Abstract
Stop codon readthrough may be promoted by the nucleotide environment or drugs. In such cases, ribosomes incorporate a natural suppressor tRNA at the stop codon, leading to the continuation of translation in the same reading frame until the next stop codon and resulting in the expression of a protein with a new potential function. However, the identity of the natural suppressor tRNAs involved in stop codon readthrough remains unclear, precluding identification of the amino acids incorporated at the stop position. We established an in vivo reporter system for identifying the amino acids incorporated at the stop codon, by mass spectrometry in the yeast Saccharomyces cerevisiae. We found that glutamine, tyrosine and lysine were inserted at UAA and UAG codons, whereas tryptophan, cysteine and arginine were inserted at UGA codon. The 5′ nucleotide context of the stop codon had no impact on the identity or proportion of amino acids incorporated by readthrough. We also found that two different glutamine tRNAGln were used to insert glutamine at UAA and UAG codons. This work constitutes the first systematic analysis of the amino acids incorporated at stop codons, providing important new insights into the decoding rules used by the ribosome to read the genetic code.
INTRODUCTION
Translation corresponds to the decoding of mRNA by the ribosome. It has four stages: initiation, elongation, termination and recycling. Translation termination occurs when a stop codon enters the A site of the ribosome and is recognized by a complex of two factors: eRF1, which interacts directly with the stop codon (1,2), and eRF3, a ribosome-dependent GTPase that stimulates eRF1 in the presence of GTP (3,4). This termination complex mediates the hydrolysis of P-site peptidyl-tRNA in the peptidyl-transferase center (PTC) of the 60S subunit (5). The recycling of post-termination complexes is mediated by ABCE1, a conserved, essential member of the ATP-binding cassette (ABC) family of proteins (6). Translation termination is an efficient process, essential for the correct expression of proteins.
Termination efficiency can be influenced by a number of factors, including the nucleotide context of the stop codon (7), the identity of the last two amino acids incorporated into the polypeptide chain (8), the P-site tRNA (9) and the presence of stimulatory elements downstream from the stop codon (10,11). These elements can greatly increase the probability of a stop codon being decoded by a tRNA rather than a release factor, leading to the ribosome synthesizing an elongated protein with potentially different biochemical properties. This event is called readthrough and corresponds to the incorporation of a near-cognate tRNA, or natural suppressor, at the stop codon, allowing translation to continue in the same frame until the ribosome reaches the next stop. This tRNA may specify the insertion of an unusual amino acid, such as selenocysteine at UGA codons (12) or pyrrolysine at UAG codons (13,14). However, in most cases, the misreading of termination codons involves various normal cellular tRNAs primary used during the reading of their cognate sense codons. The recognition of stop codons by these naturally occurring suppressor tRNAs requires unconventional base pairing between the codon and anticodon. In addition to anticodon–codon affinity, a number of intrinsic features of the suppressor tRNA contribute to the ability to read non-cognate codons. These features include the degree of base modification within the anticodon or in the vicinity of the anticodon likely to increase or decrease the efficiency of misreading (15,16).
Stop codon readthrough was first detected in Escherichia coli infected with RNA phage Qβ and it was shown that tRNATrp 3′ACC5′ stimulated the readthrough of a UGA codon at the end of the coat protein cistron that is essential for the formation of infective particles (17–19). Most of what we know about the residues inserted by readthrough at stop codons in eukaryotes or about the natural suppressor tRNAs involved in this process was gleaned from in vitro experiments in rabbit reticulocyte lysate (RRL) (20). For example, tryptophan, cysteine and arginine were identified at UGA stop codons (21) and some of these results were confirmed by the identification of the corresponding tRNAs as UGA stop codon suppressors (22,23). It has also been shown that glutamine residues may be inserted at UAA and UAG stop codons (21). The available in vivo data for yeast suppressor tRNAs concern only the two glutamine tRNAs with UUG and CUG anticodons identified as UAA and UAG suppressors, respectively (24–27). Fearon et al. has also reported the insertion of tyrosine, lysine and tryptophan at UAG stop codons in yeast Ste6p (28).
Most of the available data concerning the nature of the amino acids inserted by readthrough were obtained from in vitro studies on plant tRNAs and viral mRNAs. Very few in vivo data are available, mostly for rabbit and yeast, and these data are not always consistent with the results obtained in vitro. For example, Fearon et al. did not find the same amino acids inserted at UAG codons as Feng et al. (21,28). Moreover, the relative quantification of readthrough amino acids was never addressed. With such a diversity of approaches and of organisms studied, and the absence of a systematic survey in a single defined organism, it is currently impossible to predict which amino acids are likely to be incorporated during stop codon readthrough. The nature of the readthrough amino acids and the efficiency with which they are incorporated could strongly influence the function of proteins translated from the growing number of genes known to use programmed stop codon readthrough (29,30). This is also important for the development of medical premature termination codon (PTC) suppression strategies (31). Indeed, the restoration of protein activity depends strongly on the identity of the amino acid inserted during PTC readthrough. We decided to develop a new strategy for the systematic study of amino-acid insertion at all stop codons in a single eukaryotic organism. We set up an in vivo reporter system for the production and purification of readthrough proteins in the yeast Saccharomyces cerevisiae. For efficient identification and quantification of the amino acids incorporated at stop codons, we chose to analyze readthrough proteins by high-resolution mass spectrometry (MS), which has the potential to provide the sensitivity, accuracy and robustness required for reliable peptide sequencing and a high degree of confidence in quantification data.
By reading the yeast genetic code it is possible to predict 6, 7 and 4 natural suppressors tRNAs (i.e. unmutated tRNAs which can form at least 2 out of 3 pairing with the stop codons, also named near-cognate tRNAs) for UAA, UAG and UGA, respectively (Supplementary Figure S1). Interestingly, we showed that only a subset of these tRNAs incorporated at the various stop codons. We found that tyrosine, glutamine and lysine were incorporated at UAA and UAG codons, whereas tryptophan, cysteine and arginine were incorporated at UGA codon. The nucleotide context around the stop codon is known to be a major determinant of stop codon readthrough efficiency (7). It is possible that the nucleotide context exerts its effects by modifying the ability of some natural suppressor tRNAs to decode stop codons. We therefore tested several nucleotide contexts, but we detected no influence on the identity of the amino acids inserted at stop codons or on the efficiency of incorporation. We observed quantitative differences in the efficiencies of incorporation for tyrosine, glutamine and lysine between UAA and UAG stop codons. We showed that these differences were due to the preferential use of the two 3′GUU5′ and 3′GUC5′ Gln isoacceptor tRNAs for the decoding of UAA and UAG stop codons, respectively. These results provide new information about codon–anticodon pairing, making it possible to better understand the rules governing decoding.
MATERIALS AND METHODS
Strains and media
The [PSI+] ΔUPF1 strain, a derivative of the 74D694 strain of Saccharomyces cerevisiae (MATa ade1–14 trp1–289 leu2–3,112 his3Δ200 ura3–52 upf1::TRP1 [PSI+]) was used for the production and purification of readthrough GST proteins. The UPF1 gene was deleted by TRP1 from Kluyveromyces lactis.
