Abstract
NONO, SFPQ and PSPC1 make up a family of proteins with diverse roles in transcription, RNA processing and DNA double-strand break (DSB) repair. To understand long-term effects of loss of NONO, we characterized murine embryonic fibroblasts (MEFs) from knockout mice. In the absence of genotoxic stress, wild-type and mutant MEFs showed similar growth rates and cell cycle distributions, and the mutants were only mildly radiosensitive. Further investigation showed that NONO deficiency led to upregulation of PSPC1, which replaced NONO in a stable complex with SFPQ. Knockdown of PSPC1 in a NONO-deficient background led to severe radiosensitivity and delayed resolution of DSB repair foci. The DNA-dependent protein kinase (DNA-PK) inhibitor, NU7741, sensitized wild-type and singly deficient MEFs, but had no additional effect on doubly deficient cells, suggesting that NONO/PSPC1 and DNA-PK function in the same pathway. We tested whether NONO and PSPC1 might also affect repair indirectly by influencing mRNA levels for other DSB repair genes. Of 12 genes tested, none were downregulated, and several were upregulated. Thus, NONO or related proteins are critical for DSB repair, NONO and PSPC1 are functional homologs with partially interchangeable functions and a compensatory response involving PSPC1 blunts the effect of NONO deficiency.
INTRODUCTION
SFPQ (PSF), NONO (p54nrb) and PSPC1 (paraspeckle component 1) make up a small family of proteins that are involved in both ribonucleic acid (RNA) synthesis and deoxyribonucleic acid (DNA) repair. The defining feature of the family is a conserved Drosophila behavior human splicing (DBHS) motif, which consists of tandem RNA recognition motif domains and about 170 adjacent amino residues (1). Each family member forms heterodimeric complexes with the other two (2–4), and a recent crystal structure reveals distinctive features of this interaction (5). A variety of biological functions in RNA biogenesis have been ascribed to the DBHS family members, including pre-messenger RNA (mRNA) nuclear retention (6–9), pre-mRNA 3′-end formation (10) and transcriptional activation and repression (for example (11–15); see also (16) for an early review). Functions in RNA metabolism are mediated, in part, through association with a long, noncoding RNA to form nuclear bodies known as paraspeckles (reviewed in (17)).
Surprisingly, SFPQ and NONO are involved in DNA repair as well as RNA biogenesis. The purified SFPQ–NONO complex stimulates end joining by up to 10-fold in a reconstituted system containing recombinant nonhomologous end-joining (NHEJ) proteins (18). Repair activity involves direct binding to DNA or chromatin and appears to occur via a distinct sub-pathway of nonhomologous end-joining repair (19–22). Separate work implicates SFPQ, but not NONO, in homologous recombination repair (23,24).
There have been two prior studies of the effect of germ line mutation on mammalian DBHS family members. One was in the mouse, which focused on the role of NONO as a transcriptional regulator (12), and the other was in the zebrafish, which showed that the SFPQ homolog (whitesnake) is required for development (25). Other genetic studies have used RNA knockdown methods to achieve transient attenuation of expression in mammalian cells. These have shown that attenuation of NONO and SFPQ function leads to partial double-strand break (DSB) repair deficiencies and sensitivity to DNA damage (21–23,26). PSPC1, the third protein in the family, has been investigated primarily in the context of its function in paraspeckles and is only indirectly implicated in the DNA damage response (21).
Here, we report the investigation of DSB repair and DNA damage in MEFs derived from NONO knockout mice. Prior studies with RNA-mediated NONO knockdown have shown only partial radiosensitivity and repair deficits, and it is unclear whether this is because NONO is not essential for repair, or alternatively because there is residual NONO expression in treated cells. The mouse knockout can be used to address this question because it appears to be null for NONO expression. The knockout also provides the opportunity to investigate long-term compensatory responses, which can provide important additional information that transient knockdown experiments do not afford. Initially, results showed that NONO-deficient MEFs and their wild-type counterparts have very similar growth rates and cell cycle distributions. Moreover, the NONO-deficient MEFs were only mildly radiosensitive. Further work showed, however, that the modest phenotype was attributable to upregulation of the third DBHS family member, PSPC1. Doubly deficient cells, which lacked NONO and had reduced levels of PSPC1, were markedly radiosensitive and showed a very significant increase in unresolved DNA DSB repair foci post-irradiation. Together, results suggest that DBHS proteins play a critical role in DNA repair under the conditions tested.
MATERIALS AND METHODS
Mouse strain and MEF cell derivation
Baygenomics embryonic stem cell clone YHA266 (distributed by the Mutant Mouse Regional Resource Centers, www.mmrrc.org) has a gene trap cassette at the Nono locus, which is on the X chromosome. The clone was sequence-verified and used to generate chimeric mice by standard methods. The strain was maintained by backcrossing heterozygous females to C57/Bl6 males. MEFs were derived as described (27) and propagated in Dulbecco's Minimal Essential Medium supplemented with 10% Fetal bovine serum (FBS) at 37ºC in a 3–5% O2, 5% CO2 atmosphere. Genotyping primers were: P1, d(GGGGGTGTTGAGTCTTGCTACG); P2: d(CTTTCCCAGTCACCCCTCCAGA); P3: d(GTTTGGGGCTTCTGTTTTCTCATT); P4 d(CCCTGGGGTTCGTGTCCTAC). Polymerase chain reaction (PCR) was performed by heating to 94°C for 2 min, followed by 30 cycles of 94ºC for 45 s, 60ºC for 60 s and 72ºC for 60 s.
RNA analysis in mouse tissue was performed as follows: tissues from different organs of 4-day-old new born mice, including the skin, brain, thymus, lung, heart, liver, stomach, intestine, kidney, spleen and testis, were collected. Total RNA was extracted using Trizol (Invitrogen) followed by reverse transcription at 42°C for 30 min. PCR primers were: Nono, d(GCCAGAGGCAGTCGAGGTTAGTG) and d(TTCAGGTCAATAGTCAAGCCTTCATTCT); β-geo, d(CCGGGCAACTCTCTGGCTCAC) and d(AGGCGGTCGGGATAGTTTTCTTG); β-actin, d(CAGTTCGCCATGGATGACGATAT) and d(ACATGATCTGGGTCATCTTTTCACGGTT). For the detection of Nono and β-actin RNA, PCR was performed by heating to 94°C for 4 min, followed by 32 cycles of 94ºC for 30 s, 56.5ºC for 30 s and 72ºC for 30 s. For the detection of β-geo expression, PCR was performed by heating to 94°C for 4 min, followed by 35 cycles of 94ºC for 30 s, 58ºC for 30 s and 72ºC for 30 s.
