Abstract
Site-directed spin labeling is emerging as an essential tool to investigate the structural and dynamical features of RNA. We propose here an enzymatic method, which allows the insertion of a paramagnetic center at a specific position in an RNA molecule. The technique is based on a segmental approach using a ligation protocol with T4 RNA ligase 2. One transcribed acceptor RNA is ligated to a donor RNA in which a thio-modified nucleotide is introduced at its 5′-end by in vitro transcription with T7 RNA polymerase. The paramagnetic thiol-specific reagent is subsequently attached to the RNA ligation product. This novel strategy is demonstrated by introducing a paramagnetic probe into the 55 nucleotides long RNA corresponding to K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-Box leader RNA. The efficiency of the coupling reaction and the quality of the resulting spin-labeled RNA were assessed by Mass Spectrometry, Electron Paramagnetic Resonance (EPR) and Nuclear Magnetic Resonance (NMR). This method enables various combinations of isotopic segmental labeling and spin labeling schemes, a strategy that will be of particular interest to investigate the structural and dynamical properties of large RNA complexes by NMR and EPR spectroscopies.
INTRODUCTION
RNA molecules play crucial roles in many biological processes in all organisms. RNAs are information carriers, regulators of numerous biological pathways and also act as catalysts (1–4). Over the past decades, many diverse functional RNA architectures were discovered and their importance in many regulatory mechanisms of biological processes was established. RNA functions depend on their sequence and their striking ability to fold into complex structures capable of undergoing large structural rearrangements upon interactions with other molecular partners (5–7).
The elucidation of the mechanisms of recognition between RNAs and their interacting partners remains a major objective in molecular and structural biology. These efforts led to a significant increase of the number of high-resolution RNA structures over the past 10 years. However, thorough descriptions of RNA flexibility, an essential aspect of RNA function, remain less common. Nuclear Magnetic Resonance (NMR) provides an efficient method to unravel both structural and dynamical aspects of RNA and this approach has been successfully used to study a wide range of systems (8–11). Moreover, NMR is widely used to investigate the mechanisms involved in the interactions between RNA and other biomolecules, such as proteins (12,13). However, the study of large RNA molecules by NMR remains challenging, due to the combined effect of efficient relaxation and spectral crowding. Several methods combining isotopic labeling and optimized NMR signal acquisition schemes were recently developed to address these issues (14,15). One promising route to the application of NMR methodologies to larger RNA systems relies on the incorporation of paramagnetic probes, coupled to the measurement of the induced effects on nuclear spins (16). The paramagnetic relaxation enhancement (PRE) and chemical shift variations induced by the probe depend on: (i) the distance between the observed nucleus and the probe and (ii) the dynamics of the molecule. Despite its long-standing use in NMR, this technique was only applied in the 1990s to proteins and DNA/protein complexes (17–21). The strategy was later implemented for the study of RNA-protein complexes by adapting the paramagnetic spin label to RNA chemistry (22–24). The method was successfully used to refine 3D structures by taking advantage of supplementary long-range distances, to observe the existence of transient encounter complexes between interacting proteins or to investigate protein-DNA recognition mechanisms (16,25,26). Noteworthy, the use of spin label enabled the application of Electron Paramagnetic Resonance (EPR) approaches to study biological macromolecules and their complexes (27–29). Of particular interest is the ability of this spectroscopy to describe conformational changes within large complexes by the measurement of long-range distances between two sites bearing electronic probes (30). The development of efficient methods of incorporation of a paramagnetic center at specific positions in an RNA is therefore of primary importance for these two magnetic spectroscopies.
During recent years, several spin labeling techniques of RNA were developed to study RNA and their complexes (24,25,31–35). A recent approach describes the use of a novel 2,2,6,6-tetramethylpiperidine 1-oxyl (TEMPO) radical phosphoramidite building block whose attachment at the RNA 5′-end allows PRE measurement by NMR (25). However, the chemical synthesis of RNA is limited to short RNA oligonucleotides. Enzymatic methods were also reported for grafting a nitroxide group to RNA termini (31,32). Macosko et al. used the T7 RNA polymerase to introduce a guanosine monophosphorothioate at the 5′-end of an RNA molecule, allowing its subsequent modification by the covalent addition of the spin label (31). The T4 polynucleotide kinase was used by Grant and Qin to link a phosphorothioate group at the 5′-end of the RNA, enabling its reaction with iodomethyl derivative of nitroxide (32). A non-covalent spin labeling strategy was used by Helmling et al. to obtain long-range structural NMR restraints (35). Recently, one approach using convertible nucleosides and DNA-catalyzed RNA ligation was also suggested to insert a nitroxide spin label at specific positions for EPR studies (34).
Here, we propose a general, fully enzymatic method for the introduction of a paramagnetic probe at a specific position in an RNA sequence. Our technique is based on a segmental approach using a ligation protocol with T4 RNA ligase 2 (36–38). Two RNA fragments corresponding each to a part of the full-length RNA are designed and transcribed using T7 RNA polymerase. One transcribed acceptor RNA is ligated to a donor RNA in which a thio-modified nucleotide is introduced at its 5′-end by in vitro transcription with T7 RNA polymerase (Figure 1A). The ligation product corresponding to the full-length RNA contains a single thio-modified nucleotide at a specific position (Figure 1C). The paramagnetic spin label is subsequently attached to the RNA containing the thio-modified base at the chosen position. This method yields to a very efficient way of incorporating modified nucleotides in an RNA sequence without size limitation. This enables the study of large size RNA molecules using spectroscopic tools. The high incorporation efficiency of the paramagnetic probe will be of interest to measure distances with EPR methods. For NMR, it offers the possibility to combine segmental and selective isotopic labeling with PRE measurements, opening a novel perspective to study the dynamics and the structure of RNA molecules.
Figure 1.
