Abstract
The essential Rcl1p and Bms1p proteins form a complex required for 40S ribosomal subunit maturation. Bms1p is a GTPase and Rcl1p has been proposed to catalyse the endonucleolytic cleavage at site A2 separating the pre-40S and pre-60S maturation pathways. We determined the 2.0 Å crystal structure of Bms1p associated with Rcl1p. We demonstrate that Rcl1p nuclear import depends on Bms1p and that the two proteins are loaded into pre-ribosomes at a similar stage of the maturation pathway and remain present within pre-ribosomes after cleavage at A2. Importantly, GTP binding to Bms1p is not required for the import in the nucleus nor for the incorporation of Rcl1p into pre-ribosomes, but is essential for early pre-rRNA processing. We propose that GTP binding to Bms1p and/or GTP hydrolysis may induce conformational rearrangements within the Bms1p-Rcl1p complex allowing the interaction of Rcl1p with its RNA substrate.
INTRODUCTION
Ribosomes are molecular machines that translate mRNAs into proteins. These large nucleoprotein complexes consist of four different ribosomal RNA (rRNA) species and ∼80 ribosomal proteins in eukaryotes. Through extensive analyses in yeast, ∼200 different proteins and RNA/protein complexes necessary for the assembly and maturation of ribosome precursors have been identified (1–3). These factors act in a highly coordinated and poorly understood series of events that lead to the removal of internal and external transcribed spacers (ITS and ETS) by endo- and exonucleolytic cleavages, to folding rearrangements and chemical modification of the pre-rRNA, and to its assembly with ribosomal proteins (4,5). Altogether, these complex events aim at producing complete ribosomal particles with the least number of errors for accurate translation.
Ribosomal RNAs are generated from two independent transcription units. RNA polymerase I synthesizes a long RNA precursor, the 35S pre-rRNA, encompassing the mature 18S, 25S and 5.8S rRNA sequences in yeast. The RNA polymerase III transcript leads to the 5S rRNA. During biogenesis of the small ribosomal subunit (SSU), a series of cleavages occurring in the 5′-ETS of the RNA precursor at sites A0 and A1, and in ITS1 at site A2 liberates the 20S pre-rRNA precursor to the mature 18S rRNA, which is then exported to the cytoplasm for cleavage at site D releasing the mature 18S rRNA. The downstream product of A2 site cleavage, the 27SA2 pre-rRNA, is further processed into the mature 5.8S and 25S rRNAs of the ribosome LSU (4,6).
Yeast RCL1 is an essential gene encoding a 40 kDa protein involved in 18S rRNA processing (7). Rcl1p depletion leads to the accumulation of the 35S pre-rRNA while the 20S intermediate and the mature 18S rRNA disappear, leading to the proposal that Rcl1p is involved in A0, A1 and A2 cleavages. Importantly, accumulation of the 22S pre-rRNA upon Rcl1p depletion (7) indicates that cleavages at sites A1 and A2 are more affected than cleavage at A0. Rcl1p shares high sequence and structural homology with RNA 3′-terminal phosphate cyclase enzymes (8,9). However, the catalytic residues required for cyclization in bona fide enzymes are not conserved in Rcl1p and purified recombinant Rcl1p is not able to perform such a reaction in model substrates relevant to other cyclases (7). Recently, Rcl1p has been proposed as the endonuclease responsible for cleavage at site A2 (10). The purified protein cleaves an in vitro-transcribed yeast ITS1 fragment encompassing the A2 site and cis mutations known to inhibit A2 cleavage in vivo impair this in vitro cleavage (10). In yeast cells, Rcl1p is required for production of the mature 18S rRNA but, in contrast, cleavage at site A2 has been shown to be dispensable for production of the mature 18S rRNA (11,12). Cleavage at site A2 can therefore be bypassed through cleavages at other sites and the essential role of Rcl1p in the production of the mature 18S rRNA resides in another function. The catalytic residues of Rcl1p involved in the endonucleolytic cleavage remain mysterious so far. Amino acid substitutions in the pocket of Rcl1p homologous to the catalytic pocket of Escherichia coli RtcA cyclase (8) do not induce detectable growth defects (9). However, given that A2 cleavage is not essential, mutations specifically affecting the nuclease activity of Rcl1p may not be expected to induce growth defects. Recently, a triple amino acid substitution in the C-terminus of Rcl1p (R327A, D328A and K330A) was shown to reduce but not abolish A2 cleavage and it was proposed that these residues may not be directly involved in catalysis but rather in RNA substrate binding (10).