Immunoblotting, immunofluorescence and immunoprecipitation
Immunoblotting was performed using rabbit monoclonal anti-NONO (Epitomics, cat. # 3708-1, 1:5000), rabbit anti-SFPQ (Bethyl Laboratories, cat. # A301-321A, 1:2000 dilution), rabbit anti-PSPC1 (Bethyl Laboratories, cat. # A303-205-A, 1:2000 or Dundee Cell Products UK, cat. # AB1013, 1:500) or mouse anti-β-actin (Sigma, cat. # A5316, 1:1000) with horseradish peroxidase-conjugated goat-anti-mouse or anti-rabbit IgG secondary antibodies (GE Healthcare, cat. # NA 931 and NA 934V). Membranes were developed using Enhanced Chemiluminescence substrate (GE Healthcare) and immune complexes were visualized using X-ray film. Immunofluorescence was performed using anti-NONO (1:200), anti-PSPC1 (Bethyl, 1:1000), rabbit anti-phospho-H2AX (EMD Millipore, cat. # 07-164, 1:500), rabbit anti-53BP1 (Novus Biologicals, cat. #100-904, 1:500) and Alexa Fluor 594-conjugated goat anti-mouse or anti-rabbit antibodies (Invitrogen, cat. # A11037 or A150077, 1:500). Immunoprecipitation was performed using Dynabeads Protein G (Invitrogen, cat. # 100.04D) according to the vendor's protocol. Briefly, 1.2 mg of Dynabeads were incubated with 2 mg of the same antibodies as for immunoblotting or with control rabbit IgG (Sigma-Aldrich, cat. # I8140). After washing, beads were incubated with MEF protein extracts, washed, and immune complexes were resolved by sodium dodecyl sulphate-polyacrylamide gel electrophoresis and analyzed by immunoblotting.
Cell growth, flow cytometry and clonogenic survival assay
For growth curves, 3 x104 MEFs were seeded in 6-well plates. Triplicate samples for each time point were harvested daily for 9 days. Cell cycle distribution was measured in duplicate samples using flow cytometry with propidium iodide. Clonogenic survival assays were performed in T-25 flasks. Cells were seeded (1000–10 000/flask), allowed to attach and irradiated using a 137Cs source. Flasks were incubated for 10 days with two changes of medium, stained with 0.25% crystal violet, 3.7% formaldehyde in 80% methanol, and colonies of ≥50 cells were scored. The difference in survival between wild-type and NONO-deficient MEFs at each dose point was evaluated using Student's t-test.
microRNA treatment and green cell microcolony formation assay
A PSPC1-specific microRNA (miRNA) vector with linked EmGFP was constructed as in (21). The Pspc1 cDNA sequence (accession number BC026772) was analyzed using the BLOCK-iT RNAi designer tool (Life Technologies Corp., Grand Island, NY, USA). We prepared a double-stranded oligonucleotide representing the top scoring sequence (d(TGCTGATACGTACTTGCTTCAGGTTTGTTTTGGCCACTGACTGACAAACCTGACAAGTACGTAT) and its complement d(CCTGATACGTACTTGTCAGGTTTGTCAGTCAGTGGCCAAAACAAACCTGAAGCAAGTACGTATC)) and inserted it into linearized pcDNATM6.2-GW/EmGFP-miR vector (BLOCK-iT Pol II miR RNAi expression vector kit, Life Technologies Corp.). This or an empty control vector was introduced into MEFs by electroporation. Microcolony assays were performed essentially as described (28). MEFs (3 × 106) were suspended in 150 μl of MEF buffer 1 (MEF I Nucleofector kit, Lonza, cat. # VPD-1004), mixed with 15 μg of vector and subjected to electroporation using a Lonza Nucleofector 2b Device with program T-20. Cells were diluted into growth medium and plated at 0.25–1.0 × 105 cells/well in 6-well plates. After 2 days, the MEFs were irradiated with 0, 1, 2 or 4 Gy of γ-rays. At 5 days post-irradiation, the cells were washed with phosphate buffered saline and fixed with 4% paraformaldehyde for 15 min. For each experimental group, 200 green microcolonies were scored. Statistical analysis was performed using the 2-way repeated measures Analysis of Variance (ANOVA) method. Where noted, 2 μM of the DNA-dependent protein kinase (DNA-PK) inhibitor, NU7441 (Torcris Bioscience, cat. #3712) was added 4 h prior to irradiation and incubation was continued with the drug for the remainder of the experiment.
DNA repair foci assay
Control or PSPC1 miRNA was introduced by electroporation into wild-type or NONO-deficient MEFs as described above. Two days after electroporation, MEFs were exposed to 0 or 1 Gy of γ-rays. At 0.5 h or 4 h post-treatment, the cells were washed and fixed as above, then analyzed by indirect immunofluorescence using anti-γ-H2AX. A total of 30 GFP-positive nuclei were scored per experimental group. Data were analyzed by one-way ANOVA analysis, followed by Dunnett's Multiple Comparison Test. In separate experiments, cells were treated similarly and analyzed by indirect immunofluorescence using anti-53BP1 antibody.
Analysis of mRNA levels by quantitative PCR
MEFs were electroporated with control or PSPC1 miRNA vector. Total RNA was isolated at 48 h post-transfection using an RNeasy Mini Kit (QIAGEN Inc., Valencia, CA, USA) and reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Life Technologies). Taqman quantitative PCR assays were performed using the following probes (Life Technologies Corp.): Nono (Assay ID: Mm00834875_g1), Sfpq (Assay ID: Mm01179807_m1), Pspc1 (Assay ID: Mm00481804_m1), Prkdc (Assay ID: Mm01342967_m1), Xrcc5 (Assay ID: Mm00550142_m1), Xrcc6 (Assay ID: Mm00487458_m1), Lig4 (Assay ID: Mm01221720_m1), Nhej1 (Assay ID: Mm01259071_m1), Xrcc4 (Assay ID: Mm00459213_m1), Mre11a (Assay ID: Mm00450600_m1), Rad50 (Assay ID: Mm00485504_m1), Nbn (Assay ID: Mm00449854_m1), Rad51 (Assay ID: Mm00487905_m1), Rad51d (Assay ID: Rn01752219_m1) and internal control GAPDH (Assay ID: Mm99999915_g1). Reactions (25 μl) were performed using a StepOnePlus system (Life Technologies) with cycling parameters as follows: 50°C for 2 min, 95°C for 20 s, 40 cycles of 95°C for 1 s and 60°C for 20 s. Data were processed with StepOne Software V2.2.2 (Life Technologies) and analyzed using the ΔΔCt method (29). Data were analyzed by one-way ANOVA analysis, followed by Dunnett's Multiple Comparison Test.