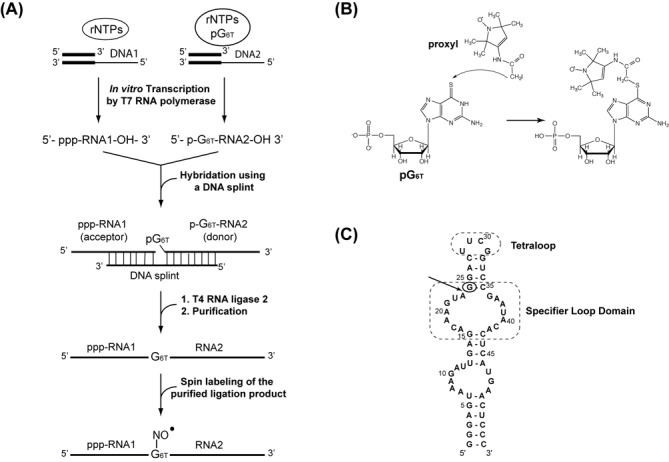
Method for the site-directed spin labeling approach. (A) Schematic representation of the synthesis of site-specific spin labeled RNA. 6-thioguanosine-5′-O-monophosphate (pG6T = 6-T-GMP) is incorporated at a specific position. (B) The proxyl-RNA coupling reaction involves the addition of 3-(2-Iodoacetamido)-proxyl on 6-thioguanosine-5′-O-monophosphate. (C) Secondary structure of the 55-nt (tyrS) RNA molecule corresponding to the specifier loop domain and K-turn sequence motif of the tyrS leader RNA. The Specifier Loop Domain and the tetraloop are boxed. The arrow indicates the chosen segmentation site and the circle indicates the thio-modified site.
The efficiency of the site-specific spin-labeling reaction and the quality of the spin-labeled RNA were monitored using NMR, mass spectrometry (MS) and EPR. This site-directed spin labeling method is demonstrated on the 55-nt (17.7 kDa) RNA corresponding to K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-Box leader RNA, whose structure was solved by NMR by Wang and Nikonowicz (PDB id: 2KZL; 39). We show that a paramagnetic center can be successfully inserted at a specific position in an RNA sequence using T4 RNA ligation techniques, with yields compatible with NMR and EPR studies.
MATERIALS AND METHODS
Synthesis of RNA segments
All RNA segments were prepared either unlabeled or 13C-15N labeled by in vitro transcription with homemade T7 RNA polymerase from oligonucleotide templates containing a 2′-O-methyl modification at position 2 (40). The DNA templates were purchased from Sigma (France) and Eurogentec (Belgium). Transcription conditions were optimized according to previous protocols (41,42). 6-thioguanosine-5′-O-monophosphate (6-T-GMP) was purchased from BIOLOG (Germany). Labeled NTPs (nucleotide triphosphate) were purchased from Eurisotop and Sigma (France). The full-length RNA and segment (G1-A23) were prepared unmodified (Figure 1C). The RNA segment (G24-C55) was modified at its 5′-end by incorporating 6-T-GMP nucleoside. Each NTP was added at a classical concentration of 4 mM, with the exception of guanosine 5'-triphosphate (GTP) whose concentration was 3 mM. 6-T-GMP was added at a concentration of 9.1 mM (32). RNAs were purified on denaturing polyacrylamide gels as described by Wyatt et al. (42). After electroelution and ethanol precipitation, RNAs were resuspended in water. The concentration of RNA samples was measured using a Nanodrop Spectrometer and calculated with molar extinction coefficients obtained from the Eurofins MWG Operon calculator (http://www.operon.com/tools/oligo-analysis-tool.aspx).
Hybridization of RNA fragments to DNA splint
The interactions of RNA segments with DNA were characterized by electrophoresis under native conditions. The samples were heated at 95°C (2 min) and snap-cooled at 4°C in the T4 RNA ligase 2 buffer (50 mM TRIS-HCl, 2 mM MgCl2, 1 mM DTT, 400 μM adenosine triphosphate (ATP) at pH 7.5). After centrifugation, at 10 000 revolutions per minute (rpm), for 5 min at room temperature, the samples were examined by electrophoresis on a 10% polyacrylamide native gel. The gel was run in 0.5X TBE at 150 V, at 4°C. Bands were visualized with toluidine blue.
Ligation of RNA segments
The (G1-A23) RNA segment was ligated to the thio-modified (G24-C55) fragment by T4 RNA ligase 2 using a complementary 43-nt DNA splint. The T4 RNA ligase 2 was purchased from New Englands Biolabs (United States). The 43-nt DNA splint (5′-TCATGAGTGTATTCGGACCGAAGTCCTACTTGTCTCAATCTTT-3′) was purchased from SIGMA (France). A series of test experiments performed in 10 μl, with varying temperatures, reaction time, RNA and enzyme concentrations was carried out in the T4 RNA ligase 2 buffer (50 mM TRIS-HCl, 2 mM MgCl2, 1 mM DTT, 400 μM ATP at pH 7.5). After optimization, a 5 ml preparative one-pot ligation reaction with equimolar strand concentrations ranging from 10 to 30 μM of each RNA segment and the DNA splint was performed in the T4 RNA ligase 2 buffer. The reaction mixture was heated at 95°C and snap-cooled at 4°C. Then 2.25 kU of T4 RNA ligase 2 were added and the mixture was incubated overnight at 37°C. The efficiency of the ligation was followed on a 12% polyacrylamide denaturing gel containing 7 M urea. The product of ligation corresponding to the full-length RNA was purified from denaturing polyacrylamide gel electrophoresis as described previously (42). The RNA was then electroeluted from the gel and recovered by ethanol precipitation. The yield of the ligation was estimated at 50% by quantification of the intensities of the stained bands on the gel.
Spin labeling of RNA
3-(2-Iodoacetamido)-proxyl (proxyl) was purchased from Sigma (France). The efficiency of the coupling reaction between the nitroxide reagent and the thio-modified nucleotide was verified by NMR (Figure 1B; Supplementary Figure S1).
In order to prevent light-induced degradation of the nitroxide, all following steps were performed in the dark.
The RNA containing one 6-T-GMP modification at a specific position, was resuspended in 400 μl of 100 mM sodium phosphate buffer at pH 8.0. A total of 40 μl of 200-fold-excess of 3-(2-Iodoacetamido)-proxyl dissolved in ethanol was added in two steps to the RNA solution. The reaction mixture was incubated at 40°C for 24 h. The integrity of the spin-labeled RNA was then verified on a 20% denaturing polyacrylamide gel (Supplementary Figure S2). In addition, after a few weeks, no cleavage or degradation of the spin-labeled RNA was observed.