Rcl1p physically interacts with Bms1p (13,14), also required for early cleavages of the pre-rRNA at sites A0, A1 and A2 and production of the mature 18S rRNA. Bms1p is a guanosine triphosphate (GTP)-binding protein whose GTPase activity is modulated by its own C-terminal domain acting as a GTPase-activating protein (GAP) domain (15). Rcl1p and Bms1p were initially found associated with U3 snoRNP (7,14) even though low levels of U3 snoRNP co-immunoprecipitate with Bms1p (14). In a later study, Baserga et al. did not detect Rcl1p or Bms1p upon TAP-tagged purification of the U3 snoRNP proteins, Mpp10p and Nop58p, two components of the SSU processosome (1). Depletion of Bms1p prevents incorporation of Rcl1p into pre-ribosomes as assessed by sedimentations on sucrose gradients (14) and based on biochemical studies, it was proposed that Bms1p acts as a loading factor for Rcl1p onto the U3 snoRNP (15) within pre-ribosomes. Consistently, both proteins are predominantly found in the nucleolus (14) and a C-terminal Nuclear Localization Signal (NLS) was identified in Bms1p (13). Karbstein et al. proposed that GTP binding by Bms1p is necessary for interaction with Rcl1p. The Bms1p-Rcl1p interaction would render Bms1p able to interact with the U3 snoRNP. Upon undefined stimuli, Bms1p would then hydrolyse its bound GTP and lose affinity for both Rcl1p and U3 snoRNP, leaving Rcl1p associated with U3 snoRNP. Several questions remain so far, including how does Rcl1p reach the nucleolus, what is the molecular basis of the Bms1p and Rcl1p interaction, does this interaction affect Rcl1p endonucleolytic activity and at which stages are Rcl1p and Bms1p recruited to and dissociated from pre-ribosomal particles?
To address some of these issues, we conducted a structural and functional study of the Rcl1p–Bms1p complex. Here, we describe the crystal structure of Bms1p in complex with Rcl1p. We show that Bms1p forms a stable complex with Rcl1p in vitro even in the absence of Bms1p GTPase and GAP domains. The structure allows identification of two conserved binding sites on Rcl1p, both necessary for the recognition of Bms1p. Interestingly, the identified interaction surface includes residues previously involved in RNA cleavage at site A2 (10), suggesting that Bms1p acts as chaperone masking Rcl1p active site. We show that Bms1p is required for the import of Rcl1p into the nucleus and that the two proteins are loaded into nascent pre-ribosomal particles at a similar stage of the assembly pathway. GTP-binding to Bms1p is not required for these steps but is crucial for pre-rRNA processing and production of the 20S pre-rRNA. We propose that GTP binding to Bms1p and/or GTP hydrolysis unveils the RNA binding surface of Rcl1p and promotes pre-rRNA cleavage.