The strain was grown in minimal medium supplemented with the appropriate amino acids for maintenance of the various plasmids. Adenine was added in excess, to prevent accumulation of the red pigment AIR, an intermediate of the adenine biosynthesis chain that competes with GST (glutathione S-transferase).
Plasmids
Readthrough proteins were produced from pYX24-GST. This vector was constructed by removing the Ecl136II restriction site from the pYX212 vector (Ingenius MBV-028–10), which has a TPI1 promoter, a 2μ origin and URA3 selectable marker, and inserting the open reading frame of the GST gene into the filled EcoRI site. Expression was optimized by inserting the GST gene (from Schistosoma japonicum) downstream from the promoter, the entire 5′ UTR, the ATG and the first four codons of the TPI1 gene, which is strongly expressed in yeast, its protein product accounting for about 2% of total soluble cellular protein in yeast (32). A single Ecl136II restriction site was created during cloning, at the junction between the TPI1 and GST sequences, to facilitate the cloning of the different readthrough sequences (Table 1). The modified alanine tRNAs with UUG or CUG glutamine anticodons were inserted into Ecl136II site of pFL44H (corresponding to pFL44L (33) in which the URA3 marker has been replaced by HIS3) and expressed in the [PSI+] ΔUPF1 yeast strain cotransformed with the pYX24-GST constructs.
Table 1. Oligonucleotide sequences derived from S. cerevisiae genes with readthrough consensus contexts.
| Sequence name | Oligonucleotide sequences |
|---|---|
| IXRI-TAA | ATAAAAAATGAATAACAAATAAACAAC |
| IXRI-TAG | ATAAAAAATGAATAGCAAATAAACAAC |
| IXRI-TGA | ATAAAAAATGAATGACAAATAAACAAC |
| SFB-TGA | ATCAACAGATGACAATCAGTC |
| TIP41-TAA | ATTCAGTTCTAACAATCAAAT |
Western blotting
Protein samples were separated by SDS–polyacrylamide gel electrophoresis and the bands were electroblotted onto PVDF membrane, at 16 V for 45 min. Monoclonal antibodies directed against GST (Santa Cruz Biotechnology) were hybridized with the membrane and the binding of these antibodies was detected by incubation with secondary antibodies coupled to alkaline phosphatase and NBT/BCIP staining.
Quantification of readthrough efficiency
The pAC derivative vectors were constructed by inserting the fragment of interest into the single MscI site of pAC99 (34). Luciferase and β-galactosidase activities were assayed in the same crude extract as previously described (35). Values are reported as box plots, with median values for six independent measurements. The significance of differences was evaluated in non-parametric Mann–Whitney tests.
Purification of readthrough proteins
For readthrough GST purification, the [PSI+] ΔUPF1 strain was transformed with pYX24-GST vectors. The pellet of a 1 l overnight culture was resuspended in 1 × PBS supplemented with 1 mM DTT, 0.5% Nonidet and 1× Complete EDTA-free protease inhibitor cocktail. Cells were disrupted in a French press and the whole-cell extract was loaded onto a 1 ml GSTrap HP column (GE Healthcare). Proteins were purified with an AKTA purifier 10 FPLC system. GST binding was achieved in PBS buffer (1 × phosphate-buffered saline supplemented with 1 mM dithiothreitol) and proteins were eluted from the affinity medium with a Tris-glutathione buffer (50 mM Tris pH 8, 10 mM glutathione).
Mass spectrometry analyses
Sample preparation
GST-purified proteins were separated by SDS-PAGE and stained with Coomassie Blue. Bands corresponding to GST proteins were excised and subjected to enzymatic digestion in the Progest robot (Genomic Solutions). Briefly, protein bands were excised and extensively washed with CH3CN and 25 mM NH4HCO3. The excised bands were treated with 100 μl 10 mM DTT at 57°C for 30 min. The DTT was removed and 100 μl of 55 mM iodoacetamide was added for cysteine carbamidomethylation. The reaction was allowed to proceed at room temperature for 30 min. The supernatant was removed, the washing procedure was repeated and the gel slices were dried. We added 20 μl of 20 ng/μl LysN (Seikagaku Biobusiness) or 10 ng/μl trypsin (Promega) diluted in 25 mM NH4HCO3, and the mixture was incubated overnight at room temperature. Peptides were extracted in 60% acetonitrile and 0.1% (v/v) formic acid, dried under vacuum and then resuspended in 0.1% n-octylglucopyranoside (Sigma–Aldrich) to prevent the loss of hydrophobic peptides before LC-MS/MS analyses.
LC-MS/MS analyses
Proteolytic peptides were analyzed with two different instruments. Nano-liquid chromatography elution conditions were very similar for both systems, with a flow rate of 300 nl/min and an acetonitrile gradient of 5–35% (v/v) acetonitrile over 40 min.
Peptides were initially identified with an LTQ Orbitrap-Velos mass spectrometer (Thermo Scientific) coupled to the EASY nanoLC HPLC system (Proxeon). MS/MS spectra were acquired by a data-dependent acquisition method involving selection of the 20 precursors giving the most intense signals, for Collision-Induced Dissociation (CID) fragmentation. Raw data were processed and analyzed with Proteome Discoverer 1.3 software, using the MASCOT algorithm.
Peptides were further identified and quantified with a Triple-TOF 4600 mass spectrometer (ABSciex) coupled to the nanoRSLC system (Thermo Fisher Scientific) equipped with a trap column (Acclaim PepMap100C18, 75 μmi.d.×2 cm, 3 μm) and an analytical column (Acclaim PepMapRSLCC18, 75 μmi.d.× 15 cm, 2 μm, 100 Å). MS/MS spectra were acquired by a data-dependent acquisition method involving selection of the 10 precursors giving the most intense signals, CID fragmentation with the Q1 quadrupole set, at low resolution to improve sensitivity. Raw data were processed with MS Data Converter software and analyzed with PeakView software (ABSciex).
Protein identification
Proteins were identified with the MASCOT algorithm and an in-house database constructed by merging Swissprot with user-generated GST sequences, each harboring one of the 20 amino acids in the readthrough site. The other search parameters were as follows: digest reagent LysN (cleavage at the N-terminal of lysine) or trypsin, cysteine carbamidomethylation was considered a complete modification and oxidation (methionine and tryptophan) was considered variable. Peptide and fragment tolerances were set at 5 ppm and 0.6 Da, respectively, for Orbitrap data and at 10 ppm and 0.01 Da, respectively, for Triple-TOF data. Only ions with a score higher than the identity threshold and a false-positive discovery rate of less than 1% (Mascot decoy option) were considered.
Relative quantification of readthrough peptides
MS extracted-ion chromatograms (XIC) were generated for the peptides harboring readthrough amino acids. The intensity of each chromatographic peak was corrected by a factor taking the ionization and digestion efficiencies of each readthrough peptide into account. These factors were calculated by producing and purifying in-frame GST proteins with the same sequences as the readthrough proteins and analyzing them by NanoLC-MS/MS, as described above. Each factor was calculated as the ratio of the intensity of the peptide peak corresponding to the readthrough peptide sequence to the mean intensity for the three most intense peaks corresponding to internal peptides. The means of four technical replicates are reported.