After ethanol precipitation and centrifugation at 12 000 rpm at 4°C for 30 min, the pellet of the spin-labeled RNA was washed with 400 μl ethanol. After removal of the supernatant, the RNA was resuspended in water and dialysed against the buffer used for NMR and EPR experiments, overnight at 4°C. The dialysis buffer was changed four times. The sample was concentrated by lyophilization and resuspended in H2O/D2O (90/10) for NMR and EPR experiments. The labeling efficiency was evaluated using NMR, MS and EPR. The concentrations of RNA samples were determined as described above.
MS experiments
Prior to MS analysis, RNA samples were dialyzed against 200 mM ammonium acetate on Amicon Ultra-4 centrifugal filter units-10000NMWL and concentrated to 30 μM final. RNA intact mass detections were performed using an ESI-TOF mass spectrometer (Q-TOF micro, Waters, MA, USA). Calibration was achieved in the negative ion mode, using phosphoric acid. Samples were diluted to about 1 μM in 1:1 water–acetonitrile containing 1% triethylamine. MaxEnt (maximum entropy deconvolution software) was used to provide a first approximate of the masses whereas exact masses were calculated manually from the multiple charged species.
The RNA modification by the spin label was characterized after RNaseA treatment on a MALDI-TOF/TOF mass spectrometer (Autoflex III Smart Beam, Bruker, Billerica, MA, USA). Dried-droplet method was used with 3-Hydroxy picolinic acid matrix. Spectra were recorded in negative-ion mode after an external calibration using a synthetic ODN (OligoDeoxiriboNucleotide) mixture. MS/MS mode was used to confirm the nucleotide sequence and the localization of the modification.
Continuous-wave EPR spectroscopy
Conventional X-band field swept EPR spectra were acquired on an EMX X-band spectrometer (EMXplus from Bruker Biopsin GmbH, Germany), equipped with a high sensitivity resonator (4119HS-W1, Bruker). RNA-labeled sample in 70 mM sodium phosphate buffer at pH 6.5 supplemented by 10% D2O was loaded into glass capillaries (Hirschmann ringcaps, 20 μl) that were sealed at both ends. Standards of known concentrations of PROXYL were used to estimate the labeling efficiency. All experiments were performed at room temperature (295 ± 1 K). Main acquisition parameters were: 0.05 mT amplitude modulation, 1.8 mW incident microwave power, 40 ms time constant, 100 ms conversion time, 1200 points/scan, 15 mT sweep width and at least 3 scans were accumulated per spectrum to achieve reasonable signal to noise (S/N) ratio. Simulations were generated under matlab environment using Easyspin Toolbox (43).
NMR experiments
NMR experiments were recorded at 500, 600 and 700 MHz on Avance Bruker spectrometers equipped with a z-gradient TBI probe (at 500 MHz) and cryoprobes. NMR data were processed using TopSpin (Bruker) and NMRpipe (44), and analyzed with Sparky software (45). NMR experiments were recorded in 70 mM sodium phosphate buffer at pH 6.5. The concentration of RNA samples was 200 μM in 150 μl, in 3 mm NMR tubes. Resonance assignments were performed on the basis of previously reported NMR data (BMRB entry: 17316; 39). All NMR spectra were acquired at 293 K. Solvent suppression was either achieved using a ‘Jump and Return’ combined with a WATERGATE or single WATERGATE pulse sequence for 1D and 2D experiments, respectively (46). 2D TOCSY (total correlation spectroscopy) spectra were acquired using a MLEV (Malcolm Levitt composite-pulse decoupling sequence) mixing sequence and a mixing time of 60 ms.
The cross-peak intensities in the homonuclear TOCSY and heteronuclear 1H-13C HSQC (Heteronuclear Single Quantum Coherence) experiments were measured using the Sparky software. Intensities were normalized using the intensity corresponding to the H5-H6 correlation peak of C54 on the TOCSY spectra and the H8-C8 correlation peak of A50 on the HSQC. Both C54 and A50 residues are located more than 50 Å away from the paramagnetic center, according to the 3D structure, and therefore are not affected by the paramagnetic relaxation. PRE was then quantified using the ratio between the intensities of the spin-labeled RNA and those of the unlabeled RNA. Observed perturbations upon the attachment of the spin-label were reported on the structure (PDB id: 2KZL; 39) visualized using the Pymol software (47).
RESULTS AND DISCUSSION
Enzymatic synthesis of site-specific thio-modified RNA
The RNA ligation site was chosen such as to optimize the transcription efficiency of the 5′ fragment by the T7 RNA polymerase that displays higher reaction yields for sequences starting with a GG dinucleotide (41). Among the possible sites, the A23-G24 linkage was chosen for its central position within the RNA (Figure 1C). In addition, the sulfur atom located in the thio-modified nucleotide needs to be accessible for the spin-labeling reaction. Consequently, it is preferable to choose a position located in a structure that could be melt at a temperature below 50°C to maintain the integrity of the nitroxide.
The RNA fragment (G1-A23) was therefore synthesized by classical in vitro transcription whereas (G24-C55) RNA was modified at its 5′-end with the incorporation of a 6-T-GMP nucleoside as described (31). After purification of RNA transcripts, optimization experiments were performed for the ligation by T4 RNA ligase 2, which displays better activity on double-stranded nucleic acids (38). Optimal conditions for the hybridization of the RNA fragments to the complementary DNA splint were first investigated to avoid the formation of side-products. After heating an equimolar mixture of DNA and RNAs at 95°C and snap cooling at 4°C, the formation of the DNA/RNA hybrids was examined by electrophoresis on a native gel (Figure 2A). The experiments revealed that the optimal ligation yield was obtained after a minimal reaction time of 5 h at 37°C, with oligonucleotides concentrations ranging from 10 to 30 μM (Figure 2B). After purification and dialysis against the buffer used for NMR and EPR studies, the ligation product containing one thio-modified guanosine (6-T-GMP), was analyzed by NMR. Figure 2C compares the imino proton region of 1D spectra of the wild-type full-length RNA with the corresponding spectrum of the (G1-A23)-(G246TG–C55) ligation product. The two spectra are nearly identical, suggesting that the incorporation of the modified guanine does not alter the hydrogen-bonding pattern. In conclusion, our NMR data clearly show that the thio-modified residue was introduced into the RNA without altering the global fold of the RNA.
