Madison, WI, USA). The protein concentration of cell lysate was determined by Bradford assay based on the measurement BSA calibration curve (Thermo, Rockford, IL, USA). Deionized H2O with a resistance of 18.2 MΩ was obtained using a Millipore water system.
2.2. Fabrication of Fe3O4 Nanoparticles (NPs) by Hydrothermal Precipitation
The nanoscale magnetic iron oxide particles were prepared by hydrothermal homogeneous coprecipitation [21–23]. Ferric chloride (FeCl3, 5.2 g) and ferrous chloride (FeCl2, 2.0 g) were dissolved with sonication in 50 mL clean glassware containing aqueous hydrochloric acid (2 M, 25 mL) at 50°C. Subsequently, the mixture was degassed continuously using a nitrogen gas cylinder. Sodium hydroxide (NaOH 150 mL, 2 M) was slowly injected drop by drop into the glassware. The color of the solution transformed from yellow to orange, and finally the color shifted to black through the NaOH dripping down. After incubation at 50°C for one hour, the supernatant was removed through a strong magnet absorbing NPs. The generated NPs were then washed with deionized water and removed by a centrifugal tube with 1,800 rpm for two minutes. Then finally the NPs were rinsed with 0.5 M HCl, deionized water and ethanol sequentially.
2.3. Characterization of Fe3O4 by Transmission Electron Microscope (TEM)
The size of the synthetic Fe3O4 NPs was characterized by a JEOL JEM 1200-EX transmission electron microscope (TEM) with the accelerating voltage 80 keV. One μL of the nanoparticles solution was dropped on a cupric grid with stabilized carbon, and subsequently the surplus solution evaporated in the atmosphere.
2.4. NRK-52E Cell Culture, the Conditions of Fe3O4 NPs Treatment, and Protein Concentration
The cells, NRK-52E (CRL-1571; American Type Culture Collection, Manassas, VA), were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich) and supplemented with 5% fetal bovine serum (FBS) and 1% penicillin (Gibco, Grand Island, NY, USA). Cells were cultured in a 100 mm dish with a 5% CO2 incubator at 37°C. Until cells reached 80% confluence, they were cultured with the absence of serum medium for 24 hours. After starvation cells were treated with 1 ng Fe3O4 NPs in each dish for 24 hours, and comparison groups were treated with dimethyl sulfoxide (DMSO). After being washed three times with phosphate-buffered saline (PBS), the cells were lysed with modified RIPA buffer containing 1% NP-40, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl, and 1 tablet/10 mL of Roche minicomplete protease inhibitor cocktail at pH 7.5. Finally, the protein concentration of NRK-52E cell lysate was determined using the Bradford assay.
2.5. Tryptic Digestion, Dimethyl Labeling, Desalting, and Fractionation
Samples of lysated protein solution 60 μL containing 100 μg of total proteins treated with Fe3O4 NPs and DMSO were reduced by reaction with 0.7 μL of 1 M DTT and 9.3 μL of 7.5% SDS at 95°C for 5 minutes. The sample bottles were put on ice and then for alkylation were treated with 8 μL of 50 mM IAM at room temperature for 30 minutes in the dark with agitation. Furthermore, sample proteins were precipitated by 52 μL of 50% trichloroacetic acid (TCA) with incubation on ice for 15 minutes. By centrifugation to remove the supernatant, the protein pellets were washed in different solutions according to priority with 150 μL of 10% TCA, 250 μL of deionized H2O, 250 μL of acetone, and two times with 250 μL of deionized H2O. After removing the supernatant, the protein pellets were digested in 2 μg of trypsin in 200 μL 50 mM NH4HCO3 (pH 8.5). An additional 2 μg of trypsin was added after 4 hours of tryptic digestion, and then the samples were digested for 14 hours at 37°C. Eventually the tryptic digestion samples were dried by vacuum centrifugation to remove the digested buffer. For labeling procedures, the lyophilized samples were redissolved in 180 μL of 100 mM sodium acetate buffer at pH 5.5. Simultaneously the treated NPs samples were labeled using 10 μL of 4% formaldehyde-D2 and then those samples treated DMSO were labeled using 10 μL of 4% formaldehyde-H2. Both of the samples were vortexed for 5 minutes, followed by treatment with 10 μL of 600 mM sodium cyanoborohydride for 1 hour. Finally, the labeled solutions were combined after adjusting to pH 2-3 using 10% TFA/H2O for desalting by a homemade C18 cartridge desalting kit. After vacuum centrifugation drying, the triplicate combined samples were fractionated with a hydrophilic interaction chromatography (HILIC) separated system. HILIC system was performed using an L-7100 pump system (Hitachi, Tokyo, Japan) connecting a TSK gel Amide-80 HILIC column (2.0 mm × 150 mm, 3 μm particle size; Tosoh Biosciences, Tokyo, Japan) with a flow rate of 200 μL/min. The mobile phase system was used for gradient elution: solvent (A) was 0.1% TFA in water; solvent (B) was 0.1% TFA in 100% MeCN. The eluted fraction from the homemade C18 cartridge was dissolved in 25 μL of solution containing 85% MeCN and 0.1% TFA and then injected into the 20 μL sample loop. Furthermore, the gradient was performed as follows: 95% (B) for 2 minutes; 95%~60% (B) for 32 minutes; 60%~5% (B) for 10 minutes; 5% (B) for 5 minutes; 5%~95% (B) for 1 minute; and 95% (B) for 10 minutes. Each fraction contained 1.2 mL buffer in the total of 10 collected fractions, and finally every fraction was dried in a vacuum centrifuged system.