MATERIALS AND METHODS
Constructs, protein expression and purification
The generation of the constructs used in this study is described in the Supplementary Material. For formation and purification of the native Bms1p–Rcl1p complexes, the constructs were co-transformed and overexpressed in E. coli BL21(DE3) Gold pLysS cells. The cultures were grown at 37°C in 2xYT medium and protein expression was induced at 15°C with 0.5 M Isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight. Cells were lysed in buffer containing 50 mM Tris pH 8.0, 150 mM NaCl. After centrifugation at 50 000 g, the clarified cell lysate was combined with His-Select Co2+ resin (Sigma) for 30 min at 4°C. After extensive wash the protein complexes were eluted with a 10–250 mM imidazole gradient. After cleavage with Tobacco Etch Virus (TEV) (1:100 w/w) overnight at 4°C, the complexes were further purified with a HiQ-Sepharose (GE Healthcare) and size exclusion chromatography over Superdex 200pg HR16/60 (GE Healthcare).
Crystallization and structure determination
Initial attempts of crystallization of Bms1p (1–705)–Rcl1p were conducted at 10–15 mg/ml in a buffer containing 10 mM Tris, pH 8.0, 100 mM NaCl, 1.0 mM DTT. The complexes were crystallized at 20°C by the sitting drop method against a reservoir containing either 10% PEG 3350 and 0.1 M Tris-HCl, pH 7.5 for the Bms1p (1–705)–Rcl1p complex or 12% PEG 8K, 0.2 M NaCl and 0.1 M Hepes, pH 7.5 for both the native and Se-methionine substituted Bms1p (523–670)–Rcl1p complex. Crystals were transferred to a cryo protectant (25% ethylene glycol, 10% PEG 8K and 0.1 M Hepes, pH 8.0), flash-frozen in liquid nitrogen, and maintained at 100 K in a nitrogen cryostream during data collection.
Crystals are in the space group P212121 with unit cell dimensions a = 69.13 Å b = 88.30 Å c = 102.14 Å and contain one molecule each of Bms1p and Rcl1p in the asymmetric unit. The structure of Bms1p (523–670)–Rcl1p complex was solved by Single Wavelength Anomalous Dispersion (SAD) using Se-substituted protein crystals. Data were collected on the SOLEIL Proxima-1 beamline. Data sets were reduced using XDS (16). Seven Se sites were located with SHELXCD (17) and refined with autoSHARP (18). The initial model was built using Arp/wARP (19). The final model was refined with BUSTER (20) and contains Rcl1p residues 7–361, Bms1p residues 547–569 and 606–636, 159 water molecules and has an R-factor of 20.02% and a free-R factor of 21.66% (Supplementary Table SI).
Co-precipitation experiments
BL21(DE3) cells were co-transformed with pET-15b derived Bms1p constructs and with a pET-28b derivative containing Rcl1p. Cells grown at 37°C were induced with 1 mM ITPG. After an overnight incubation at 15°C, cells were harvested by centrifugation and re-suspended in a buffer containing 1.5× phosphate buffered saline, 1 mM Mg Acetate, 0.1% NP-40, 20 mM imidazole, 10% glycerol. The cell pellet was sonicated for 5 s at 7 W. A fraction of crude extract was saved at this stage and boiled in Laemmlli buffer, the remaining was centrifuged and the supernatant was incubated with cobalt-affinity resin for 30 min at 4°C. Beads were washed three times with 1 ml of the lysis buffer. About 5% of the crude extract and 15% of the bound fraction were analysed on a 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were analysed by western blot using mouse monoclonal antibodies directed against the His-tag (GE Healthcare) or rabbit polyclonal antibodies directed against Rcl1p.
Yeast strains and media
The yeast strains used in this study were generated from the BY4741 or BY4743 wild-type strains or from strains expressing different TAP-tagged proteins, which are all derivatives of Saccharomyces cerevisiae strain S288C [MATa;his3Δ1;leu2Δ0;met15Δ0;ura3Δ0]. Construction of all the strains is described in Supplementary Materials and Methods.
S. cerevisiae strains were grown either in YP medium [1% yeast extract, 1% peptone] (Becton–Dickinson) supplemented with 2% galactose or 2% glucose as the carbon source (rich medium) or in YNB medium [0.17% yeast nitrogen base (MP Biomedicals), 0.5% (NH4)2SO4] supplemented with 2% galactose or 2% glucose and the required amino acids (minimal medium). Selection of the kanR transformants was achieved by addition of G418 at a final concentration of 0.2 mg/ml.