Figure 2.
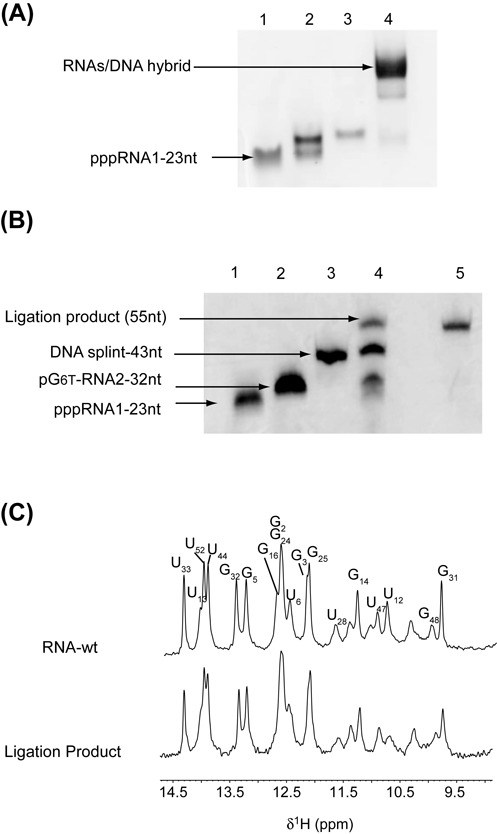
Synthesis of site-specific spin-labeled RNA. (A) Native gel electrophoresis on 10% polyacrylamide. RNA segments and DNA splint were loaded in the T4 RNA ligase 2 buffer: lane 1, (pG246T-C55); lane 2, (pppG1-G23); lane 3, DNA splint; lane 4, (pppG1-G23) + (pG246T-C55) + DNA splint. (B) Denaturing 12% polyacrylamide gel. Lane 1: RNA fragment (G1-G23) acceptor; lane 2: RNA fragment (pG246T-C55) donor; lane 3: DNA splint (43-nt); lane 4: preparative ligation; lane 5: purified ligation product. (C) Imino-proton region of 1D spectra recorded at 20°C of the wild-type RNA full-length (top) and the ligation product (bottom). All imino protons were assigned via sequential Nuclear Overhauser Effects (NOEs) observed in 2D-NOESY experiments, with the exception of the resonances at 10.37, 11.09 and 11.43 p.p.m. that could not be identified unambiguously.
Coupling reaction of the spin label to the thio-modified RNA
Next, optimal reaction conditions to couple the nitroxide derivative to the RNA were determined using the (G1-A23)-(G246TG–C55) ligation product as substrate for the reaction. Optimal yields were obtained by adding 200-fold excess of the proxyl reagent over 24 h at pH 8.0 and 40°C. This temperature was chosen to partially melt the structure in which the G246TG is involved in order to enhance its accessibility to the spin label.
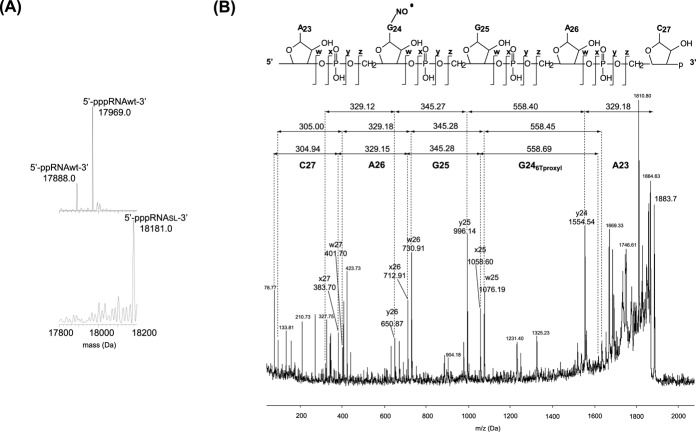
The coupling reaction efficiency as well as the integrity of the resulting spin-labeled RNA were checked using Electro Spray Inject Mass Spectrometry. Exact masses for the wild-type RNA and the spin-labeled RNA were found at 17967.5 ± 0.5 Da and 18180.5 ± 0.9 Da, respectively (Table 1). The corresponding mass difference (213 Da) corresponds to the addition of the proxyl group and the substitution of an oxygen by a sulfur atom on the guanine as expected. Furthermore, as shown in Figure 3A, the spin-labeled RNA was obtained as a major product and no significant signal corresponding to the unmodified RNA could be detected, indicating that the coupling reaction was effective. The modification was further characterized using an RNaseA treatment prior MS analysis in order to confirm the nucleotide sequence and the localization of the modification (Figure 3B). The RNase A cleaves RNA sequence after pyrimidine nucleotides. The fragment 5′-A23G24SLG25A26C27 p -3′ whose monoisotopic mass is 1884.1 Da, contains the thio-modified G bearing the nitroxide modification. MS fragmentation of this modified oligonucleotide leads to typical fragment ions depicted in Figure 3B (Supporting Table S1; 48). Mass values of 329.1, 345.3, 329.1 and 305.0 were found respectively for A23, G25, A26 and C27 while the mass of the spin-labeled G24 nucleotide was found at 558.4 Da, demonstrating that the RNA is fully modified at the desired position.
Table 1. Molecular weight determined by MS.
| m.w. calculated (Da) | m.w. found (Da) | |
|---|---|---|
| wt-RNA | 17967.8 | 17967.5 ± 0.5 |
| Spin-labeled RNA | 18181.1 | 18180.5 ± 0.9 |
Figure 3.