2.6. Protein Identification by LC-MS/MS Analysis
The vacuum dried lyophilized samples were redissolved in 10 μL of 0.1% FA in H2O and analyzed using a Thermo LTQ Orbitrap XL (Thermo Fisher Scientific, San Jose, CA). A total of 10 μL of sample was injected onto a C18 capillary pretrapped column (0.3 mm × 5 mm, 5 μm particle size, Agilent Zorbax XDB); HPLC loading pump and separation was performed by a C18 column (i.d. 75 μm × 150 mm, 3 μm particle size, Micro Tech, Fontana, CA). A LC-MS/MS separation was performed using an Agilent 1200 series Nanoflow pump (Agilent Technologies, Santa Clara, CA). The flow rate of loading pump was set at 5 μL/min, and the gradient pump was set at 300 nL/min. The mobile phases consisted of (A) 0.1% FA in water and (B) 0.1% FA in 100% MeCN. The linear gradient was as follows: 2% (B) in 2 minutes, 2%~40% (B) in 40 minutes, 40%~95% (B) in 8 minutes, 95% (B) in 2 minutes, 95%~2% (B) in 1 minute, and 2% (B) in 7 minutes. The digested peptides were detected by a voltage of 1.8 kV in the positive detection ion mode. The separated system with full scanning mode conditions was set at m/z 400–1600 Da with resolution = 30,000. The data-dependent mode was set according to a priority to select peptides which were detected with 5 high-intensity signals in the MS mode and transferred into collision chamber for fragmentation with a collision energy of 35 eV. The fragmented ions were analyzed and detected at second MS analyzer with a mass range of m/z 100–2000 Da. To exclude ions with similar m/z and avoid interferences, the data-dependent mode was also set a repeat duration of 30 seconds.
2.7. Mascot Database Search and Mascot Distiller Quantitation
The software Xcalibur (version 2.0.7, Thermo Scientific Inc., San Jose, CA) was utilized to control Orbitrap XL and to acquire the MS and fragmentation data. The raw data including MS and MS/MS spectra were converted to a suitable file type by the Mascot Distiller software (version 2.5.1.0 (64 bits), Matrix Science Ltd., London, UK) to perform protein identification and quantitation. The parameters of the Mascot Distiller were set as follows: “Orbitrap_res_MS2” (default parameter setting) for peak list transformation; “Rattus” for the taxonomy in the Swiss-Prot databank of the Mascot search engine; zero allowable missed cleavages for tryptic digestion; dimethylation (MD) for quantitation; fixed modification was selected carbamidomethyl for cysteine modification; peptide tolerance of 10 ppm with precursor ions; and 0.8 Da tolerance for MS/MS. Peptides charge was selected when they had charges of 1+, 2+, and 3+, and the instrument was set to “ESI-trap.” Finally, the protein quantitative result was listed by the heavy-labeled/light-labeled (D/H) ratios from the Mascot Distiller.