RESULTS
Derivation and validation of NONO-deficient MEFs
The knockout mice used in these studies were genetically similar to those in a recent study (12) but were independently derived. They contain a gene trap insertion in the second intron in the X-linked Nono locus (Figure 1A). As the first two exons are noncoding, the predicted hybrid Nono- β-geo transcript does not contain any of the native Nono open reading frame. Mice were produced from the ES cells by standard techniques. DNA repair and radiosensitivity studies used mice that were backcrossed for >10 generations into the C57/Bl6 background.
Figure 1.
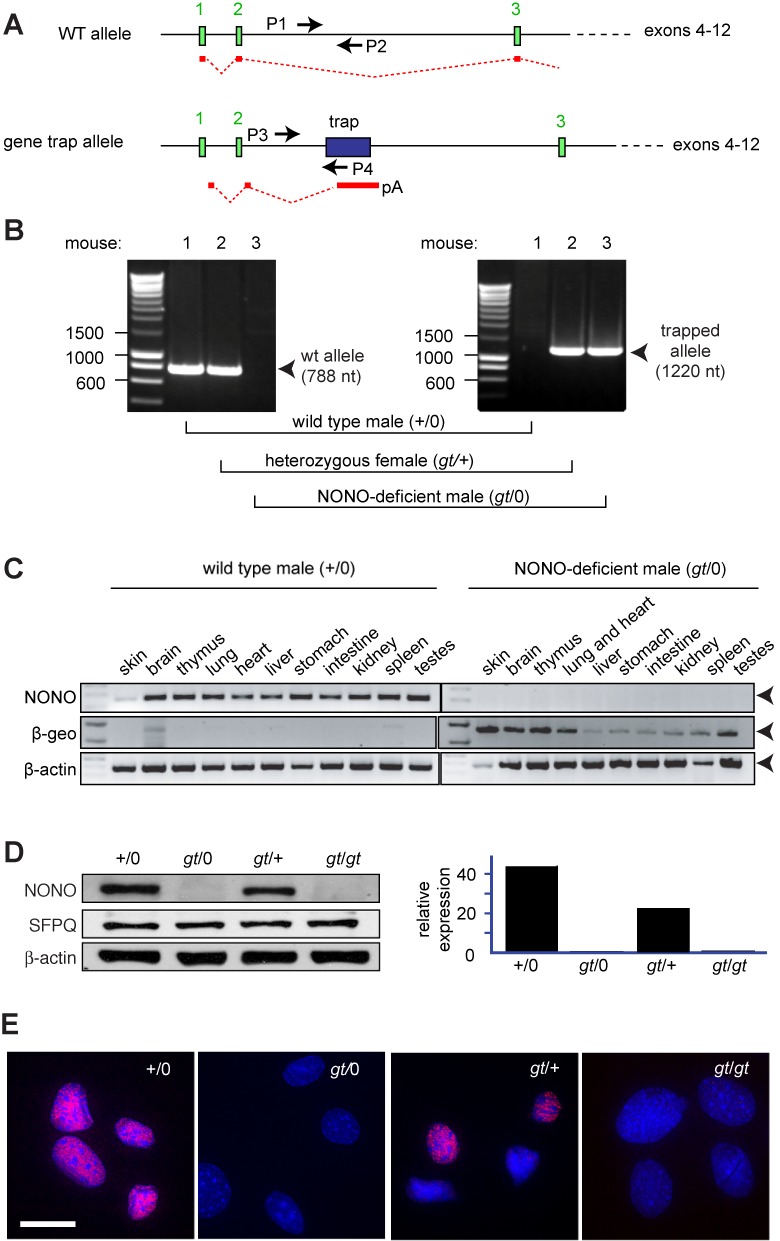
Derivation and validation of NONO-deficient MEFs. (A) Portion of mouse X chromosome depicting the Nono locus. Positions of gene trap within second intron and primers used for genotyping are shown. The mRNA product of the gene trap allele, which does not contain any of the Nono open reading frames, is shown in red. (B) Representative image showing genotyping by PCR. Left, analysis using primers P1 and P2 to detect wild-type allele; right, analysis using primers P3 and P4 to detect gene trap allele. In each panel, lane 1, wild-type male (+/0); lane 2, heterozygous female (gt/+); lane 3, Nono-gene trap male (gt/0). (C) Expression of NONO and β-geo gene trap mRNAs in tissues of mice sacrificed at postnatal day 4. Image shows PCR products with β-actin as a loading control. (D) NONO, SFPQ and internal reference protein (β-actin) expression in MEF isolates from mice bearing wild type (+) or gene trap (gt) alleles. Total cell lysates were probed with indicated antibodies. Left panel, gel image; right panel, quantification. (E) Immunostaining of the same four MEF isolates with anti-NONO antibody. Scale bar denotes 10 μm.
Representative genotyping, using PCR primers specific for the wild-type (wt) and gene trap (gt) allele, is shown in Figure 1B. Analysis of mRNA by reverse-transcriptase PCR indicated that the Nono transcript was undetectable in any tissue examined and that it was replaced by a β-geo transcript encoded by the gene trap cassette (Figure 1C). Hemizygous knockout males were smaller than wild-type littermates and bred poorly, particularly after they were fully backcrossed to the C57/Bl6 background (S. Li, unpublished data). The mice otherwise did not show gross defects.