Molecular weight determined by MS. (A) ESI-MS spectra of wild-type RNA (top) and of the spin-labeled RNA after MaxEnt deconvolution showing a first approximate of the masses. Exact masses were next calculated manually from the multiply charged species (Table 1). (B) MSMS spectrum of the spin-labeled RNA after RNaseA treatment. The fragment 5′-A23G24SLG25A26C27 p-3′ whose parent mass is 1884.1 Da, contains the thio-modified G bearing the nitroxide modification. Possible fragmentations are indicated by w, x, y and z series.
Characterization of spin-labeled RNA by EPR spectroscopy
EPR spectroscopy was then used to provide a quantitative assessment of the nitroxide labeling efficiency. The proxyl coupling to the modified RNA resulted into significant changes of the EPR spectrum (Figure 4). While the free unbound PROXYL exhibits three sharp lines of almost equal amplitudes (upper spectrum), larger and heterogeneous line widths were found for the spin-labeled RNA spectrum, indicative of slow and restricted rotational motions (lower spectrum). Further line-shape analysis provided us a correlation time in the range of a few nanoseconds compatible with the RNA molecular size, while it is of ca. 100 picoseconds for the free spin label (43). It is worth noting that no significant trace of free proxyl was found in the spin-labeled RNA spectrum, indicating that all spins are coupled to RNA. The EPR signal was further used to quantify the coupling reaction yield using PROXYL standards of known concentrations. The spin concentration was found to be ca. 150 ± 15 μM from the double integration of the EPR spectrum. This value was in very good agreement with the RNA concentration obtained by ultraviolet absorption (147 ± 15 μM), suggesting that a proxyl-RNA coupling efficiency greater than 95% was achieved. Such a high coupling efficiency results from the separation of the enzymatic RNA ligation and the spin label coupling reactions by a purification step in order to remove the residual DTT. Indeed, while efficient ligation has already been achieved using spin-labeled RNA substrates, significant loss of EPR signal was observed due to the nitroxide reduction under ligation conditions (34).
Figure 4.
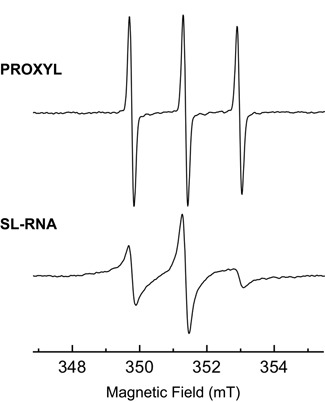
Characterization of spin-labeled RNA by EPR spectroscopy. EPR spectra of covalently labeled RNA (bottom) compared to free PROXYL (top). Both spectra were recorded in 70 mM sodium phosphate buffer (pH = 6.5), supplemented by 10% v/v D2O, and acquired with identical instrumental parameters. In this case the concentration of spin-labeled RNA was found to be 150 ± 15 μM, whereas the concentration of the free proxyl spectrum corresponds to 50 μM.
Characterization of spin-labeled RNA by NMR spectroscopy
The impact of the spin-labeled guanosine on the RNA 3D structure was then further investigated using NMR. The pattern of imino-proton frequencies observed for the wild-type and the spin-labeled RNA was very similar, except for few residues that displayed reduced intensities, such as G24, indicating that the global fold of the RNA is not affected by the presence of the spin label (Figure 5A). In particular, the unperturbed resonance of G31 imino proton indicates that the UUCG tetraloop fold is not altered in the spin-labeled RNA. A semi-quantitative analysis of the enhanced relaxation due to the paramagnetic proxyl was conducted by monitoring intensities of the cross-peaks between non-exchangeable H5 and H6 protons in MLEV experiments (Figure 5B). Semi-quantitative analysis of PRE is in agreement with the global RNA 3D structure (Figure 6A and B). Indeed, residues C18, U22, C27, U33 and C35 display low values of PRE (IproxylRNA/IRNA), consistent with their positions in the vicinity of the spin label while C54 and C55, which are located more than 50 Å away from the labeling site, were not affected (Figures 5B and 6A). Noteworthy, larger PRE values are found for residues U39, C41 and C43 located in the Specifier Loop Domain of the RNA, opposite to the tetraloop, suggesting that the proxyl is oriented toward this region. It should be noted that dynamical behaviors of specific regions of an RNA molecule may considerably affect the paramagnetic relaxation, as reported for the paramagnetic 5′-end labeled HIV-1 TAR RNA study where significant differences between experimental and modeled distances were observed (25). Interestingly, PRE values measured using homonuclear H5-H6 correlations are larger than those measured using heteronuclear experiments (compare PRE values of C6-H6 with H5-H6 of C27 in Figure 6A and C). Further experimental and modeling studies will be necessary to establish accurate quantification procedures enabling accurate distances to be obtained from PRE data in RNA. Our efficient and selective RNA spin labeling protocol provides unprecedented perspectives in this direction.
Figure 5.
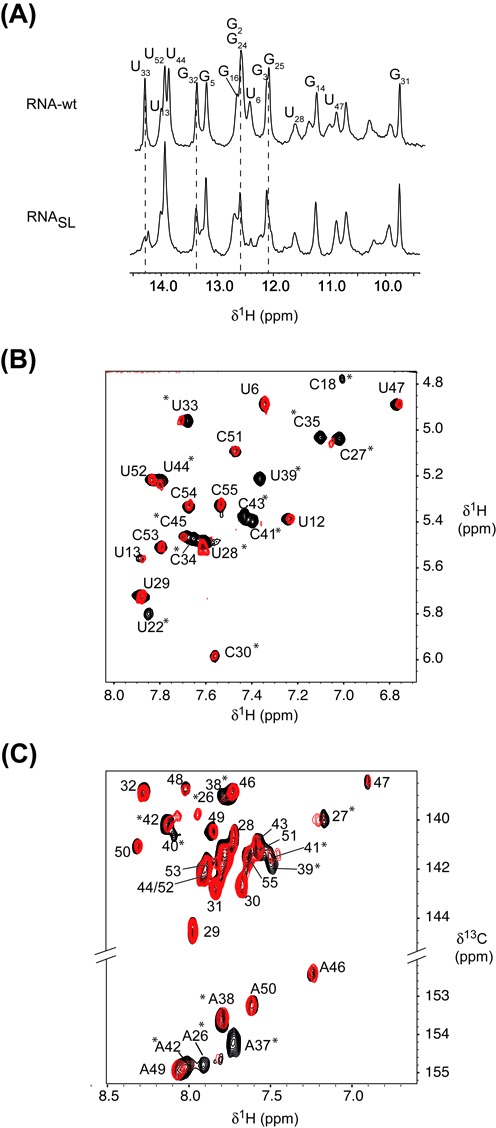
NMR of site-specific spin-labeled RNA. Imino-proton region of 1D spectra, recorded at 20°C, of the wild-type RNA (top) and the spin-labeled RNA (bottom). (B) H6-H5 region of MLEV experiments, recorded at 20°C, of the free RNA (black) and the spin-labeled RNA (red). Stars indicate residues that undergo variations upon nitroxide attachment. (C)1H-13C HSQC spectra showing the aromatic H2-C2 (bottom) and H6/8-C6/8 (top) of the free RNA (black) and the spin-labeled RNA (red). Stars indicate residues that undergo variations upon nitroxide attachment.