RESULTS
A new reporter system for identifying the amino acids incorporated at stop codons during readthrough
Several reporter systems have been used to identify amino acids incorporated during stop codon readthrough in various organisms (21,28,36). However, none of these systems is appropriate for the systematic analysis of different readthrough sequences in a single organism. Our first objective was, thus, to set up a system for the efficient purification, in a single step, of a protein synthesized during a stop codon readthrough event. We chose to use the GST protein, which can be specifically and efficiently purified in a single step, by affinity capture chromatography. The coding sequence of GST was modified by the insertion of a restriction site four codons downstream from the initiator AUG, for the insertion of stop codon sequences (see the Materials and Methods). GST was thus produced only if the ribosomes read through the inserted stop codon.
The next step in the establishment of the reporter system was the choice of the most appropriate yeast strain to maximize the yield of GST without affecting the pool of tRNA, as our main objective was to determine which natural tRNAs could read through stop codons. At least two major factors can make it difficult to obtain high levels of GST production. First, the introduction of a stop codon close to the AUG would be expected to induce degradation of the mRNA via nonsense-mediated mRNA decay (NMD) (37). We overcame this obstacle by using a strain from which the UPF1 gene, encoding a major element of the NMD pathway (38), had been deleted. In the absence of UPF1, NMD is completely abolished, and mRNAs carrying PTCs are stable.
The second major potential drawback concerns the efficiency of stop codon readthrough. Indeed, the efficiency of unprogrammed stop codon readthrough is generally very low (< 0.3%) in yeast (10). We increased this efficiency by using a strain carrying the [PSI+] prion, which corresponds to the aggregated form of eukaryotic release factor 3 (eRF3). The sequestration of eRF3 in [PSI+] aggregates impairs the termination activity of eRF3, thereby increasing the efficiency of stop codon suppression (39). However, this increase is not sufficient to attain readthrough levels sufficiently high for the production of large amounts of GST. We previously identified a consensus readthrough motif (CA(A/G) N(U/C/G)A) that increased stop codon readthrough efficiency when located immediately downstream from the stop codon (10). In Saccharomyces cerevisiae, at least three genes (SFB2, TIP41, IXR1) naturally harbor this motif downstream from their stop codon. We first checked that these sequences actually promoted high levels of readthrough in [psi−] and [PSI+] strain, when used in a dual reporter system (Figure 1).
Figure 1.
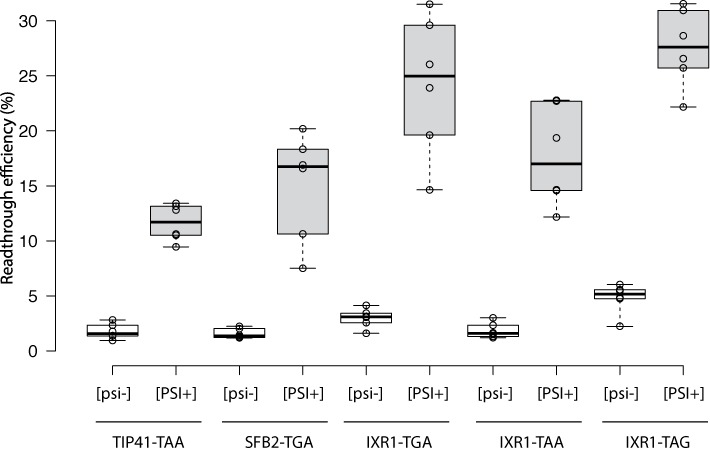
Readthrough efficiency for the sequences used for the GST reporter system. The lines in the center of the boxes indicate the median values; the box limits indicate the 25th and 75th percentiles calculated with R software; the whiskers extend to the minimum and maximum values; data points are plotted as open circles. n = 6 sample points. [psi−] values are indicated in white boxes, [PSI+] values are indicated in gray boxes.
The IXR1-TAG sequence was found to be the most efficient (25% of stop codon readthrough) and was therefore used in the GST purification protocol. Cell lysates were prepared as described in the Materials and Methods and GST proteins were purified on glutathione affinity columns. For GST-IXR1-TAG purification, a single peak was eluted from the column at the same position as the in-frame GST (Figure 2a). This peak corresponded to a single band at the position of the in-frame GST in the elution fractions, as revealed by SDS-PAGE and Coomassie Blue staining (Figure 2b). Using an antibody specific for GST, we confirmed that the single band observed was indeed GST (Figure 2c). These data demonstrate that this reporter system is suitable for the purification of readthrough GST proteins.
Figure 2.
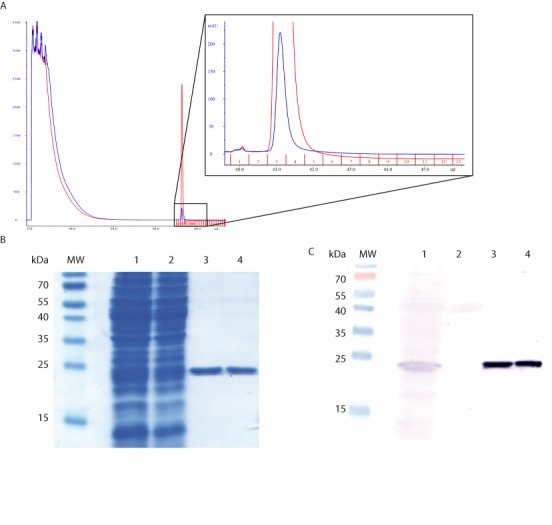
Purification and verification of readthrough GST proteins. (A) The chromatograms show absorbance at 280 nm. The red curve corresponds to in-frame GST purification; the blue curve corresponds to GST-IXRI-TAG purification. The elution peaks appears in fraction 3. (B) SDS-PAGE gel stained by Coomassie Blue. Lane 1: whole-cell extract; lane 2: flowthrough after binding of the extract; lane 3: in-frame GST, and lane 4: purified readthrough protein, GST-IXRI-TAG. For lanes 3 and 4, 1.8 μg of protein was loaded on the gel. (C) The presence of the GST protein was checked by western blotting with an antibody against GST. Lane 1 corresponds to the whole-cell extract, lane 2 corresponds to the flowthrough after binding of the extract, lane 3 to in-frame GST, and lane 4 to purified readthrough protein, GST-IXRI-TAA. For lanes 3 and 4, 0.5 μg of protein was loaded on the gel.
Nucleotides upstream of stop codons do not influence the identity of amino acids inserted during readthrough
We began our systematic analysis in the context of IXR1, which has been shown to promote stop codon readthrough highly efficiently, regardless of the stop codon considered (Figure 1). After the purification and enzymatic digestion of readthrough GST proteins, the peptides obtained were analyzed by LC-MS/MS. We found that the same three amino acids, glutamine, tyrosine and lysine, were inserted at the UAA and UAG codons (Table 2 and Supplementary Figures S2a–S2c), whereas tryptophan, cysteine and arginine were inserted at the UGA codon (Table 2 and Supplementary Figures S2d–S2f).