Further studies were performed using MEFs derived from e13.5-day embryos. We derived MEFs independently from several embryos of each genotype. To minimize endogenous oxidative stress and DNA damage, we maintained the cells in a 3–5% oxygen atmosphere (30). Under these conditions, populations could be maintained for at least several weeks without the onset of senescence. Immunoblotting showed an essentially complete loss of NONO protein in MEF-derived hemizygous males (gt/0) or homozygous gene trap females (gt/gt). There was about a 50% decline in overall NONO protein expression in MEFs from heterozygous (gt/+) females, reflecting mosaic X chromosome inactivation (Figure 1D). Results were confirmed by immunofluorescence, which showed mosaic expression in MEFs from heterozygous females and no expression in MEFs from hemizygous males or homozygous-deficient females (Figure 1E).
Effect of NONO deficiency on cell growth and radiosensitivity
We compared the growth of wild-type and NONO gene trap MEFs, using cells isolated separately from three different embryos of each genotype. Although there were small differences between independently derived cell populations, there was no consistent effect of genotype on growth rate (Figure 2A). We chose two isolates that had approximately equal growth rates under 3% oxygen conditions (WT-2 and gt-2) and tested their growth in atmospheric (21%) oxygen. Although all cells grew more slowly in 21% oxygen, both genotypes were affected approximately equally (Supplementary Figure S1). We also compared the cell cycle distribution in WT-2 and gt-2 MEFs. The distributions were similar (Figure 2B and Supplementary Figure S1).
Figure 2.
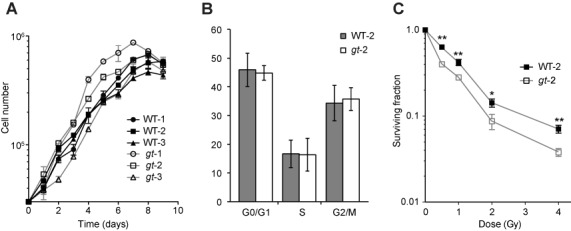
Effect of NONO deficiency on cell growth and radiosensitivity. (A) Growth curves for six MEF populations derived from different embryos of indicated genotypes. Cells were seeded at 3 × 104 cells per well in 6-well plates and individual wells were harvested and counted daily. Each point represents the mean of triplicate cultures. Error bars denote standard deviation. Genotypes are indicated. (B) Cell cycle distribution of WT-2 and gt-2 populations determined by flow cytometry using propidium iodide staining. Data reflect mean and standard deviations from three independent experiments. (C) Clonogenic survival assays performed as described in the Materials and Methods section using indicated MEF populations. Each point represents pooled data from two independent experiments, each performed in triplicate. Data were normalized to the surviving fraction in non-irradiated control cells in each experiment. Mean and standard deviations are shown. Doses of 137Cs gamma rays are indicated. (*P < 0.05 and **P < 0.01).
To evaluate radiosensitivity, we performed clonogenic survival assays. The gt-2 MEFs were significantly more radiosensitive than wild type, although the difference was ≤3-fold at all doses tested. The concave shape of the survival curves differs from the linear-quadratic survival curves typically seen with human adult fibroblasts and reflects greater radiosensitivity in the 0.5–2-Gy range. At this time, we do not have an explanation as to why the curves do not conform to a linear-quadratic model, but we note that the concave shape of the curves was genotype independent.
Increased levels of PSPC1 and formation of SFPQ–PSPC1 complex in NONO-deficient cells
One hypothesis to account for the mildness of the radiosensitivity observed in NONO-deficient MEFs is that compensatory upregulation other genes might have occurred, either in the course of mouse development or subsequently in culture. One candidate was PSPC1, because of its extensive homology with NONO.
We investigated PSPC1 expression by immunofluorescence and immunoblotting using anti-PSPC1 antibody (Figure 3A and B). PSPC1 was present at low levels, near the limit of detection, in wild-type MEFs (+/0). Expression was strongly increased in NONO-deficient MEFs (gt/0 and gt/gt). Thus, it appears there is a reciprocal relationship between loss of NONO and gain of PSPC1 expression.
Figure 3.
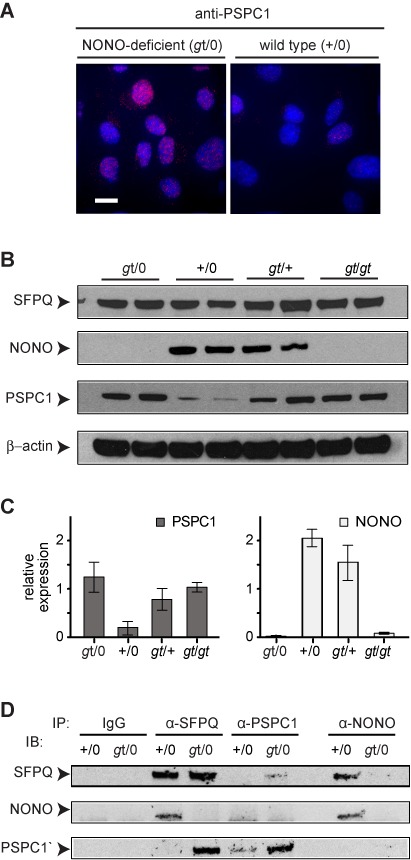
Increased levels of PSPC1 and SFPQ–PSPC1 complex in NONO-deficient cells. (A) Immunostaining of WT-2 and gt-2 MEF isolates using anti-PSPC1 antibody. Scale bar, 10 μm. (B) Immunoblotting to determine levels of SFPQ, NONO and PSPC1 proteins in cells of indicated genotype. Two independent MEF populations, derived from different embryos, were analyzed for each type. Arrowheads denote proteins as indicated. (C) Quantification of data from panel (B). Values are normalized to β-actin. Error bars reflect standard deviation of values from independent MEF populations. (D) Immunoprecipitation (IP), followed by immunoblotting (IB), to protein–protein complexes in MEFs of indicated genotypes.
The situation is a bit more complicated with the MEFs derived from heterozygous females (gt/+). Because of random X-chromosome inactivation, these populations are mixtures of phenotypically wild-type and NONO-deficient cells, and the proportion may vary between embryos. It appears that there is more NONO and less PSPC1 in one of the (gt/+) samples and that the reverse is true in the other, likely reflecting this mosaicism. In both samples, there is somewhat more PSPC1 than can be accounted for by loss of NONO, and we cannot rule out the possibility that cross-talk between cells in the mixed population influences PSPC1 expression.
Reciprocal co-immunoprecipitation showed that the overexpressed PSPC1 formed stable complexes with SFPQ (Figure 3C). We interpret the upregulation of PSPC1 and incorporation into complexes with SFPQ in terms of selective pressure arising from loss of NONO, and we suggest that this phenomenon arises either during mouse development or during subsequent growth of the MEFs.