Figure 6.
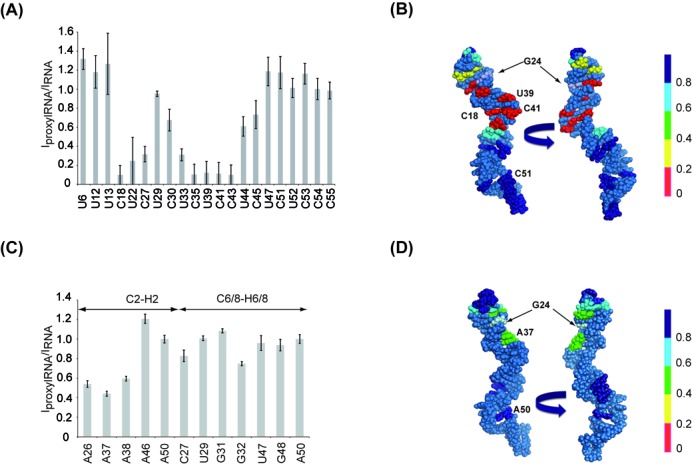
Effect of site-specific spin-labeled RNA analyzed by NMR. (A) Bar graph of intensity ratio between the spin-labeled RNA (IproxylRNA) and the free form RNA (IRNA), normalized to C54, extracted from MLEV experiments, versus the primary sequence of the RNA. The quantification of residues C18, U22, C35, U39, C41 and C43 was not feasible due to the disappearance of the corresponding resonances. The corresponding ratios were estimated by measuring values at the same frequencies as the resonances observable in the wild-type RNA. (B) Sphere representation of the RNA structure colored according to the intensity ratios extracted from MLEV experiments. The spin-labeled G24 residue is highlighted in light blue. (C) Bar graph of intensity ratio between the (G1-A23)-(G246TGproxyl–C55) RNA and the free form RNA, normalized to A50, extracted from HSQC experiment, versus the primary sequence of the RNA. (D) Sphere representation of the RNA structure colored according to the intensity ratios extracted from HSQC experiments. The spin-labeled G24 residue is highlighted in light blue.
Combining Site-specific spin-labeling with segmental isotope labeling
The use of NMR to study the structure and dynamics of RNA larger than 50 nucleotides requires generally the use of dedicated isotopic labeling strategies to address both the spectral crowding and signal broadening problems. One particularly appealing strategy for RNA is the segmental labeling, which allows the selective observation of a specific labeled domain within the full-length RNA (36). Since this labeling scheme requires an enzymatic ligation, it may be useful to combine both segmental isotopic labeling and site selective incorporation of a spin label. In order to establish the feasibility of such an approach, the fragment (G246TG–C55) was transcribed with 13C labeled nucleotides, with the exception of the thio-modified G24, and then ligated to the unlabeled (G1-A23) segment. After addition of the proxyl spin label, the intensities of C2H2, C6H6 and C8H8 correlation peaks were measured in a 1H-13C HSQC spectra and compared to those obtained from a 13C labeled RNA lacking the spin label (Figures 5C and 6C). The correlations were assigned using previously reported NMR data (BMRB entry: 17316; 39). Upon nitroxide attachment to the G24 residue, the C2H2 correlations corresponding to A26 and A37 disappear, the intensities of A38 and A42 cross-peaks show a significant decrease while A50 and A46 remain almost unaffected (Figures 5C and 6C and D). The C8H8 cross-peaks corresponding to A26, A38, U39, A40, C41 and A42 display attenuations and frequency shifts upon the nitroxide incorporation (Figures 5C and 6C). It is noteworthy that the HSQC experiment displays a weaker sensitivity to paramagnetic effect in comparison to the MLEV experiments (Figure 6A and C), as illustrated by residue C27. This is due to the combined effects of a longer evolution time characterizing the MLEV mixing sequence and the higher gyromagnetic ratio of the proton. Our data clearly show that the site-specific spin labeling can be successfully combined with segmental isotope labeling, a strategy that allows to combine NMR and EPR spectroscopies to investigate both local and global aspects of RNA plasticity.
CONCLUSION
We provide a flexible enzymatic protocol for the site-directed spin-labeling of long RNA yielding enough product for NMR studies while maintaining the advantage of selective and segmental isotope labeling with nitrogen-15 and carbon-13. In contrast with approaches relying on partial or complete solid-phase chemical synthesis methods, our enzymatic approach can be applied to large RNA molecules. Such feature is of particular interest for EPR studies when two specific sites have to be labeled in order to monitor the dipolar interaction between the two electronic spins, allowing inter-nitroxide distances measurement (49). Since the coupling between the nitroxide and RNA occurs at the last step of the labeling protocol, our labeling strategy preserves the quality of the spin label as shown by EPR results. This method enables various combinations of isotopic segmental labeling and spin labeling schemes, a strategy that will be of particular interest to investigate the structural and dynamical properties of large RNA complexes by NMR and EPR spectroscopies.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
The authors thank Claude Ling for technical assistance at the NMR spectrometer. We are also grateful to Robert Drillen for critical reading of the manuscript.