Table 2. Identification of the amino acids at readthrough sites.
| Sequence name | Stop condon | Identified readthrough peptides | Amino acids | Peptide mass (Da) | Retention time (min) |
|---|---|---|---|---|---|
| IXRI | UAA | KNEYQINNLSPILGYW | Y | 1950.98 | 39.3 |
| IXRI | UAA | KNEQQINNLSPILGYW | Q | 1915.97 | 36.7 |
| IXRI | UAA | KQINNLSPILGYW | K | 1544.83 | 35.3 |
| IXRI | UAG | KNEYQINNLSPILGYW | Y | 1950.98 | 39.4 |
| IXRI | UAG | KNEQQINNLSPILGYW | Q | 1915.98 | 36.7 |
| IXRI | UAG | KQINNLSPILGYW | K | 1544.83 | 35.3 |
| IXRI | UGA | KNEWQINNLSPILGYW | W | 1974.00 | 40.5 |
| IXRI | UGA | KNECQINNLSPILGYW | C | 1947.95 | 36.8 |
| IXRI | UGA | KNERQINNLSPILGYW | R | 1944.02 | 32.7 |
| SFB | UGA | AREINRWQSVLSPILGYW | W | 2187.15 | 38.9 |
| SFB | UGA | AREINRCQSVLSPILGYW | C | 2161.11 | 34.2 |
| SFB | UGA | AREINRRQSVLSPILGYW | R | 2157.18 | 29.2 |
| TIP41 | UAA | AREIQFYQSNLSPILGYW | Y | 2184.10 | 23.06 |
| TIP41 | UAA | AREIQFQQSNLSPILGYW | Q | 2149.09 | 22.25 |
| TIP41 | UAA | KQSNLSPILGYW | K | 1404.74 | 20.93 |
This table summarizes the readthrough peptides unambiguously identified by LC-MS/MS analyses for the IXRI, SFB and TIP41 contexts. All the cysteine residues found were carbamidomethylated. Mass accuracy was ≤ 1 ppm for all mass measurements.
We then investigated the influence of the nucleotides surrounding the stop codon on the identity of the amino acids inserted by readthrough. High readthrough levels are essential to guarantee high protein purification yields, and it was therefore not possible to use random sequences. We decided to analyze the incorporation of amino acids at the other two sequences naturally found in the S. cerevisiae genome: SFB2, with a UGA stop codon, and TIP41, with a UAA stop codon. These sequences differ principally in terms of their 5′ nucleotides (Table 1), as they all have a favorable readthrough consensus motif 3′ to the stop codon. LC-MS/MS analyses led to the identification of glutamine, tyrosine and lysine residues at the UAA codon in the TIP41 context (Table 2) and of tryptophan, cysteine and arginine residues at the UGA codon in the SFB2 context (Table 2). These results suggest that the main determinant of amino-acid incorporation by readthrough is the sequence of the stop codon rather than the 5′ nucleotide context in which that stop codon is found, which appears to have no effect on the choice of tRNA for stop codon decoding.
Relative quantification of the readthrough amino acids at the three stop codons
Following the identification of the readthrough amino acids, we investigated whether these amino acids were incorporated at the same rate at the various stop codons. We set up a robust protocol for quantifying the relative proportions of peptides harboring the various readthrough amino acids. Intrinsic biochemical properties of peptide sequences, such as hydrophobicity, net charge or enzymatic digestion efficiency may strongly affect MS intensity measurements. We adjusted MS data with corrective factors calculated from purified in-frame GST proteins containing each of the amino acids identified in the IXR1 and SFB2 contexts (Materials and Methods; Supplementary Table S1), to prevent misquantification.
In the IXR1 context, the same amino acids were found to be incorporated at the UAA and UAG stops, but their relative proportions differed between the two types of stop codon (Figure 3A and B). Indeed, tyrosine and glutamine residues were incorporated at similar frequencies at UAA stop codons (54 and 44%, respectively), whereas lysine was incorporated much less frequently (Figure 3A and E). In contrast, tyrosine was the main amino acid incorporated (92%) at the UAG stop codon, whereas glutamine and lysine residues were incorporated at similar, low frequencies (5% and 3%, respectively) (Figure 3B and E). In the TIP41 context, at the UAA stop codon, tyrosine and glutamine residues were incorporated at similar frequencies (54% and 42%) whereas lysine was incorporated less frequently (4%). While readthrough efficiency of the TIP41-UAA context was significantly lower than that of the IXR1-UAA context (Figure 1), incorporation efficiency of readthrough aminoacids was almost identical for both contexts (Supplementary Figures S3 and Figure 3A). At the UGA stop codon in the IXR1 context, tryptophan was the main amino acid inserted (82%), followed by cysteine (14%) and then arginine (Figure 3C and E). Very similar results were obtained for the UGA stop codon in the SFB2 context (Figure 3D and E).
Figure 3.
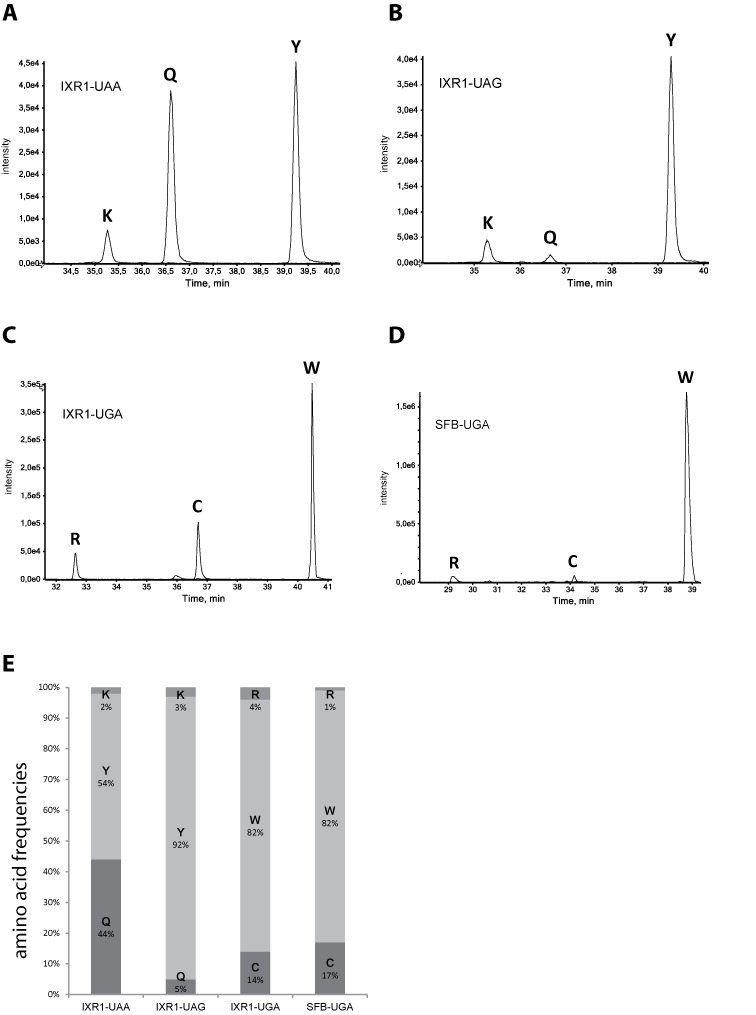
Relative quantification of readthrough peptides. (A)–(D) MS extracted-ion chromatograms (XIC) of readthrough peptides in the IXR1 and SFB contexts. The X- and Y-axis correspond to LC elution time and absolute MS signal intensity, respectively. (E) Relative frequencies of the readthrough amino acids incorporated at UAA, UAG and UGA stop codons in the IXR1 and SFB2 contexts. Quantification data were processed and adjusted as described in the Materials and Methods.