Fluorescence and immunofluorescence microscopy experiments
In Figure 5, strain RCL1::GFP was grown in a glucose-containing YP medium and strain [GAL::BMS1, RCL1::GFP] was shifted from a galactose- to a glucose-containing YP medium and grown for 11 h. In Figure 7, strain [GAL::BMS1, RCL1::3HA] transformed with vectors expressing wild-type Bms1p, Bms1pK82A or with the empty vector as a control were shifted from a galactose- to a glucose-containing synthetic medium (with all the required amino acids except histidine and leucine) and grown for 20 h to deplete the chromosome-encoded Bms1p protein. Fluorescence and immunofluorescence microscopy experiments were performed as described (21) with some minor modifications described in Supplementary Materials and Methods.
Figure 5.
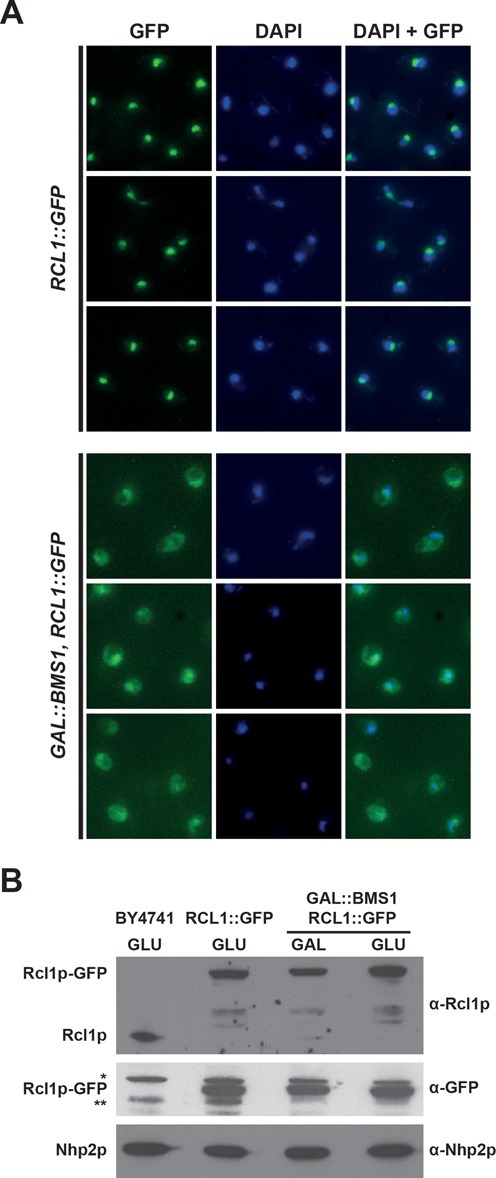
Bms1p depletion results in cytoplasmic accumulation of Rcl1p in yeast cells. (A) Subcellular localization of Rcl1p-GFP in the presence or absence of Bms1p. Upper 3 rows: yeast cells expressing a chromosomal GFP-tagged version of Rcl1p were grown in a glucose-containing rich medium. Lower 3 rows: yeast cells expressing Rcl1p-GFP and harbouring a chromosomal GAL1::BMS1 construct were shifted from a galactose- to a glucose-containing medium and grown for 11 h. In both cases, cells were harvested and processed for fluorescence microscopy. From left to right: Rcl1p-GFP fluorescence signal (green), DAPI staining (blue), merged images. (B) Accumulation levels of Rcl1p-GFP in the presence or absence of Bms1p. Yeast cells expressing Rcl1p-GFP and harbouring the chromosomal GAL1::BMS1 construct were shifted from a galactose- to a glucose-containing medium and grown for 11 h. Cells were harvested, total proteins were extracted and analysed by western blot using the indicated antibodies. The RCL1::GFP and BY4741 strains were grown on a glucose-containing medium and processed in parallel to determine the accumulation levels of Rcl1p-GFP and the endogenous Rcl1p protein, respectively, in these conditions. The nucleolar protein Nhp2p was detected as a loading control. * and **, non-specific proteins detected with the anti-GFP antibodies.