FUNDING
Agence Nationale de la Recherche [Grant RNASPIN ANR-2010-JCJC-1507–01 to I.L.]; French Infrastructure for Integrated Structural Biology (FRISBI) [ANR-10-INSB-05–01]; INSTRUCT, part of the European Strategy Forum on Research Infrastructures (ESFRI) and through national member agreements; CNRS, University of Strasbourg and INSERM. Funding for open access charge: Agence Nationale de la Recherche.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bevilacqua P.C., Blose J.M. Structure, kinetics, thermodynamics, and biological functions of RNA hairpins. Annu. Rev. Phys. Chem. 2008;59:79–103. doi: 10.1146/annurev.physchem.59.032607.093743. [DOI] [PubMed] [Google Scholar]
- 2.Serganov A., Patel D.J. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat. Rev. Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker R.R. Riboswitches and the RNA world. Cold Spring Har. Perspect. Biol. 2012;4 doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groβhans H., Filipowicz W. The expanding world of small RNAs. Nature. 2008;451:414–416. doi: 10.1038/451414a. [DOI] [PubMed] [Google Scholar]
- 5.Brunel C., Marquet R., Romby P., Ehresmann C. RNA loop-loop interactions as dynamics functional motifs. Biochimie. 2002;84:925–944. doi: 10.1016/s0300-9084(02)01401-3. [DOI] [PubMed] [Google Scholar]
- 6.Dethoff E.A., Chugh J., Mustoe A.M., Al-Hashimi H.M. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reining A., Nozinovic S., Schlepckow K., Buhr F., Fürtig B., Schwalbe H. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499:355–359. doi: 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- 8.Allen M., Varani L., Varani G. Nuclear magnetic resonance methods to study structure and dynamics of RNA-protein complexes. Methods Enzymol. 2001;339:357–376. doi: 10.1016/s0076-6879(01)39322-9. [DOI] [PubMed] [Google Scholar]
- 9.Latham M.P., Brown D.J., McCallum S., Pardi A. NMR methods for studying the structure and dynamics of RNA. Chem. Bio. Chem. 2005;6:1492–1505. doi: 10.1002/cbic.200500123. [DOI] [PubMed] [Google Scholar]
- 10.Fürtig B., Buck J., Manoharan V., Bermel W., Jäschke A., Wenter P., Pitsch S., Schwalbe H. Time-resolved NMR studies of RNA folding. Biopolymers. 2007;86:360–383. doi: 10.1002/bip.20761. [DOI] [PubMed] [Google Scholar]
- 11.Getz M., Sun X., Casiono-Negroni A., Zhang Q., Al-Hashimi H.M. NMR studies of RNA dynamics and structural plasticity using NMR residual dipolar couplings. Biopolymers. 2007;86:384–402. doi: 10.1002/bip.20765. [DOI] [PubMed] [Google Scholar]
- 12.Varani G., Chen Y., Leeper T.C. NMR studies of protein-nucleic acid interactions. Methods Mol. Biol. 2004;278:289–312. doi: 10.1385/1-59259-809-9:289. [DOI] [PubMed] [Google Scholar]
- 13.Wu H., Finger L.D., Feigon J. Structure determination of protein/RNA complexes by NMR. Methods Enzymol. 2005;394:525–545. doi: 10.1016/S0076-6879(05)94022-6. [DOI] [PubMed] [Google Scholar]
- 14.Mackereth C.D., Simon B., Sattler M. Extending the size of Protein-RNA complexes studied by nuclear magnetic resonance spectroscopy. Chem. Bio. Chem. 2005;6:1578–1584. doi: 10.1002/cbic.200500106. [DOI] [PubMed] [Google Scholar]
- 15.Foster M.P., McElroy C.A., Amero C.D. Solution NMR of large molecules and assemblies. Biochemistry. 2007;46:331–340. doi: 10.1021/bi0621314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai S., Zhu L., Zhang Z., Chen Y. Determination of the three-dimensional structure of the Mrf2-DNA complex using paramagnetic spin labeling. Biochemistry. 2007;46:4943–4950. doi: 10.1021/bi061738h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter D.E., Cohn M. Changes in tertiary structure accompanying a single base change in transfer RNA. Proton magnetic resonance and amynoacylation studies of Escherichia coli tRNAmetf1 and tRNAmetf3 and their spin-labeled (S4U8) derivatives. Biochemistry. 1976;15:3917–3924. doi: 10.1021/bi00663a003. [DOI] [PubMed] [Google Scholar]
- 18.Folkers P.J.M., van Duynhoven J.P.M., van Lieshout H.T.M., Harmsen B.J.M., van Boom J.H., Tesser G.I., Konings R.N.H., Hilbers C.W. Exploring the DNA binding domain of gene V protein encoded by bacteriophage M13 with the Aid of Spin-labeled oligonucleotides in combination with 1H NMR. Biochemistry. 1993;32:9407–9416. doi: 10.1021/bi00087a020. [DOI] [PubMed] [Google Scholar]
- 19.Gillepsie J.R., Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. I. Paramagnetic relaxation enhancement by nitroxide spin labels. J. Mol. Biol. 1997;268:158–169. doi: 10.1006/jmbi.1997.0954. [DOI] [PubMed] [Google Scholar]
- 20.Gillepsie J.R., Shortle D. Characterization of long-range structure in the denatured state of staphylococcal nuclease. II. Distance restraints from paramagnetic relaxation and calculation of an ensemble of structures. J. Mol. Biol. 1997;268:170–184. doi: 10.1006/jmbi.1997.0953. [DOI] [PubMed] [Google Scholar]
- 21.Dunham S.U., Dunham S.U., Turner C.J., Lippard S.J. Solution structure of a DNA duplex containing a nitroxide spin-labeled platinum d(GpG) intrastrand cross-link refined with NMR-derived long range electron-proton distance restraints. J. Am. Chem. Soc. 1998;120:5395–5406. [Google Scholar]
- 22.Edwards T.E., Sigurdsson, Th S. Site-specific incorporation of nitroxide spin-labels into 2’-positions of nucleic acids. Nat. Prot. 2007;2:1954–1962. doi: 10.1038/nprot.2007.273. [DOI] [PubMed] [Google Scholar]