Together, these results indicate that readthrough amino acids are not incorporated with the same frequency at each stop codon and that the 5′ nucleotide context has no effect on the nature of the tRNAs used to decode stop codons or on their proportions at the stop codon.
Anticodon identity is the principal determinant of the amino acid used to decode the stop codon
We then investigated the molecular mechanism underlying the different incorporation rates of tyrosine and glutamine at UAA and UAG stops. Our results could potentially be accounted for by differences in the decoding capacities of natural suppressor tRNAs or the use of different isoacceptor tRNAs.
There are two possible explanations for the differences in the rates of incorporation of glutamine and tyrosine between UAA and UAG codons. First, tyrosine is more efficiently incorporated at the UAG codon than at the UAA codon. In the genetic code of S. cerevisiae, there is only one tyrosine tRNA, with a 3′AΨG5′ anticodon. This tRNA can engage in partial base pairing with the two stop codons, with a G-A and a G-G mismatch at the wobble position for UAA and UAG, respectively. None of these mismatches is particularly favorable, and there is no obvious reason for this tRNATyr preferentially base pairing with UAG rather than UAA.
The second possibility is that glutamine is less efficiently inserted at the UAG codon than at the UAA codon. There are two tRNAGln in yeast (40), both of which are potential natural suppressor tRNAs: a major tRNA with a 3′GUU*5′ anticodon (the second U is highly modified), and a minor tRNA (present as a single copy) with a 3′GUC5′ anticodon. Regardless of the stop codon considered, both these tRNAs engage in the same unconventional G-U base pairing at the first position of the codon. However, for the two remaining positions, the major tRNAGln base pairs perfectly with UAA and the minor tRNAGln base pairs perfectly with UAG. This may account for our findings, as glutamine was more frequently incorporated at the UAA codon than at the UAG codon.
The simplest way to test this hypothesis would be to delete the single copy of the minor tRNAGln. Unfortunately, this single copy is essential for yeast viability (27). We therefore investigated the importance of anticodon identity for the decoding of UAA and UAG by tRNAGln, by replacing the anticodon of a non-suppressor tRNA with the anticodons of the two tRNAGln and monitoring the incorporation of the amino acid loaded by the modified non-suppressor tRNA at the UAA and UAG codons. This required the use of a tRNA for which aminoacylation is independent of the identity of the anticodon. We chose to use the alanine tRNA (3′CGU5′), because the aminoacylation of this tRNA is dependent exclusively on G3-U70 pairing (41). Modification of the anticodon of this tRNA has no impact on the efficiency of alanine loading (42), a major advantage when assessing the ability of this modified tRNA to decode stop codons.
The modified tRNAs were coexpressed with readthrough GST in the [PSI+] ΔUPF1 yeast strain. LC-MS/MS analyses were performed and showed that alanine was incorporated at the UAA codon only if the tRNAAla carried the 3′GUU5′ anticodon (Figure 4A and B and Supplementary Figure S4a) Thus, this tRNAAla can decode UAA codons only if it carries this anticodon. At the UAG codon, alanine was detected only in the presence of the modified tRNAAla 3′GUC5′ (Figure 4C and D and Supplementary Figure S4b). These results clearly demonstrate that anticodon identity is the principal determinant of stop codon decoding and that the observed differences in glutamine incorporation are due to differences in the relative abundance of tRNAs able to decode the various stop codons.
Figure 4.
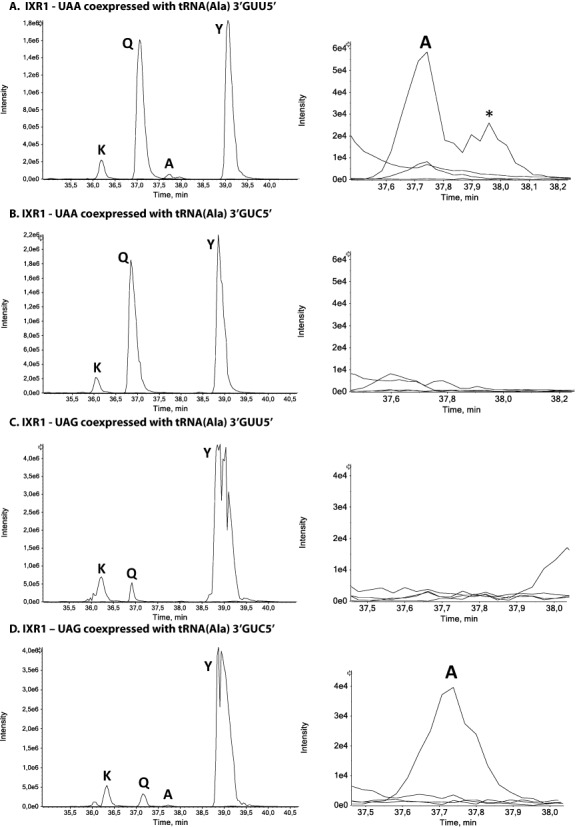
Alanine insertion at stop codons after the coexpression of IXR1-UAA and IXR1-UAG readthrough proteins with modified 3′GUU5′ and 3′GUC5′ tRNAsAla. MS extracted-ion chromatograms (XIC) of readthrough peptides with lysine, glutamine, tyrosine and alanine incorporated at the UAA and UAG stop codons are shown in left panels. To better visualize alanine incorporation at UAA and UAG stop codons, a zoom of the elution region is shown on the right side of each XIC. The alanine incorporated readthrough peptide (KNEAQINNLSPILGYW) is detected only when IXR1-UAA and IXR1-UAG are coexpressed with tRNAAla3′GUU5′ (panel A) or tRNAAla3′GUC5′ (panel D), respectively. The asterisk (*) indicates a peak corresponding to the alanine isobaric peptide with alanine incorporated at the glutamine site downstream from the stop codon (KNEQAINNLSPILGYW).