Effect of attenuation of PSPC1 expression on radiosensitivity
To investigate whether the spontaneous upregulation of PSPC1 expression in NONO-deficient MEFs blunts the DNA damage sensitivity phenotype, we used an MEF-specific electroporation protocol to introduce PSPC1 or control miRNA vectors. We estimated the transfection efficiency to be 30–40%, based on co-expression of an Emerald Green Fluorescent Protein (EmGFP) marker encoded by the miRNA vector. Cells expressing EmGFP showed essentially complete loss of PSPC1 expression as determined by immunofluorescence (Figure 4A).
Figure 4.
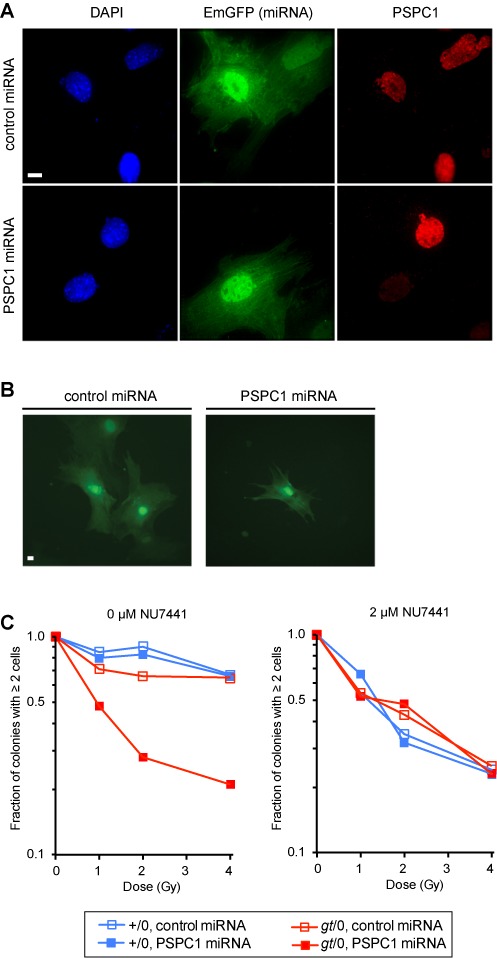
Green cell microcolony formation assay. (A) Verification of miRNA-mediated PSPC1 knockdown. Vectors encoding control or PSPC1 miRNA were introduced by electroporation into NONO-deficient (gt/0) MEFs. A linked EmGFP gene serves as a marker for miRNA expression. Cells were fixed and analyzed by indirect immunofluorescence using anti-PSPC1 primary and red secondary antibodies. Cells were counterstained for DNA with DAPI. Each row depicts blue (DAPI), green (EmGFP) and red (anti-PSPC1) channels for the same field. Scale bar, 10 μm. (B) Representative images of multi-cell and 1-cell microcolonies after 5 days. Scale bar, 10 μm. (C) Clonogenic survival. Colonies of ≥2 cells were expressed as a proportion of total colonies. Data are normalized to fraction of colonies with ≥2 cells in non-irradiated control populations, which did not vary significantly with genotype. For each genotype and radiation dose, 200 green microcolonies were scored. Left panel, microcolony formation in the absence of DNA-PK inhibitor; right panel, microcolony formation in the presence of 2-μM NU7441.
Because only a fraction of the population expressed the transfection marker, we turned to single-cell assays, which allowed us to investigate the behavior of EmGFP-positive cells, rather than the total population. We used a microcolony assay, which measures the first 1–3 cell divisions following plating. This is an established and validated assay for clonogenic growth (28,31) and is particularly suited for following marked cells in a mixed population. The microcolony assay does, however, tend to give somewhat higher surviving fractions than a classic colony formation assay, presumably because some cells that form microcolonies would not go on to form colonies of ≥50 cells.
For the assays, cells were transfected, plated and irradiated, and the size distribution of green cell clusters was analyzed at 5 days post-irradiation. Reproductively inactivated cells remain as isolated cells on the plate, whereas surviving cells form microcolonies. Results are scored as the fraction of microcolonies of ≥2 cells relative to the total (EmGFP-expressing microcolonies plus single EmGFP-expressing cells). Examples of a microcolony of green cells, versus a single non-dividing green cell, are shown in Figure 4B.
We compared four experimental groups for radiation sensitivity using the microcolony assay (wild-type or NONO-deficient, transfected with control or PSPC1 miRNAs) (Figure 4C). Single deficiency in NONO had only a modest effect on survival, as expected based on results of the conventional clonogenic survival assay in Figure 2. Treatment of wild-type MEFs with PSPC1 also had little effect, consistent with the fact that wild-type cells express very little of this protein. However, treatment of NONO-deficient MEFs with PSPC1 miRNA had a strong radiosensitizing effect, based on the small fraction of miRNA positive microcolonies. The results presented here are for microcolonies of ≥2 cells. The doubly deficient cells were even more radiosensitive when colonies of ≥3 cells were scored (Supplementary Figure S2), although relative radiosensitivity, comparing the different groups, was the same. We conclude that PSPC1 and NONO functionally substitute for one another in a pathway that is essential for radiation survival.
It was of interest to determine the relationship between this pathway and the canonical, DNA-PK-dependent pathway of NHEJ. To investigate this, we performed the same experiment in the presence of the DNA-PK inhibitor, NU7441. The inhibitor led to radiosensitization of the wild-type and singly deficient cells, but had no further effect on the already-radiosensitive NONO/PSPC1 doubly deficient cells. Results suggest that NONO/PSPC1 and DNA-PK are in the same epistasis group and thus function in the same pathway.