3. Results and Discussion
3.1. Fe3O4 NPs Synthesis and Characterization
Bare and unmodified Fe3O4 NPs were synthesized by hydrothermal precipitation and NPs were fabricated by chemical method to have a uniform particle size. In Figure 1, TEM images showed that we took three photos of Fe3O4 NPs with 20 nm, 50 nm, and 100 nm scale bars. It is considered that NPs with a higher surface area are easier to aggregate with other NPs; therefore through the aggregation in Fe3O4 NPs there were Fe3O4 particles generated above 100 nm if no additional protective agent was modified onto Fe3O4 (Figure 1(c)). We considered that Fe3O4 NPs with dispersive diameter sizes are suitable for cytotoxicity evaluation due to the fact that NPs found in the environment have random sizes.
Figure 1.

TEM images of Fe3O4 NPs with different sizes as follows: (a) 20 nm, (b) 50 nm, and (c) 100 nm scale bars.
3.2. Schematic Representation of Samples Pretreatments, Fractionation, Protein Identification, and Protein Quantitation
For protein quantitation, the procedures and using reagents including formaldehyde-H2, formaldehyde-D2, and sodium cyanoborohydride (NaBCNH3) were obeyed as following the previous literature [19]. By utilization of dimethyl labeling, the differences of protein expressions in treatment with and without NPs was enabled to be determined. In the mass spectrum, the proteins of NRK-52E treated NPs were labeled as formaldehyde-D2 and proteins of untreated NPs were labeled as formaldehyde-H2. In the same sequential peptides, however, different treatments had a mass difference 4 Da or 8 Da depending on the use of the labeling agent. Off-line HILIC fractionation was utilized to decrease the complication of the mixed samples [16, 18]. Through LC-MS/MS, the proteins identification and quantitative ratios of D/H peptides were identified and calculated using the bioinformatic Mascot Distiller software to finally generate a protein list. The schematic flow chart of the process is shown in Figure 2 from sample pretreatment and protein identification to protein quantitation.
Figure 2.
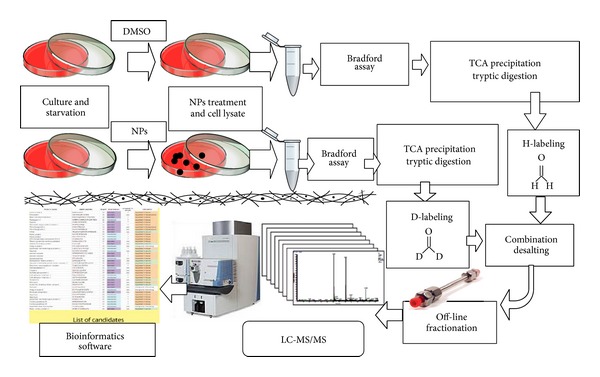
Simplified flow chart of sample pretreatment, fractionation, and analysis using LC-MS/MS.
3.3. Quantitative Results of NRK-52E Proteins Associated with Fe3O4 NPs
From the statistical results we identified 435 proteins, of which 311 proteins have a D/H ratio and 124 proteins which we were unable to specific values. For example, complement component 1 Q subcomponent-binding protein (C1qBP) was identified with an arithmetic average D/H ratio of 4.23. Figure 3 showed the peptide belonging to C1qBP having sequences GVDNTFADELVELSTALEHQEYITFLEDLK which was fragmentized by collision induced dissociation (CID), and its MS/MS spectrum was illustrated by y-ions and b-ions. From a quantitative standpoint in support of the Mascot Distiller in bioinformatics software, the result of protein list showed that the D/H ratio belonging to GVDNTFADELVELSTALEHQEYITFLEDLK peptide was 4.23. Simultaneously the raw data from LC-MS/MS that we extracted from the labeled peptide of C1qBP by m/z 1165.92 (3+, H-labeled) and 1168.60 (3+, D-labeled) presented signals of peak area and peak height (Figure 4(a)) and showed the isotope pattern of H-labeled peptide and D-labeled peptide (Figure 4(b)) which demonstrated the changes in different labeling. The statistical results showed that peak area was 4.42-fold in D/H ratio and peak height was 4.22-fold; the statistical data was calculated and shown in Table 1.
Figure 3.