Figure 7.
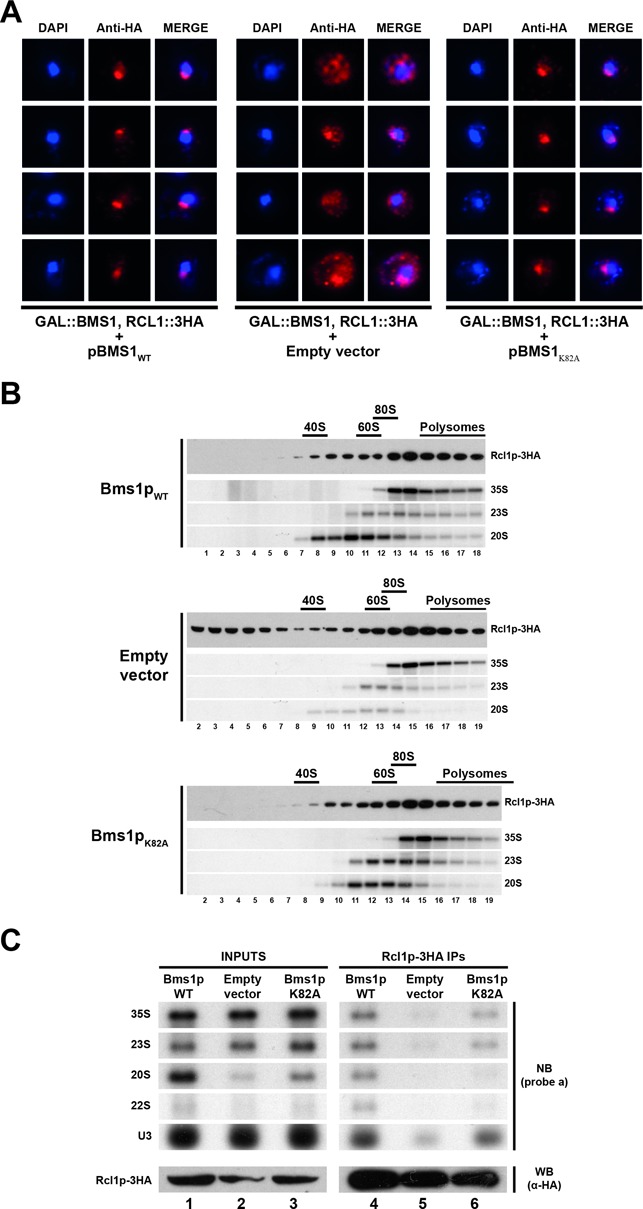
GTP binding to Bms1p is not required for incorporation of Rcl1p into pre-ribosomes. (A) Subcellular localization of Rcl1p-3HA in cells expressing Bms1pK82A or in ‘wild-type’ or Bms1p-depleted cells as controls. A yeast strain expressing Rcl1p-3HA and harbouring the chromosomal GAL::BMS1 construct was transformed with vectors expressing wild-type Bms1p (left panel), Bms1pK82A (right panel) or with the empty vector as a control (central panel). The resulting strains were shifted from a galactose- to a glucose-containing medium and grown for 20 h. Cells were harvested and processed for immunofluorescence microscopy using anti-HA antibodies. From left to right: DAPI (4',6'-diamidino-2-phénylindole) staining (blue), Rcl1p-3HA immunofluorescence signal (red) and the merged images. (B) Sucrose gradient sedimentation profile of Rcl1p using extracts from cells expressing ‘wild-type’ Bms1p (upper panel), Bms1pK82A (lower panel) or from Bms1p-depleted cells (central panel). The yeast strains grown as described in panel A were treated with cycloheximide and harvested. Total extracts were prepared and sedimented through 4.5–45% sucrose gradients. Twenty fractions were collected from which RNAs and proteins were extracted. Rcl1p-3HA was detected in the protein samples by western blot using anti-HA antibodies. The 35S, 23S and 20S pre-rRNAs were detected by northern blot using probe ‘a’. (C) Pre-rRNAs associated with Rcl1p-3HA in cells expressing Bms1pK82A or in ‘wild-type’ or Bms1p-depleted cells as controls. The pre-ribosomal particles containing Rcl1p-3HA were immunoprecipitated from extracts corresponding to the different strains grown as described in (A). The associated RNAs were analysed by northern blot using probe ‘a’ and probe ‘U3’.