DISCUSSION
We performed a systematic analysis of the amino acids incorporated at stop codons in vivo during natural readthrough in eukaryotes (i.e in the absence of a mutated suppressor tRNA). We achieved this by setting up an in vivo reporter system for the production and purification of proteins from readthrough in the yeast Saccharomyces cerevisiae. To this aim we used a strain carrying the prion [PSI+]. This prion stimulates stop codon readthrough by partially depleting cells of termination complex, without affecting tRNA abundance. Therefore we believe that identity and relative incorporation frequencies of the amino acids inserted at stop codons will be the same in both the [PSI+] and wild type strains. The proteins were analyzed by MS to identify the readthrough peptides and the residues inserted at the stop codon. We first investigated insertions at the three stop codons in the same nucleotide context, that of the IXR1 gene. We identified the same three amino acids—glutamine, tyrosine and lysine—at both UAA and UAG codons. This is the first time that tyrosine and lysine have been found at a UAA stop codon. At the UGA stop codon, we observed the insertion of tryptophan, cysteine and arginine, confirming previous results obtained in vitro (RRL) (21) (Table 2).
Previous reports have reported the surrounding nucleotide context to have a major effect on termination efficiency (7,9,10,43). One interesting possibility was the mediation of this effect by the selection of specific suppressor tRNAs. Indeed, high-resolution X-ray structures of the prokaryotic ribosome have revealed interactions between the elbow base U47 of P-site tRNA and the elbow base D16 of A-site tRNA (44), raising the possibility that the P-site tRNA plays a role in the selection of the near-cognate tRNA incorporated at the stop codon. We tested this hypothesis by using two other contexts, differing principally in terms of their 5′ nucleotides. The results obtained (Table 2) clearly indicated that the same amino acids were inserted at a given type of stop codon, regardless of the 5′ nucleotide context. We then investigated whether the nucleotide context affected the relative frequencies with which the different residues were inserted at the stop codon. We set up a rigorous method of relative quantification by MS and showed that, for a given stop codon, readthrough aminoacids were inserted in similar proportions in different nucleotide environments (Figure 3E and Supplementary Figure S3). Thus, 5′ nucleotide context has no impact on the nature of the amino acids inserted by readthrough or their proportion. These results suggest that upstream nucleotides are not involved in the selection of the near-cognate tRNA.
We found that tyrosine, glutamine and lysine residues were incorporated at both UAA and UAG stop codons. We quantified the insertion of these three amino acids at these two stop codons. We found that tyrosine and glutamine were inserted with similar frequencies (54% and 44%) at UAA codons, whereas lysine incorporation rates at this stop codon were very low (2%). However, the proportions were different for UAG stop codons, at which tyrosine was the main amino acid inserted (92%), with glutamine incorporated much less frequently (5%) than at UAA codons. Lysine was also incorporated at UAA codons, but at a low frequency (3%). We can, therefore, conclude that the identity of the stop codon affects not only the nature of the residues incorporated by readthrough, but also their relative frequencies at stop codons. In yeast, glutamine is carried by two tRNAs (40). One bears a 3′GUU5′ anticodon and is present as nine gene copies; the other has a 3′GUC5′ anticodon and is present as a single gene copy that is essential for yeast viability (27). We investigated the roles of these two tRNAs in the insertion of glutamine at UAA and UAG codons, by developing a strategy for assessing the impact of codon–anticodon pairing in stop codon suppression. For this experiment, we needed to use a tRNA that is not naturally able to recognize stop codons, with an easily modifiable anticodon and carrying an amino acid with a mass significantly different from the amino acids usually incorporated by readthrough. We selected the alanine tRNA 3′GUC5′, which satisfies all these criteria and for which anticodon modification has no significant impact on alanine loading (41). We replaced the anticodon of this tRNA with anticodons from both tRNAGln. At the UAA codon, the incorporation of alanine was detected only when tRNAAla 3′GUU5′ was present in the cell. At the UAG codon, alanine was incorporated in the presence of the tRNAAla with the 3′GUC5′ anticodon.
Thus, the major tRNAGln with the 3′GUU5′ anticodon is responsible for the insertion of glutamine at UAA codons, whereas the minor tRNAGln with the 3′GUC5′ anticodon is principally responsible for the insertion of glutamine at UAG codons. These results are consistent with the possibilities for base pairing between these anticodons and the two stop codons (Figure 5A).
Figure 5.
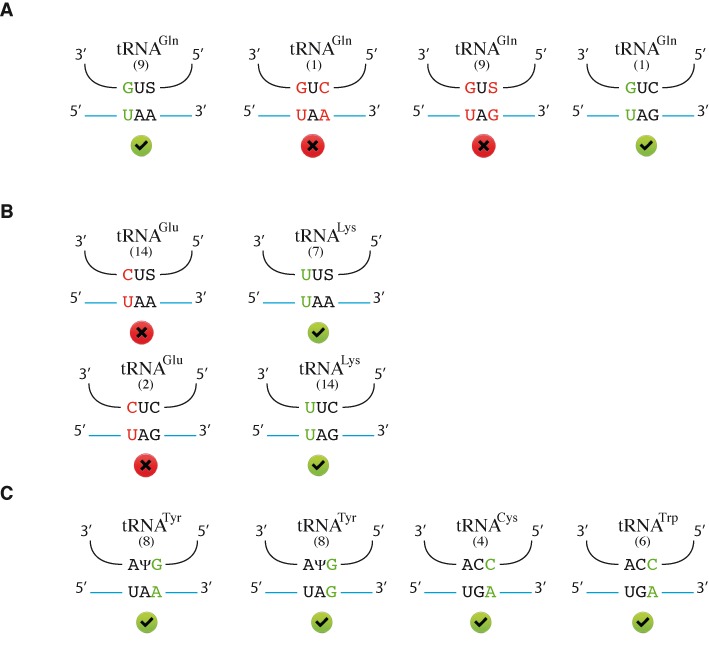
Proposed rules for stop codon decoding. This figure shows matching between potential suppressor tRNAs and stop codons. For each tRNA, the number of gene copies is indicated in brackets. The non-canonical base pairings appear in color green when they are accepted, red when they are not. Panel A corresponds to glutamine tRNAs, panel B to glutamate and lysine tRNAs, panel C to tyrosine, cysteine and tryptophan tRNAs. S represents a uridine modified with mcm5S2 and Ψ represents a uridine converted into a pseudouridine.
This work clearly demonstrates that not all near-cognate tRNAs are actually used in vivo for the decoding of UAA and UAG codons in yeast. Indeed, we found no glutamate at UAA or UAG sites, despite the presence of 14 and 2 copies of the tRNA bearing 3′CUS5′ and 3′CUC5′, respectively. Modifications of tRNAs, which are frequently associated with various decoding properties, cannot explain these results, as all the potential near-cognate tRNAs with a mismatch at the first position of the codon involved in the decoding of UAA codons carry the same mcm5S2 U34 modification. The two tRNAGlu not used during stop codon readthrough have a single non-standard Watson–Crick pair (C-U) at the first position of the codon, whereas the tRNA used to decode UAA and UAG stop codons have either a G-U (for Gln) or a U-U (for Lys) non-standard Watson–Crick pair at the first position of the codon (Figure 5A and B). We cannot rule out the possibility that glutamate is incorporated at levels below the detection threshold. However, our findings may reflect a more fundamental property of genetic decoding, according to which the non-standard Watson–Crick pair C-U in the first position is more detrimental than U-U base pairing. Recent studies based on the crystallization of the prokaryotic ribosome with a near-cognate tRNA at the A-site have revealed that the most important parameter for the acceptance of the near-cognate tRNA by the ribosome is not the number of hydrogen bonds, but the shapes of the base pairs (45–47). We therefore suggest that the geometry of the C-U pair is not accepted by the ribosome, whereas the geometry of the U-U pair is acceptable (albeit with a low efficiency).