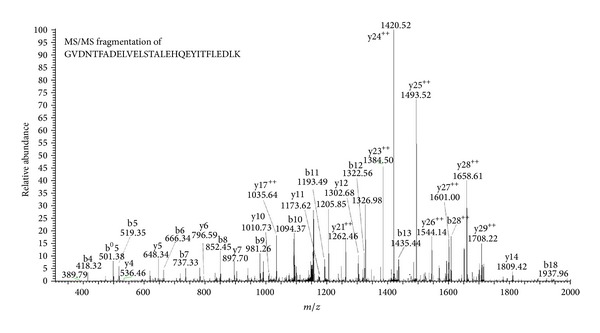
MS/MS spectrum of the peptide sequence GVDNTFADELVELSTALEHQEYITFLEDLK (m/z 1168.60, 3+), which is a peptide of complement component 1 Q subcomponent-binding protein (C1qBP).
Figure 4.
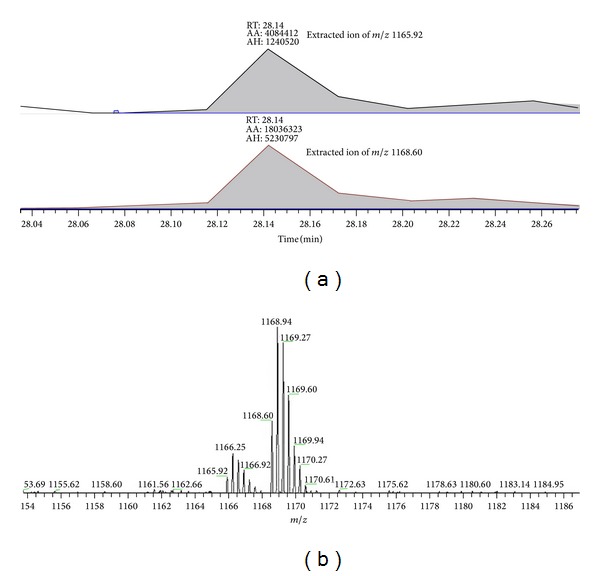
(a) Extracted ions of m/z 1165.92 (H-labeled) and m/z 1165.92 (D-labeled) showing signals of peak area (AA) and peak height (AH). (b) MS spectrum of GVDNTFADELVELSTALEHQEYITFLEDLK which are labeled formaldehyde-H2 and formaldehyde-D2. These two labeled peptides are shown in a coeluted isotopic pattern.
Table 1.
The MS intensities of peptide GVDNTFADELVELSTALEHQEYITFLEDLK labeled H (m/z 1165.92) and labeled D (m/z 1168.60) in peak area and peak height.
| m/z | Peak area | Peak height | Ratio of area | Ratio of height |
|---|---|---|---|---|
| 1165.92 | 4084412 | 1240520 | 4.42 | 4.22 |
| 1168.60 | 18036323 | 5230797 |
3.4. Differentially Expressed Proteins of NRK-52E by Treatment with Fe3O4 NPs
In this study, we used Fe3O4 NPs to treat NRK-52E and used bioinformatics software to generate proteins identification and proteins quantitation with a protein list (the protein list is shown in the electronic supplementary information available online in the Supplementary Materials at http://dx.doi.org/10.1155/2014/754721). In the protein list, we observed that when Fe3O4 NPs were treated, ras-related proteins were expressed including ras-related protein Rab-7L1 (RAB7L_RAT, 3.2-folds), ras-related protein Rab-1A (RAB1A_RAT, 2.3-folds), ras-related protein Rab-2A (RAB2A_RAT, 1.6-folds), ras-related C3 botulinum toxin substrate 1 (RAC1_RAT, 1.2-folds), ras-related protein Rap-1A (RAP1A_RAT), and ras-related protein Rab-7a (RAB7A_RAT). According to previous studies, ras-related proteins were associated with cancer cell metastasis [24, 25], and ras-related C3 botulinum toxin substrate 1 was associated with differential roles such as cell proliferation which could be inhibited by miR-101 [26] and signal transduction to upregulate in esophageal squamous cell carcinoma and esophageal adenocarcinoma [27].
We also identified related proteins in cell death and apoptosis, such as apoptotic protease-activating factor 1 (APAF_RAT, 780.9-folds), programmed cell death protein 10 (PDC10_RAT, 233.2-folds), galectin-1 (LEG1_RAT, 1.75-folds), and programmed cell death 6-interacting protein (PDC6I_RAT). Apoptotic protease-activating factor 1 activated procaspase-9 and modulated cellular apoptosis [28]. However, programmed cell death protein 10 promotes cell proliferation and protects malignant T cells from apoptosis [29]. The galectin family played different roles in cell apoptosis such as galectin-7 enhancing apoptosis, but galectin-1 has the opposite effect [30]. In accordance with previous studies and the proteins quantitative list, we considered that the treatment of Fe3O4 NPs caused an antagonistic effect. Fe3O4 NPs induced expression of ras-related proteins and induced programmed cell death protein 10, galectin 1 to promote cell metastasis, proliferation, and progression. However, the apoptotic protease-activating factor 1 promoted cell to go to apoptosis.