Effect of attenuation of PSPC1 expression on resolution of DSB repair foci
To determine whether the radiosensitivity arose from a deficit in DSB repair per se, we again used a single-cell assay. We transfected with miRNA, irradiated and scored γ-H2AX foci, a marker of unrepaired DSBs, in cells expressing the EmGFP transfection marker. We irradiated at 0 or 1 Gy, then scored cells in various treatment groups at 0.5 h post-irradiation, to examine the ability to form γ-H2AX foci, and at 4 h post-irradiation, to examine the ability to recover. There was a low background of spontaneous foci in the non-irradiated cells (Figure 5). This increased to about 30 foci/per cell at 30 min following treatment with 1 Gy of 137Cs γ-rays, consistent with the predicted number of DSBs at this dose assuming a diploid mouse genome. The increase was similar in all four experimental groups. After 4 hours, most of the foci resolved in the wild-type genetic background (with or without PSPC1 knockdown). Most of the foci also resolved in the NONO-deficient cells transfected with control miRNA. However, dual deficiency in NONO and PSPC1 resulted in near absence of recovery at the 4-h time point. Taken together, results indicate that deficiency in NONO/PSPC1 function had no effect on the formation of γ-H2AX foci, which is a very early step in the cascade of events involved in DSB repair. Deficiency did affect the resolution of γ-H2AX foci, which is a very late step.
Figure 5.
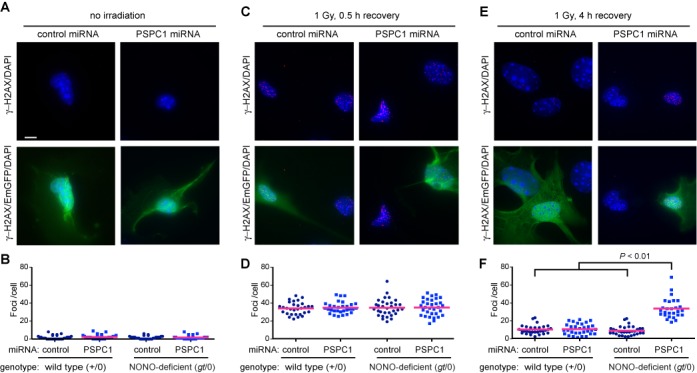
Delayed resolution of repair foci. Vectors encoding control or PSPC1 miRNA were introduced by electroporation into wild-type (+/0) or NONO-deficient (gt/0) MEFs. Cells were mock irradiated or exposed to 1 Gy of 137Cs γ-rays. At indicated times following irradiation, cells were fixed and analyzed by indirect immunofluorescence using anti-γ-H2AX primary and red secondary antibody and DAPI counterstain. (A, C, E) Representative fields showing γ-H2AX/DAPI channels (top row) or γ-H2AX/EmGFP/DAPI-merged images in NONO-deficient (gt/0) MEFs. Images were collected for non-irradiated control cells or for cells that were irradiated and allowed to recover for indicated times. Cell morphology and foci appearance were indistinguishable in wild-type (+/0) and NONO-deficient (gt/0) MEFs; only the latter are shown in the figure. Scale bar denotes 10 μm. (B, D, F) Foci per cell in populations corresponding to panels (A, C, E). A total of 30 nuclei were scored per experimental group. Blue symbols depict score for individual cells; red bar depicts mean. Treatment of NONO-deficient cells with PSPC1 RNA resulted in a significant increase in residual foci at 4 h post-irradiation (P < 0.01).
To narrow down the stage at which NONO or PSPC1 is involved, it was of interest to investigate the formation of 53BP1 foci, which form after γ-H2AX foci, but before DNA end joining. Also, NONO and PSPC1 are RNA-binding proteins, and there has been a report that small, DSB-induced RNAs (diRNAs) are required for 53BP1 foci formation (32), raising the possibility that NONO and PSPC1 are effectors of diRNA function. We performed immunostaining for 53BP1 foci in wild-type, NONO-deficient, PSPC1-deficient and doubly deficient cells. We observed no differences in 53BP1 foci induction at 30 min post-irradiation (Supplementary Figure S3), indicating that NONO- and PSPC1-containing complexes function at a stage of repair that occurs after 53BP1 foci formation. This is consistent with prior biochemical experiments showing that purified SFPQ–NONO complexes promote DNA end joining in a reconstituted system containing no chromatin proteins or RNA (18,19).
Effect of attenuation of PSPC1 on the expression of other DSB repair genes
PSPC1, NONO and SFPQ are multi-functional proteins that have been implicated in many steps of RNA synthesis and processing. Although dual PSPC1, NONO deficiency affects DSB repair, it was important to investigate whether this might occur indirectly, by decreasing levels of mRNAs encoding other repair proteins. To investigate this, we measured the relative mRNA levels for 11 other genes chosen to represent the canonical nonhomologous end-joining pathway and the two pathways that require end resection (alternative-NHEJ and classical homologous recombination).
Results, shown in Figure 6, indicate that the single greatest effect of NONO deficiency was upregulation of PSPC1, consistent with observations at the protein level. The upregulation of PSPC1 was partially reversed by transfection with PSPC1 miRNA, although suppression is incomplete, likely because the population consists of a mixture of transfected and non-transfected cells. There was also a smaller, but significant, upregulation of SFPQ mRNA, although these had not been seen at the protein level in Figure 3.
Figure 6.
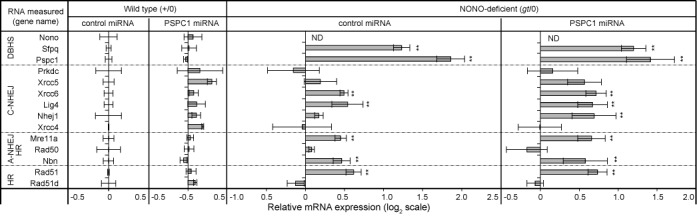
Relative mRNA expression for other DSB repair genes. Vectors encoding control or PSPC1 miRNA were introduced by electroporation into wild-type (+/0) or NONO-deficient (gt/0) MEFs. Expression was measured using Taqman probes with Gapdh as an internal reference. Data are normalized to expression in control miRNA-transfected wild-type MEFs. Values are mean of three independent biological replicates with standard deviation shown. Genes are grouped according to the presence of a DBHS motif, participation in canonical NHEJ (C-NHEJ), participation in alternative NHEJ (A-NHEJ) or participation in homology-directed repair (HR), respectively (**P < 0.01 and *P < 0.05).
Interestingly, five other mRNAs encoding repair factors also showed increases in the NONO-deficient background. These included Xrcc6 (which encodes the 70-kDa Ku subunit), Lig4 (which encodes DNA ligase IV), Mre11a and Nbn (which encode subunits of the MRE11•RAD50•NBS1 complex) and Rad51 (which encodes RAD51, a protein central to homologous recombination). All of these mRNAs showed small additional increases in NONO-deficient MEFs transfected with PSPC1 miRNA. Importantly, there were no significant declines in any of the repair-related mRNAs tested, and the results thus lend no support to the model NONO and PSPC1 affect repair by an indirect mechanism involving repression of other genes at the RNA level.