Tyrosine is the only amino acid incorporated at UAA and UAG codons by a tRNA with a non-standard Watson–Crick pair (G-A or G-G) at the wobble position of the codon–anticodon triplet (in yeast there is only one tyrosine tRNA with a 3′AΨG5′ anticodon). Tyrosine is the amino acid most frequently incorporated at UAA and UAG codons. The same observation is true for cysteine and tryptophan, which were both efficiently incorporated at UGA codon, despite the G-A and C-A pairs, respectively, in the wobble position (Figure 5C). This implies that G-A, G-G and C-A pairs are well accepted by the ribosome when in the third position of the codon–anticodon triplet. However, this is probably not true for all codons, as it would imply, for example that the tRNAHis 3′GUG5′ would also be able to decode CAA or CAG glutamine codons. We cannot formally exclude the possibility of very low levels of histidine incorporation at glutamine codons, but this seems unlikely, as the only reported case of such substitution is related to E. coli in conditions of glutamine starvation and tRNAHis overexpression (48). The main difference between the incorporation of tyrosine at stop codons and that of histidine at glutamine codons is that, in the first case, there is no cognate tRNA to compete with the near-cognate tRNA. As demonstrated in several studies, translation termination is slower than elongation (11,49). The cognate tRNA therefore probably decodes its codon more efficiently than release factors decode stop codon. We suggest that these differences in the kinetics of the two reactions result in better incorporation of the near-cognate tRNA at a stop codon than at a sense codon.
Moreover, tRNAGln 3′GUC5′ did not decode the UAA codon, despite the presence of a C-A pair in the wobble position (Figure 5A). This indicates that the ribosome cannot tolerate two unconventional pairings. If a G-U pair is already present in the first position of the codon, then a second unconventional pair will not be accepted. This hypothesis also seems to be confirmed for the other tRNAGln 3′GUU5′, which is not accepted by the ribosome at the UAG codon (Figure 5A). It appeared possible that two G-U pairs (one in the first position, and the other in the third position of the codon) might be accepted by the ribosome, due to the similarity in the shapes of this unconventional pair and a G-C pair. However, this was found not to be the case, suggesting that the geometry of the G-U pair exerts too strong a constraint on the codon-anticodon interaction, preventing the formation of a second G-U pair. We identified no amino acids corresponding to a tRNA recognizing stop codons with a non-standard Watson–Crick pair in the second position of the codon (leucine, serine for UAA and UGA, and leucine, serine and tryptophan for UAG). This strongly suggests that the second position of the codon is under very tight selection by the ribosome for the acceptance or rejection of the tRNA from the A site.
These conclusions highlight the power of our approach and its usefulness for further studies on the decoding properties of tRNAs in vivo in yeast. It will be particularly interesting to focus on the role of tRNA modifications in the fidelity of decoding for stop and sense codons. This system will also be useful for the rationalization of translational PTC suppression strategies. Indeed, such therapeutic approaches are dependent on the re-expression of an active protein through the use of aminoglycosides. This activity may be strongly affected by the identity of the amino acid inserted at the stop codon. Using this system, we should be able to determine whether the amino acids inserted in the presence of aminoglycosides are the same as those inserted in the absence of this drug.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
English usage was corrected by Alex Edelman & Associates. We would like to thank Marc Mirande and Henri Grosjean for their help in the design of the experiment for identifying the tRNA incorporating glutamine at stop codons. This work benefited from the facilities and expertise of the SICaPS platform of IMAGIF (Centre de Recherche de Gif).
Footnotes
Present address: Argentini Manuela, INSERM, U968, Paris, F-75012, France; Sorbonne Universités, UPMC Univ Paris 06, UMR_S 968, Institut de la Vision, Paris, F-75012, France; CNRS, UMR_7210, Paris, F-75012, France.
FUNDING
This work received the support of the French foundation ARC [SFI20101201647 and PJA 20131200234]; French association ‘Ligue Nationale contre le cancer’ [3FI10167LVCY to O.N.]; Fellowship from the ARC [to S.B.]. Funding for open access charge: Fondation ARC pour la Recherche sur le Cancer, contract N°PJA 20131200234.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bertram G., Bell H.A., Ritchie D.W., Fullerton G., Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chavatte L., Seit-Nebi A., Dubovaya V., Favre A. The invariant uridine of stop codons contacts the conserved NIKSR loop of human eRF1 in the ribosome. EMBO J. 2002;21:5302–5311. doi: 10.1093/emboj/cdf484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhouravleva G., Frolova L., Le Goff X., Le Guellec R., Inge-Vechtomov S., Kisselev L., Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frolova L., Le Goff X., Zhouravleva G., Davydova E., Philippe M., Kisselev L. Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase. RNA. 1996;2:334–341. [PMC free article] [PubMed] [Google Scholar]
- 5.Loh P.G., Song H. Structural and mechanistic insights into translation termination. Curr. Opin. Struct. Biol. 2010;20:98–103. doi: 10.1016/j.sbi.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Pisarev A.V., Skabkin M.A., Pisareva V.P., Skabkina O.V., Rakotondrafara A.M., Hentze M.W., Hellen C.U., Pestova T.V. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonetti B., Fu L., Moon J., Bedwell D.M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 8.Janzen D.M., Frolova L., Geballe A.P. Inhibition of translation termination mediated by an interaction of eukaryotic release factor 1 with a nascent peptidyl-tRNA. Mol. Cell Biol. 2002;22:8562–8570. doi: 10.1128/MCB.22.24.8562-8570.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mottagui-Tabar S., Tuite M.F., Isaksson L.A. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem. 1998;257:249–254. doi: 10.1046/j.1432-1327.1998.2570249.x. [DOI] [PubMed] [Google Scholar]
- 10.Namy O., Hatin I., Rousset J.P. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertram G., Innes S., Minella O., Richardson J., Stansfield I. Endless possibilities: translation termination and stop codon recognition. Microbiology. 2001;147:255–269. doi: 10.1099/00221287-147-2-255. [DOI] [PubMed] [Google Scholar]