A variety of glutathione-related proteins with high expression were identified, such as glutathione reductase (Fragment), glutathione S-transferase Mu 1, glutathione S-transferase Mu 2, glutathione S-transferase P, and glutathione S-transferase alpha-3. We thought that Fe3O4 NPs induced reactive oxygen species (ROS) to cause overexpression of glutathione-related proteins. After one day of Fe3O4 NPs treatment, a strong magnet was utilized to recover the residual NPs, but there were no NPs to be recycled. We considered that the NPs had transferred into ferric ions (Fe3+) and ferrous ions (Fe2+) and that endogenous hydrogen peroxide (H2O2) reacted with ferrous ions to compose hydroxide free radicals (Fenton reaction) [31, 32]. A variety of reports show that ROS effects are exhibited when NPs were treated [7, 14]. However, ferrous ion (Fe2+) is an initiator in the Fenton reaction, and ferrous ions will react with hydrogen peroxide to produce hydroxide free radicals.
3.5. Chaperone Proteins Overexpression and STRING Networks Establishment
We also characterized chaperone proteins such as heat shock proteins, serpin H1, protein disulfide-isomerase, endoplasmin, and endoplasmic reticulum resident protein.
Eventually, we used the STRING (vision 9.1, http://string-db.org/) database to establish the interactions between chaperone proteins and related proteins to demonstrate the relationships between Fe3O4 NPs and the associated proteins [20]. The relationships between chaperone proteins, ras-related proteins, glutathione related proteins, and cell death and apoptosis related proteins are listed in Table 2 and illustrated by STRING in Figure 5. Heat shock proteins connected with each other and related ras-related C3 botulinum toxin substrate 1 and then combined to glutathione related proteins.
Table 2.
Statistic classification of related proteins in NRK-52E cell lines with Fe3O4 NPs treatment.
| UNIPROT accessiona | UNIPROT accession numbera |
Protein identification | Molecular mass (kDa)b | Number of peptidesb | Ratioc |
|---|---|---|---|---|---|
| Cell death and apoptosis related proteins | |||||
| APAF_RAT | Q9EPV5 | Apoptotic protease-activating factor 1 | 146.1 | 2 | 780.9 |
| PDC10_RAT | Q6NX65 | Programmed cell death protein 10 | 25.0 | 2 | 233.2 |
| LEG1_RAT | P11762 | Galectin-1 | 15.5 | 5 | 1.75 |
| PDC6I_RAT | Q9QZA2 | Programmed cell death 6-interacting protein | 99.2 | n.d. | n.d. |
|
| |||||
| Ras-related proteins | |||||
| RAB7L_RAT | Q63481 | Ras-related protein Rab-7L1 | 23.7 | 2 | 3.2 |
| RAB1A_RAT | Q6NYB7 | Ras-related protein Rab-1A | 23.4 | 13 | 2.3 |
| RAB2A_RAT | P05712 | Ras-related protein Rab-2A | 24.1 | 3 | 1.6 |
| RAC1_RAT | Q6RUV5 | Ras-related C3 botulinum toxin substrate 1 | 22.4 | 2 | 1.2 |
| RAP1A_RAT | P62836 | Ras-related protein Rap-1A | 21.8 | n.d. | n.d. |
| RAB7A_RAT | P09527 | Ras-related protein Rab-7a | 24.3 | n.d. | n.d. |
|
| |||||
| Glutathione related proteins | |||||
| GSHR_RAT | P70619 | Glutathione reductase (Fragment) | 47.8 | 2 | 3.1 |
| GSTM1_RAT | P04905 | Glutathione S-transferase Mu 1 | 26.7 | 2 | 2.8 |
| GSTM2_RAT | P08010 | Glutathione S-transferase Mu 2 | 26.5 | 3 | 2.4 |