DISCUSSION
Here we characterize the effect of genetic insufficiency for NONO protein in mouse cells. Prior work, based primarily on knockdown studies in cell culture, suggests roles of NONO and its partners in RNA processing and transport, transcriptional regulation and DNA DSB repair. Despite the many functions ascribed to NONO, knockout mice develop normally and are viable as adults. We show here that, in the absence of genotoxic stress, NONO-deficient fibroblasts grow normally and have a cell cycle distribution similar to that of wild type. They are somewhat sensitive to radiation. This modest phenotype is attributable to compensatory changes in regulation of other genes, notably overexpression of PSPC1. When PSPC1 expression is attenuated, the cells become markedly radiosensitive and show delayed DNA DSB repair. Together, results show that NONO or related proteins play a critical role in DSB repair under the conditions tested.
In principle, there are several mechanisms by which a deficiency in a multifunctional RNA- and DNA-binding protein could influence radiation sensitivity and delay DSB repair. Prior studies favor a model where NONO protein interacts with the repair substrate, based on the ability of the SFPQ and NONO complex to relocalize to damaged DNA sites in cells (20–22) and to bind directly to DNA and stimulate end joining in vitro (18,19). The results of the NU7441 experiment in Figure 4, which showed that NONO/PSPC1 and DNA-PK are epistatic, also favor the model where NONO and its partners interact directly with the repair substrate.
We saw no evidence for an alternative mechanism where NONO deficiency resulted in lower expression of other genes involved directly in nonhomologous end joining or homologous recombination; indeed a number of genes showed modestly elevated expression consistent with a compensatory response. Although the list of genes measured is not exhaustive, our results are consistent with a previous study that showed no effect of attenuation of SFPQ expression on levels of DNA repair gene expression (23).
Another possibility, which is not mutually exclusive with a direct role in repair, is that NONO influences cell cycle checkpoints via a transcriptional mechanism. There has been a report that NONO cooperates with the Per gene product to regulate p16-INK4A transcription, coupling circadian rhythm to cell cycle control (12). However, under the conditions of our experiments, NONO deficiency did not affect cell cycle distribution, either under low oxygen conditions, where growth is optimal (Figure 1), or at atmospheric oxygen concentrations (Supplementary Figure S1), which are known to promote accumulation of DNA damage in MEFs (30). The reduced number of multicellular colonies formed following irradiation in Figure 4 is further evidence against a loss of DNA-damage-dependent cell cycle checkpoint control. Our data do not, however, exclude an effect of NONO on cell cycle control under other conditions or in other cell types, and further investigation may be warranted.
NONO and related proteins in the DBHS family are of particular interest because they provide a connection between RNA and DNA metabolism. There has been a recent surge of interest in the role of RNAs and RNA-binding proteins in the DNA damage response (reviewed in (33,34)). It appears that RNAs may influence repair by several mechanisms, some involving R-loops (35–38) and others involving small, DICER-dependent RNAs (32,39–41). Prior reports describe retroelement and human noncoding RNAs that bind to isolated SFPQ in vitro and promote release of SFPQ from transcriptional promoter DNA (42,43). It will be of interest to determine if these or other RNAs also interact with SFPQ–NONO or SFPQ–PSPC1 complexes and modulate their repair activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Ms Farlyn Hudson of Georgia Regents University for expert technical assistance and Dr Guanghu Wang of Georgia Regents University for advice on the derivation of MEF cells.
Footnotes
Present address: Andrew C. Phillips, Abbvie, North Chicago, IL 60064-6098, USA.
FUNDING
National Institutes of Health [2 R01 CA98239]. W. Dynan also received support as an Eminent Scholar of the Georgia Research Alliance. Funding for open access charge: National Institutes of Health [2 R01 CA98239].
Conflict of interest statement. None declared.
REFERENCES
- 1.Dong B., Horowitz D.S., Kobayashi R., Krainer A.R. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng R., Dye B.T., Perez I., Barnard D.C., Thompson A.B., Patton J.G. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA. 2002;8:1334–1347. doi: 10.1017/s1355838202022070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox A.H., Bond C.S., Lamond A.I. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol. Biol. Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuwahara S., Ikei A., Taguchi Y., Tabuchi Y., Fujimoto N., Obinata M., Uesugi S., Kurihara Y. PSPC1, NONO, and SFPQ are expressed in mouse Sertoli cells and may function as coregulators of androgen receptor-mediated transcription. Biol. Reprod. 2006;75:352–359. doi: 10.1095/biolreprod.106.051136. [DOI] [PubMed] [Google Scholar]
- 5.Passon D.M., Lee M., Rackham O., Stanley W.A., Sadowska A., Filipovska A., Fox A.H., Bond C.S. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl Acad. Sci. U.S.A. 2012;109:4846–4850. doi: 10.1073/pnas.1120792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L.L., Carmichael G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prasanth K.V., Prasanth S.G., Xuan Z., Hearn S., Freier S.M., Bennett C.F., Zhang M.Q., Spector D.L. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Z., Carmichael G.G. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 9.Zolotukhin A.S., Michalowski D., Bear J., Smulevitch S.V., Traish A.M., Peng R., Patton J., Shatsky I.N., Felber B.K. PSF acts through the human immunodeficiency virus type 1 mRNA instability elements to regulate virus expression. Mol. Cell. Biol. 2003;23:6618–6630. doi: 10.1128/MCB.23.18.6618-6630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaneko S., Rozenblatt-Rosen O., Meyerson M., Manley J.L. The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3’ processing and transcription termination. Genes Dev. 2007;21:1779–1789. doi: 10.1101/gad.1565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura K., Imamachi N., Akizuki G., Kumakura M., Kawaguchi A., Nagata K., Kato A., Kawaguchi Y., Sato H., Yoneda M., et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell. 2014;53:393–406. doi: 10.1016/j.molcel.