Immunoprecipitation experiments
Immunoprecipitations of HA- and TAP-tagged proteins were performed as described previously (22) with minor modifications, using either EZview Red Anti-HA Affinity Gel (Sigma, E6779) or immunoglobulin G Sepharose 6 fast flow (GE Healthcare, 17–0969–02), respectively. The detailed protocols are described in Supplementary Materials and Methods.
Sedimentations on sucrose gradients
Strains [GAL::BMS1, RCL1::3HA] containing plasmids encoding Bms1pK82A, Bms1pWT or the empty vector as a control were grown as described for immunofluorescence microscopy. Total extracts prepared as described in Supplementary Materials and Methods were loaded on 4.5–45% sucrose gradients and centrifuged at 39 000 revolutions per minute for 150 min on a Beckman–Coulter Optima L-100 XP ultracentrifuge using a SW41 rotor. Fractions were collected using a Foxy Jr. gradient collector (Teledyne Isco).
RESULTS
The GTPase and GAP domains of Bms1p are not required for the interaction with Rcl1p in vitro
As shown in Figure 1A, the GTP binding domain of Bms1p covers residues 69–320 while the GAP domain (residues 737–1120) and two coiled coil regions, extending between residues 705–737 and 1120–1159, can be identified using the HHPRED server (23). Previous studies have shown that the Bms1p GAP domain is dispensable for the interaction with Rcl1p (15). In addition, yeast two hybrid experiments suggested that a minimal fragment of Bms1p, encompassing residues 535–661, interacts with Rcl1p (14). To confirm and further characterize these interactions, several truncated versions of Bms1p have been generated and tested for their ability to interact with Rcl1p (Figure 1B). No background Rcl1p binding is detected upon incubation with affinity resin while formation of the His-Bms1p (1–705)-Rcl1p complex is visualized (lane 1). In contrast, the GTPase domain (residues 1–433) is not able to retain Rcl1p (lane 4). The 433–705 region of Bms1p has been reduced to a shorter fragment through the observation and the characterization of proteolytic fragments generated during the purification procedure. The fragment encompassing residues 523–670 is sufficient to retain Rcl1p upon pull down (lane 5).
Figure 1.