- 23.Edwards T.E., Okonogi T.M., Robinson B.H., Sigurdsson S.T. Site-specific incorporation of nitroxide spin-labels into internal sites of the TAR RNA; structure-dependent dynamics of RNA by EPR spectroscopy. J. Am. Chem. Soc. 2001;123:1527–1528. doi: 10.1021/ja005649i. [DOI] [PubMed] [Google Scholar]
- 24.Ramos A., Varani G. A new method to detect long-range protein-RNA contacts: NMR detection of electron-proton relaxation induced by nitroxide spin-labeled RNA. J. Am. Chem. Soc. 1998;120:10992–10993. [Google Scholar]
- 25.Wunderlich C.H., Huber R.G., Spitzer R., Liedl K.R., Kloiber K., Kreutz C. A novel paramagnetic relaxation enhancement tag for nucleic acids: A tool to study structure and dynamics of RNA. ACS Chem. Biol. 2013;8:2697–2706. doi: 10.1021/cb400589q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwahara J., Clore G.M. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- 27.Hubbell W.L., Cafiso D.S., Altenbach C. Identifying conformational changes with site-directed spin labeling. Nat. Struct. Biol. 2000;7:735–739. doi: 10.1038/78956. [DOI] [PubMed] [Google Scholar]
- 28.Qin P.Z., Dieckmann T. Application of NMR and EPR methods to the study of RNA. Curr. Opin. Struct. Biol. 2004;14:350–359. doi: 10.1016/j.sbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Fanucci G.E., Cafiso D.S. Recent advances and applications of site-directed spin labeling. Curr. Opin. Struct. Biol. 2006;16:644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 30.Schiemann O., Weber A., Edwards T.E., Prisner T.F., Sigurdsson S.T. Nanometer distance measurements on RNA using PELDOR. J. Am. Chem. Soc. 2003;125:3334–3335. doi: 10.1021/ja0274610. [DOI] [PubMed] [Google Scholar]
- 31.Macosko J.C., Pio M.S., Tinoco JR. I., Shin Y.K. A novel 5’ displacement spin-labeling technique for electron paramagnetic resonance spectroscopy of RNA. RNA. 1999;5:1158–1166. doi: 10.1017/s1355838299990830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grant G.P.G., Qin P.Z. A facile method for attaching nitroxide spin label at the 5’ terminus of nucleic acids. Nucleic Acids Res. 2007;35:e77. doi: 10.1093/nar/gkm240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cekan P., Smith A.L., Barhate N., Robinson B.H., Sigurdsson S.Th. Rigid spin-labeled nucleoside Ç: a nonperturbing EPR probe of nucleic acid conformation. Nucleic Acids Res. 2008;36:5946–5954. doi: 10.1093/nar/gkn562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buttner L., Seikowski J., Wawrzyniak K., Ochmann A., Höbartner C. Synthesis of spin-labeled riboswitch RNAs using convertible nucleosides and DNA-catalyzed RNA ligation. Bioorganic Med. Chem. 2013;21:6171–6180. doi: 10.1016/j.bmc.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Helmling C., Bessi I., Wacker A., Schnorr K.A., Jonker H.R.A., Richter C., Wagner D., Kreibich M., Schwalbe H. Non-covalent spin labelling of ribiswitch RNAs to obtain long-range structural NMR restraints. Chem. Biol. 2014;10 doi: 10.1021/cb500050t. [DOI] [PubMed] [Google Scholar]
- 36.Nelissen F.H.T., van Gammeren A.J., Tessari M., Girard F.C., Heus H.A., Wijmenga S.S. Multiple segmental and selective isotope labelling of large RNA for NMR structural studies. Nucleic Acids Res. 2008;36:e89. doi: 10.1093/nar/gkn397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romaniuk P.J., Uhlenbeck O.C. Joining of RNA molecules with RNA ligase. Methods Enzymol. 1983;100:52–59. doi: 10.1016/0076-6879(83)00045-2. [DOI] [PubMed] [Google Scholar]
- 38.Bullard D.R., Bowater R.P. Direct comparison of nick-joining activity of the nucleic acids ligases from bacteriophage T4. Biochem. J. 2006;398:135–144. doi: 10.1042/BJ20060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Nikonowicz E.P. Solution structure of the K-turn and specifier loop domains from the Bacillus subtilis tyrS T-box leader RNA. J. Mol. Biol. 2011;408:99–117. doi: 10.1016/j.jmb.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kao C., Zheng M., Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 42.Wyatt J.R., Chastain M., Puglisi J.D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991;11:764–769. [PubMed] [Google Scholar]
- 43.Stoll S., Schweiger A. EasySpin, a comprehensive software package for spectral simulation and analysis in EPR. J. Magn. Reson. 2006;178:42–55. doi: 10.1016/j.jmr.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 44.Delaglio F., Grzesiek S., Vuister G.W., Zhu G., Pfeifer J., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–295. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Goddard T.D., Kneller D.G. SPARKY3. San Francisco: University of California; 2004. [Google Scholar]
- 46.Lebars I., Martinez-Zapien D., Durand A., Coutant J., Kieffer B., Dock-Bregeon A.C. HEXIM1 targets a repeated GAUC motif in the riboregulator of transcription 7SK and promotes base pair rearrangements. Nucleic Acids Res. 2010;38:7749–7763. doi: 10.1093/nar/gkq660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLano W.L. The PyMOL Molecular Graphics System. South San Francisco, CA, USA: Delano Scientific LLC; 2005. [Google Scholar]
- 48.Wu J., McLuckey S.A. Gas-phase fragmentation of oligonucleotides ions. Intl. J. Mass Spectroscopy. 2004;237:197–241. [Google Scholar]
- 49.Nguyen P., Qin P.Z. RNA dynamics: perspectives from spin labels. Wiley Interdiscip. Rev. 2012;3:62–72. doi: 10.1002/wrna.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.