| GSTP1_RAT | P04906 | Glutathione S-transferase P | 24.1 | 6 | 1.5 |
| GSTA3_RAT | P04904 | Glutathione S-transferase alpha-3 | 25.9 | 4 | 1.2 |
|
| |||||
| Chaperone proteins | |||||
| HSBP1_RAT | Q8K3X8 | Heat shock factor-binding protein 1 | 8.7 | 2 | 2.8 |
| HS90B_RAT | P34058 | Heat shock protein HSP 90-beta | 86.0 | 32 | 2.7 |
| SERPH_RAT | P29457 | Serpin H1 | 47.7 | 30 | 2.5 |
| HS90A_RAT | P82995 | Heat shock protein HSP 90-alpha | 87.7 | 31 | 2.4 |
| HSP74_RAT | O88600 | Heat shock 70 kDa protein 4 | 97.0 | 2 | 2.2 |
| HSP7C_RAT | P63018 | Heat shock cognate 71 kDa protein | 72.8 | 40 | 1.9 |
| PDIA4_RAT | P38659 | Protein disulfide-isomerase A4 | 75.1 | 9 | 1.8 |
| CH60_RAT | P63039 | 60 kDa heat shock protein, mitochondrial | 62.6 | 29 | 1.6 |
| CH10_RAT | P26772 | 10 kDa heat shock protein, mitochondrial | 11.2 | 3 | 1.6 |
| GRP78_RAT | P06461 | 78 kDa glucose-regulated protein | 74.4 | 19 | 1.6 |
| ENPL_RAT | Q66HD0 | Endoplasmin | 95.4 | 17 | 1.6 |
| PDIA1_RAT | P04785 | Protein disulfide-isomerase | 59.1 | 24 | 1.6 |
| ERP29_RAT | P52555 | Endoplasmic reticulum resident protein 29 | 29.3 | 3 | 1.6 |
| PDIA6_RAT | Q63081 | Protein disulfide-isomerase A6 | 49.6 | 4 | 1.5 |
| PDIA3_RAT | P11598 | Protein disulfide-isomerase A3 | 58.6 | 17 | 1.3 |
aProtein accession and protein accession number were received from the UNIPROT database available online: http://www.uniprot.org/uniprot (accessed on May 25, 2014).
bMolecular weight and number of protein peptides according to the Swiss-Prot database in the Mascot search engine.
cRatio values of each protein according to protein list in the Mascot Distiller.
Figure 5.
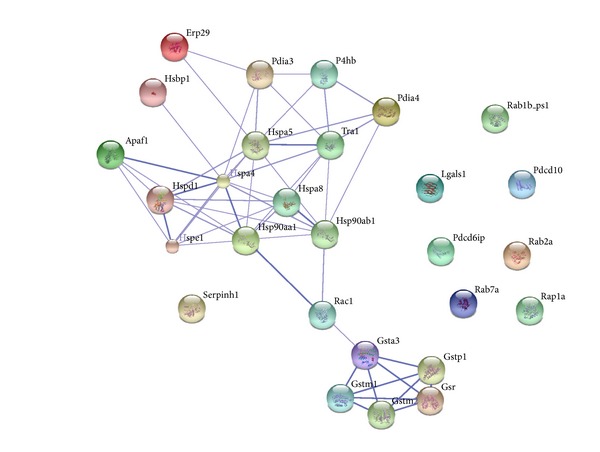
Schematic representation of the networks between cell death and apoptosis related proteins, ras-related proteins, glutathione related proteins, and chaperone proteins.
4. Conclusion
This is a pioneer experiment to show the cellular responses through Fe3O4 NPs treatment. We hypothesized an antagonistic effect in NRK-52E cell lines which is presented as cell death, apoptosis, and cancerization via programmed cell death protein and ras-related proteins; however there are protective mechanism proteins in NRK-52E such as chaperone proteins and glutathione related proteins. In future work, we shall generate or purchase Fe3O4 NPs in a smaller size to confirm the size effect of NPs, and we also need more evidence to prove the ROS effect induced via ferrous ions and to explain the tendency towards apoptosis or to avoid cell death.
Supplementary Material
The supplementary information is the raw data generated all proteins in quantitative proteomics which are listed according to the D/H ratio priority.
Acknowledgments
This work was supported by a grant from the Kaohsiung Medical University Research Foundation (KMU-Q110023), with cooperation between the National Sun Yat-Sen University and Kaohsiung Medical University (NSYSUKMU 103-P010) and the National Science Council (NSC 102-2113-M-037-013-MY2), Taipei, Taiwan. The authors are thankful for the assistance of the Center for Resources, Research and Development of Kaohsiung Medical University.