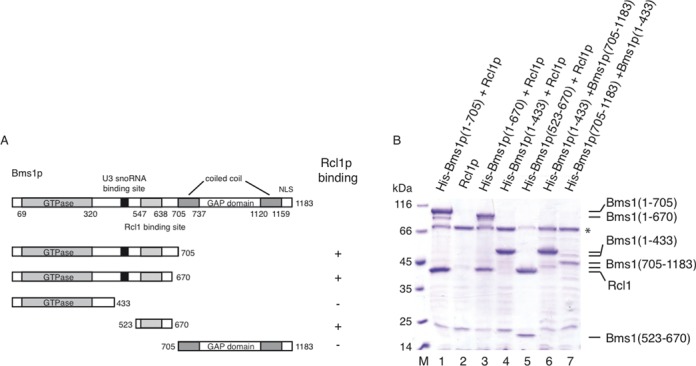
Interaction mapping of Rcl1p binding site on Bms1p. (A) Bms1p harbours from the N-terminus to the C-terminus a GTPase domain, a U3 snoRNA binding site (15), two coiled coil domains as defined by HHPRED server flanking a GAP domain and a C-terminal NLS. Several truncated versions of Bms1p have been produced in order to define the Rcl1p binding site on Bms1p. (B) Truncated derivatives of Bms1p tagged with 6 histidines at the N-terminus have been co-expressed with untagged Rcl1p. Cell lysates were incubated with cobalt-affinity resin and the bound fractions were analysed by SDS-PAGE and Coomassie-blue staining. (*) represents a known E. coli contaminating protein (Bifunctional polymyxin resistance protein Arn).
Bms1p shares homologies with the SelB translation elongation factor (23,24), the N-terminus of Bms1p (GTPase domain, residues 69–320) being homologous to SelB domain I and Bms1p C-terminus (GAP domain; residues 810–1010) being homologous to domains III–IV of SelB (13). The Bms1p GAP domain has been proposed to stimulate the GTPase activity of the GTP-binding domain (15). According to the structural homology with SelB the two domains should interact. We show that, when co-expressed in bacteria, the GTPase and GAP domains of Bms1p (residues 1–433 and 705–1185, respectively) interact with each other regardless of which protein is His-tagged (Figure 1B, lanes 6 and 7). Interestingly, a model computed from the homology with SelB shows that the P888 residue mutated to L in the thermosensitive strain bms1–1 in which 20S pre-rRNA production is almost completely blocked at non-permissive temperature (13), as well as the R833H substitution (25), are located close to the interface of the GTPase and the GAP domains (Supplementary Figure S1A).
In conclusion, the GTPase and GAP domains of Bms1p are dispensable for the interaction with Rcl1p in vitro. The minimal region of Bms1p interacting with Rcl1p can be restricted to a region encompassing residues 523–670, with no apparent specific motif. Finally, we show that the GTPase and GAP domains of Bms1p interact in trans.
Structure of the Bms1p–Rcl1p complex
The complex between full-length S. cerevisiae Rcl1p and Bms1p residues 1–705 was prepared and crystallized as described in the Materials and Methods section. Bms1p proteolysis occurred during sample preparation as visualized by Coomassie-blue staining of the purified and crystallized sample. Phases were obtained from a SAD data collection on Se-Met substituted Rcl1p (Supplementary Table S1 and Materials and Methods section). The structure was solved and refined to 20.18%/23.73% R-factor/R-free values at a resolution of 2.02 Å (Figure 2A).
Figure 2.
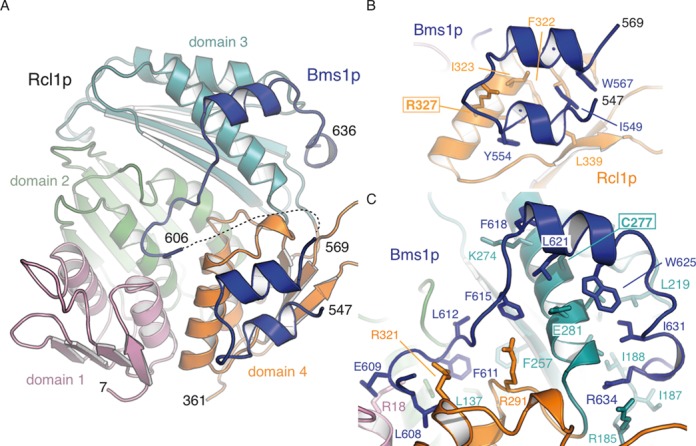
Overall structure of Rcl1p–Bms1p core interacting domain. (A) Rcl1p can be described as three repeated domains (orange, pink and green) linked to a fourth distinct one (cyan). The Bms1p fragment is shown in deep blue. This and all subsequent structural figures were generated with PyMOL ((http://www.pymol.org). (B) Residues 547–569 of Bms1p form two short α-helices bound at the surface of Rcl1p domain 4. (C) Residues 606–636 of Bms1p extend over the four domains of Rcl1p and provide multiple contacts with Rcl1p.