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Kowalska E., Ripperger J.A., Hoegger D.C., Bruegger P., Buch T., Birchler T., Mueller A., Albrecht U., Contaldo C., Brown S.A. NONO couples the circadian clock to the cell cycle. Proc. Natl Acad. Sci. U.S.A. 2013;110:1592–1599. doi: 10.1073/pnas.1213317110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roepcke S., Stahlberg S., Klein H., Schulz M.H., Theobald L., Gohlke S., Vingron M., Walther D.J. A tandem sequence motif acts as a distance-dependent enhancer in a set of genes involved in translation by binding the proteins NonO and SFPQ. BMC Genomics. 2011;12:624. doi: 10.1186/1471-2164-12-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G., Cui Y., Zhang G., Garen A., Song X. Regulation of proto-oncogene transcription, cell proliferation, and tumorigenesis in mice by PSF protein and a VL30 noncoding RNA. Proc. Natl Acad. Sci. U.S.A. 2009;106:16794–16798. doi: 10.1073/pnas.0909022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong X., Sweet J., Challis J.R., Brown T., Lye S.J. Transcriptional activity of androgen receptor is modulated by two RNA splicing factors, PSF and p54nrb. Mol. Cell. Biol. 2007;27:4863–4875. doi: 10.1128/MCB.02144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shav-Tal Y., Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett. 2002;531:109–114. doi: 10.1016/s0014-5793(02)03447-6. [DOI] [PubMed] [Google Scholar]
- 17.Bond C.S., Fox A.H. Paraspeckles: nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bladen C.L., Udayakumar D., Takeda Y., Dynan W.S. Identification of the polypyrimidine tract binding protein-associated splicing factor•p54(nrb) complex as a candidate DNA double-strand break rejoining factor. J. Biol. Chem. 2005;280:5205–5210. doi: 10.1074/jbc.M412758200. [DOI] [PubMed] [Google Scholar]
- 19.Udayakumar D., Bladen C.L., Hudson F.Z., Dynan W.S. Distinct pathways of nonhomologous end joining that are differentially regulated by DNA-dependent protein kinase mediated phosphorylation. J. Biol. Chem. 2003;278:41631–41635. doi: 10.1074/jbc.M306470200. [DOI] [PubMed] [Google Scholar]
- 20.Salton M., Lerenthal Y., Wang S.Y., Chen D.J., Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- 21.Ha K., Takeda Y., Dynan W.S. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair (Amst) 2011;10:252–259. doi: 10.1016/j.dnarep.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krietsch J., Caron M.C., Gagne J.P., Ethier C., Vignard J., Vincent M., Rouleau M., Hendzel M.J., Poirier G.G., Masson J.Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–10301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajesh C., Baker D.K., Pierce A.J., Pittman D.L. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011;39:132–145. doi: 10.1093/nar/gkq738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morozumi Y., Takizawa Y., Takaku M., Kurumizaka H. Human PSF binds to RAD51 and modulates its homologous-pairing and strand-exchange activities. Nucleic Acids Res. 2009;37:4296–4307. doi: 10.1093/nar/gkp298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowery L.A., Rubin J., Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev. Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 26.Li S., Kuhne W.W., Kulharya A., Hudson F.Z., Ha K., Cao Z., Dynan W.S. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009;37:6746–6753. doi: 10.1093/nar/gkp741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garfield A.S. Derivation of primary mouse embryonic fibroblast (PMEF) cultures. Methods Mol Biol. 2010;633:19–27. doi: 10.1007/978-1-59745-019-5_2. [DOI] [PubMed] [Google Scholar]
- 28.Mo X., Dynan W.S. Subnuclear localization of Ku protein: functional association with RNA polymerase II elongation sites. Mol. Cell. Biol. 2002;22:8088–8099. doi: 10.1128/MCB.22.22.8088-8099.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Parrinello S., Samper E., Krtolica A., Goldstein J., Melov S., Campisi J. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 2003;5:741–747. doi: 10.1038/ncb1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timchenko N.A., Wilde M., Nakanishi M., Smith J.R., Darlington G.J. CCAAT/enhancer-binding protein alpha (C/EBP alpha) inhibits cell proliferation through the p21 (WAF-1/CIP-1/SDI-1) protein. Genes Dev. 1996;10:804–815. doi: 10.1101/gad.10.7.804. [DOI] [PubMed] [Google Scholar]
- 32.Francia S., Michelini F., Saxena A., Tang D., de Hoon M., Anelli V., Mione M., Carninci P., d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Montecucco A., Biamonti G. Pre-mRNA processing factors meet the DNA damage response. Front. Genet. 2013;4:102. doi: 10.3389/fgene.2013.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dutertre M., Lambert S., Carreira A., Amor-Gueret M., Vagner S. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem. Sci. 2014;39:141–149. doi: 10.1016/j.tibs.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Stirling P.C., Chan Y.A., Minaker S.W., Aristizabal M.J., Barrett I., Sipahimalani P., Kobor M.S., Hieter P. R-loop-mediated genome instability in mRNA cleavage and polyadenylation mutants. Genes Dev. 2012;26:163–175. doi: 10.1101/gad.179721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamperl S., Cimprich K.A. The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability. DNA Repair (Amst) 2014;19:84–94. doi: 10.1016/j.dnarep.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos-Pereira J.M., Herrero A.B., Moreno S., Aguilera A. Npl3, a new link between RNA-binding proteins and the maintenance of genome integrity. Cell Cycle. 2014;13:1524–1529. doi: 10.4161/cc.28708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morales J.C., Richard P., Rommel A., Fattah F.J., Motea E.A., Patidar P.L., Xiao L., Leskov K., Wu S.Y., Hittelman W.N., et al. Kub5-Hera, the human Rtt103 homolog, plays dual functional roles in transcription termination and DNA repair. Nucleic Acids Res. 2014;42:4996–5006. doi: 10.1093/nar/gku160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michalik K.M., Bottcher R., Forstemann K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012;40:9596–9603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wei W., Ba Z., Gao M., Wu Y., Ma Y., Amiard S., White C.I., Rendtlew Danielsen J.M., Yang Y.G., Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Yamanaka S., Siomi H. diRNA-Ago2-RAD51 complexes at double-strand break sites. Cell Res. 2014;24:511–512. doi: 10.1038/cr.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song X., Sui A., Garen A. Binding of mouse VL30 retrotransposon RNA to PSF protein induces genes repressed by PSF: effects on steroidogenesis and oncogenesis. Proc. Natl Acad. Sci. U.S.A. 2004;101:621–626. doi: 10.1073/pnas.0307794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li L., Feng T., Lian Y., Zhang G., Garen A., Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl Acad. Sci. U.S.A. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