Conflict of Interests
The all authors declare that they have no conflict of interests in this study.
References
- 1.You C, Han C, Wang X, et al. The progress of silver nanoparticles in the antibacterial mechanism, clinical application and cytotoxicity. Molecular Biology Reports. 2012;39(9):9193–9201. doi: 10.1007/s11033-012-1792-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Travan A, Pelillo C, Donati I, et al. Non-cytotoxic silver nanoparticle-polysaccharide nanocomposites with antimicrobial activity. Biomacromolecules. 2009;10(6):1429–1435. doi: 10.1021/bm900039x. [DOI] [PubMed] [Google Scholar]
- 3.Mathew TV, Kuriakose S. Photochemical and antimicrobial properties of silver nanoparticle- encapsulated chitosan functionalized with photoactive groups. Materials Science and Engineering C. 2013;33(7):4409–4415. doi: 10.1016/j.msec.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 4.Ding W, Guo L. Immobilized transferrin Fe3O4@SiO2 nanoparticle with high doxorubicin loading for dual-targeted tumor drug delivery. International Journal of Nanomedicine. 2013;8(1):4631–4639. doi: 10.2147/IJN.S51745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Burnum KE, Luna ML, et al. Quantitative proteomics analysis of adsorbed plasma proteins classifies nanoparticles with different surface properties and size. Proteomics. 2011;11(23):4569–4577. doi: 10.1002/pmic.201100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simberg D, Park J, Karmali PP, et al. Differential proteomics analysis of the surface heterogeneity of dextran iron oxide nanoparticles and the implications for their in vivo clearance. Biomaterials. 2009;30(23-24):3926–3933. doi: 10.1016/j.biomaterials.2009.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faria M, Navas JM, Soares AM, Barata C. Oxidative stress effects of titanium dioxide nanoparticle aggregates in zebrafish embryos. Science of the Total Environment. 2014;470-471:379–389. doi: 10.1016/j.scitotenv.2013.09.055. [DOI] [PubMed] [Google Scholar]
- 8.Trouiller B, Reliene R, Westbrook A, Solaimani P, Schiestl RH. Titanium dioxide nanoparticles induce DNA damage and genetic instability in vivo in mice. Cancer Research. 2009;69(22):8784–8789. doi: 10.1158/0008-5472.CAN-09-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das P, Xenopoulos MA, Metcalfe CD. Toxicity of silver and titanium dioxide nanoparticle suspensions to the aquatic invertebrate, Daphnia magna. Bulletin of Environmental Contamination and Toxicology. 2013;91(1):76–82. doi: 10.1007/s00128-013-1015-6. [DOI] [PubMed] [Google Scholar]
- 10.Verano-Braga T, Miethling-Graff R, Wojdyla K, et al. Insights into the Cellular Response Triggered by Silver Nanoparticles Using Quantitative Proteomics. ACS Nano; 2014. [DOI] [PubMed] [Google Scholar]
- 11.Mirzajani F, Askari H, Hamzelou S, et al. Proteomics study of silver nanoparticles toxicity on Bacillus thuringiensis . Ecotoxicology and Environmental Safety. 2014;100:122–130. doi: 10.1016/j.ecoenv.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Jiang C, Hsu-Kim H, et al. Silver nanoparticle behavior, uptake, and toxicity in Caenorhabditis elegans: effects of natural organic matter. Environmental Science & Technology. 2014;48(6):3486–3495. doi: 10.1021/es404444n. [DOI] [PubMed] [Google Scholar]
- 13.AshaRani PV, Mun GLK, Hande MP, Valiyaveettil S. Cytotoxicity and genotoxicity of silver nanoparticles in human cells. ACS Nano. 2009;3(2):279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 14.Singh RP, Ramarao P. Cellular uptake, intracellular trafficking and cytotoxicity of silver nanoparticles. Toxicology Letters. 2012;213(2):249–259. doi: 10.1016/j.toxlet.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 15.Kim W, Kim J, Park JD, Ryu HY, Yu IJ. Histological study of gender differences in accumulation of silver nanoparticles in kidneys of Fischer 344 rats. Journal of Toxicology and Environmental Health A. 2009;72(21-22):1279–1284. doi: 10.1080/15287390903212287. [DOI] [PubMed] [Google Scholar]