- 12.Hatfield D.L., Gladyshev V.N. How selenium has altered our understanding of the genetic code. Mol. Cell Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hao B., Gong W., Ferguson T.K., James C.M., Krzycki J.A., Chan M.K. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan G., James C.M., Krzycki J.A. Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 15.Lin C.A., Ellis S.R., True H.L. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell Biol. 2010;30:354–363. doi: 10.1128/MCB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouadloun F., Srichaiyo T., Isaksson L.A., Bjork G.R. Influence of modification next to the anticodon in tRNA on codon context sensitivity of translational suppression and accuracy. J. Bacteriol. 1986;166:1022–1027. doi: 10.1128/jb.166.3.1022-1027.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofstetter H., Monstein H.J., Weissmann C. The readthrough protein A1 is essential for the formation of viable Q beta particles. Biochim. Biophys. Acta. 1974;374:238–251. doi: 10.1016/0005-2787(74)90366-9. [DOI] [PubMed] [Google Scholar]
- 18.Weiner A.M., Weber K. A single UGA codon functions as a natural termination signal in the coliphage q beta coat protein cistron. J. Mol. Biol. 1973;80:837–855. doi: 10.1016/0022-2836(73)90213-1. [DOI] [PubMed] [Google Scholar]
- 19.Hirsh D. Tryptophan transfer RNA as the UGA suppressor. J. Mol. Biol. 1971;58:439–458. doi: 10.1016/0022-2836(71)90362-7. [DOI] [PubMed] [Google Scholar]
- 20.Beier H., Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 2001;29:4767–4782. doi: 10.1093/nar/29.23.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Y.X., Copeland T.D., Oroszlan S., Rein A., Levin J.G. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag-pol junction of Moloney murine leukemia virus. Proc. Natl. Acad. Sci. U.S.A. 1990;87:8860–8863. doi: 10.1073/pnas.87.22.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urban C., Beier H. Cysteine tRNAs of plant origin as novel UGA suppressors. Nucleic Acids Res. 1995;23:4591–4597. doi: 10.1093/nar/23.22.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerfass K., Beier H. The leaky UGA termination codon of tobacco rattle virus RNA is suppressed by tobacco chloroplast and cytoplasmic tRNAs(Trp) with CmCA anticodon. EMBO J. 1992;11:4167–4173. doi: 10.1002/j.1460-2075.1992.tb05510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J.P., Aker M., Sitney K.C., Mortimer R.K. First position wobble in codon-anticodon pairing: amber suppression by a yeast glutamine tRNA. Gene. 1986;49:383–388. doi: 10.1016/0378-1119(86)90375-6. [DOI] [PubMed] [Google Scholar]
- 25.Pure G.A., Robinson G.W., Naumovski L., Friedberg E.C. Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast tRNAGln gene. J. Mol. Biol. 1985;183:31–42. doi: 10.1016/0022-2836(85)90278-5. [DOI] [PubMed] [Google Scholar]
- 26.Yoshinaka Y., Katoh I., Copeland T.D., Oroszlan S. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. U.S.A. 1985;82:1618–1622. doi: 10.1073/pnas.82.6.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss W.A., Friedberg E.C. Normal yeast tRNA(CAGGln) can suppress amber codons and is encoded by an essential gene. J. Mol. Biol. 1986;192:725–735. doi: 10.1016/0022-2836(86)90024-0. [DOI] [PubMed] [Google Scholar]
- 28.Fearon K., McClendon V., Bonetti B., Bedwell D.M. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J. Biol. Chem. 1994;269:17802–17808. [PubMed] [Google Scholar]
- 29.Dunn J.G., Foo C.K., Belletier N.G., Gavis E.R., Weissman J.S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Namy O., Duchateau-Nguyen G., Hatin I., Hermann-Le Denmat S., Termier M., Rousset J.P. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:2289–2296. doi: 10.1093/nar/gkg330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bidou L., Allamand V., Rousset J.P., Namy O. Sense from nonsense: therapies for premature stop codon diseases. Trends Mol. Med. 2012;18:679–688. doi: 10.1016/j.molmed.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Scott E.W., Baker H.V. Concerted action of the transcriptional activators REB1, RAP1, and GCR1 in the high-level expression of the glycolytic gene TPI. Mol. Cell Biol. 1993;13:543–550. doi: 10.1128/mcb.13.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bonneaud N., Ozier-Kalogeropoulos O., Li G.Y., Labouesse M., Minvielle-Sebastia L., Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7:609–615. doi: 10.1002/yea.320070609. [DOI] [PubMed] [Google Scholar]
- 34.Namy O., Hatin I., Stahl G., Liu H., Barnay S., Bidou L., Rousset J.P. Gene overexpression as a tool for identifying new trans-acting factors involved in translation termination in Saccharomyces cerevisiae. Genetics. 2002;161:585–594. doi: 10.1093/genetics/161.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl G., Bidou L., Rousset J.P., Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res. 1995;23:1557–1560. doi: 10.1093/nar/23.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chittum H.S., Lane W.S., Carlson B.A., Roller P.P., Lung F.D., Lee B.J., Hatfield D.L. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry. 1998;37:10866–10870. doi: 10.1021/bi981042r. [DOI] [PubMed] [Google Scholar]
- 37.Shoemaker C.J., Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lelivelt M.J., Culbertson M.R. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol. Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebman S.W., Sherman F. Extrachromosomal psi+ determinant suppresses nonsense mutations in yeast. J. Bacteriol. 1979;139:1068–1071. doi: 10.1128/jb.139.3.1068-1071.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Percudani R., Pavesi A., Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 41.Trezeguet V., Edwards H., Schimmel P. A single base pair dominates over the novel identity of an Escherichia coli tyrosine tRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1991;11:2744–2751. doi: 10.1128/mcb.11.5.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K., Asahara H., Himeno H., Hasegawa T., Shimizu M. Identity elements of Escherichia coli tRNA(Ala) J. Mol. Recognit. 1991;4:129–132. doi: 10.1002/jmr.300040404. [DOI] [PubMed] [Google Scholar]
- 43.Tate W.P., Poole E.S., Dalphin M.E., Major L.L., Crawford D.J., Mannering S.A. The translational stop signal: codon with a context, or extended factor recognition element. Biochimie. 1996;78:945–952. doi: 10.1016/s0300-9084(97)86716-8. [DOI] [PubMed] [Google Scholar]
- 44.Yusupov M.M., Yusupova G.Z., Baucom A., Lieberman K., Earnest T.N., Cate J.H., Noller H.F. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 45.Westhof E., Yusupov M., Yusupova G. Recognition of Watson-Crick base pairs: constraints and limits due to geometric selection and tautomerism. F1000prime Rep. 2014;6:19. doi: 10.12703/P6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G. New structural insights into the decoding mechanism: translation infidelity via a G.U pair with Watson-Crick geometry. FEBS Lett. 2013;587:1848–1857. doi: 10.1016/j.febslet.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Demeshkina N., Jenner L., Westhof E., Yusupov M., Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484:256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 48.Ulrich A.K., Li L.Y., Parker J. Codon usage, transfer RNA availability and mistranslation in amino acid starved bacteria. Biochim. Biophys. Acta. 1991;1089:362–366. doi: 10.1016/0167-4781(91)90177-n. [DOI] [PubMed] [Google Scholar]
- 49.Freistroffer D.V., Kwiatkowski M., Buckingham R.H., Ehrenberg M. The accuracy of codon recognition by polypeptide release factors. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2046–2051. doi: 10.1073/pnas.030541097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