- 16.Kuo C-J, Liang S-S, Hsi E, Chiou S-H, Lin S-D. Quantitative proteomics analysis of varicose veins: Identification of a set of differentially expressed proteins related to ATP generation and utilization. Kaohsiung Journal of Medical Sciences. 2013;29(11):594–605. doi: 10.1016/j.kjms.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Ellias MF, Ariffin SHZ, Karsani SA, Rahman MA, Senafi S, Wahab RMA. Proteomic analysis of saliva identifies potential biomarkers for orthodontic tooth movement. The Scientific World Journal. 2012;2012:6 pages. doi: 10.1100/2012/647240.647240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ficarro SB, Zhang Y, Carrasco-Alfonso MJ, et al. Online nanoflow multidimensional fractionation for high efficiency phosphopeptide analysis. Molecular & Cellular Proteomics. 2011;10(11) doi: 10.1074/mcp.O111.011064.O111.011064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu J, Huang S, Chow N, Chen S. Stable-isotope dimethyl labeling for quantitative proteomics. Analytical Chemistry. 2003;75(24):6843–6852. doi: 10.1021/ac0348625. [DOI] [PubMed] [Google Scholar]
- 20.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41(1):D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen W-Y, Chen Y-C. Functional Fe3O4@ZnO magnetic nanoparticle-assisted enrichment and enzymatic digestion of phosphoproteins from saliva. Analytical and Bioanalytical Chemistry. 2010;398(5):2049–2057. doi: 10.1007/s00216-010-4174-x. [DOI] [PubMed] [Google Scholar]
- 22.Lin H, Chen W, Chen Y. Iron oxide/tantalum oxide core-shell magnetic nanoparticle-based microwave-assisted extraction for phosphopeptide enrichment from complex samples for MALDI MS analysis. Analytical and Bioanalytical Chemistry. 2009;394(8):2129–2136. doi: 10.1007/s00216-009-2890-x. [DOI] [PubMed] [Google Scholar]
- 23.Woo K, Hong J, Choi S, et al. Easy synthesis and magnetic properties of iron oxide nanoparticles. Chemistry of Materials. 2004;16(14):2814–2818. [Google Scholar]
- 24.Yin YX, Shen F, Pei H, et al. Increased expression of Rab25 in breast cancer correlates with lymphatic metastasis. Tumor Biology. 2012;33(5):1581–1587. doi: 10.1007/s13277-012-0412-5. [DOI] [PubMed] [Google Scholar]
- 25.Adams HC, III, Chen R, Liu Z, Whitehead IP. Regulation of breast cancer cell motility by T-cell lymphoma invasion and metastasis-inducing protein. Breast Cancer Research. 2010;12(5, article R69) doi: 10.1186/bcr2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Guan H, Li H, et al. miR-101 inhibits cell proliferation by targeting Rac1 in papillary thyroid carcinoma. Bioscience Reports. 2014;2(1):122–126. doi: 10.3892/br.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bashir M, Kirmani D, Bhat HF, et al. P66shc and its downstream Eps8 and Rac1 proteins are upregulated in esophageal cancers. Cell Communication and Signaling. 2010;8, article 13 doi: 10.1186/1478-811X-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mondragón L, Orzáez M, Sanclimens G, et al. Modulation of cellular apoptosis with apoptotic protease-activating factor 1 (Apaf-1) inhibitors. Journal of Medicinal Chemistry. 2008;51(3):521–529. doi: 10.1021/jm701195j. [DOI] [PubMed] [Google Scholar]
- 29.Lauenborg B, Kopp K, Krejsgaard T, et al. Programmed cell death-10 enhances proliferation and protects malignant T cells from apoptosis. APMIS. 2010;118(10):719–728. doi: 10.1111/j.1600-0463.2010.02669.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barkan B, Cox AD, Kloog Y. Ras inhibition boosts galectin-7 at the expense of galectin-1 to sensitize cells to apoptosis. Oncotarget. 2013;4(2):256–268. doi: 10.18632/oncotarget.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyamu EW, Perdew H, Woods GM. Cysteine-iron promotes arginase activity by driving the Fenton reaction. Biochemical and Biophysical Research Communications. 2008;376(1):116–120. doi: 10.1016/j.bbrc.2008.08.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toyokuni S. Iron and carcinogenesis: from Fenton reaction to target genes. Redox Report. 2002;7(4):189–197. doi: 10.1179/135100002125000596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The supplementary information is the raw data generated all proteins in quantitative proteomics which are listed according to the D/H ratio priority.