Rcl1p harbours a fold similar to the one of its orthologue in K. lactis and the E. coli RNA 3′-terminal phosphate ligase (Supplementary Figure S1B and S1C) (root-mean-square deviation (r.m.s.d.) on Cα carbon of 0.5 Å over 345 residues and 2.3 Å over 310 residues, respectively) (8,9) with three repeats of a folding unit, namely, domains 1, 2 and 4 (pink, green and orange, respectively) and a fourth domain (domain 3, cyan) (Figure 2A). As described for K. lactis Rcl1, S. cerevisiae Rcl1p does not harbour the required conserved residues (H309 in E. coli Rtc replaced by R321 in Rcl1p and R319 in kl Rcl1) required for RNA cyclase or auto-adenylation activities (7,9).
Two Bms1p fragments encompassing residues 547–636 connected by a disordered loop (residues 570–605) have been modelled within the electron density map (Figure 2). Residues 547–569 of Bms1p form the first Rcl1p binding site. This interaction is mainly driven by hydrophobic contacts with domain 4 of Rcl1p (Figure 2B) and is stabilized by contribution from hydrogen bonding between R327 of Rcl1p and the Bms1p backbone and a water molecule bridging the Bms1p Y554 hydroxyl group to Rcl1p Cα backbone. Interestingly, the bms1–7 mutant, in which residues 557–559 were replaced by alanines, was previously identified as greatly impaired for the interaction with Rcl1p (15). The second binding site on Bms1p comprises residues 606–636 and extends over all four domains of Rcl1p (Figure 2C). The Cα backbone of residues 606–614 of Bms1p make extensive contacts with Rcl1p side chains while F611 of Bms1p is deeply buried into an aliphatic cavity contributed to by all four Rcl1p domains and provided by side chains of residues A128, L137 and F257 (Figure 2C). Even though the location of F611 overlaps with the position of the covalent adenylate moiety found in RNA 3′ terminal cyclase (8,26), these aforementioned residues and those of Rcl1p they interact with, have not been conserved during evolution. Residues 615–636 interact with Rcl1p through extensive hydrophobic contacts in addition to polar side chain interactions. Notably, the α-helix formed by Bms1p residues 618–625 contacts α-helix 5 of Rcl1p. The Cα backbone between residues L621 and A623 provides the centre of the interaction with Rcl1p C277 residue (Figure 2C).
Identification of Rcl1p point mutations impairing the interaction with Bms1p in vitro
On the basis of the structure, we identified and mutated some Rcl1p residues located at the interface with Bms1p. The C277R and R327A substitutions altering one or the other binding site and the double mutation C277R/R327A were designed (Figure 2B and C). The various point mutants were co-expressed in bacteria with His-fused Bms1p (1–705) and pull-down experiments using a cobalt affinity resin were performed and analysed by SDS-PAGE (Figure 3). Interaction between wild-type Rcl1p and Bms1p as well as control experiments in which either Bms1p or Rcl1p was omitted are shown in lanes 1–3. The C277R single point mutation does not detectably affect Bms1p–Rcl1p interaction (lane 5), whereas, in contrast, the R327A mutation reduces the amount of Rcl1p pulled down by Bms1p (lane 4). The combination of C277R and R327A mutations seems to affect more drastically the interaction between the proteins (lane 6). Altogether we show that both Bms1p binding sites on Rcl1p contribute to the interaction with Bms1p and their modification affects the interaction between Bms1p and Rcl1p in vitro.
Figure 3.
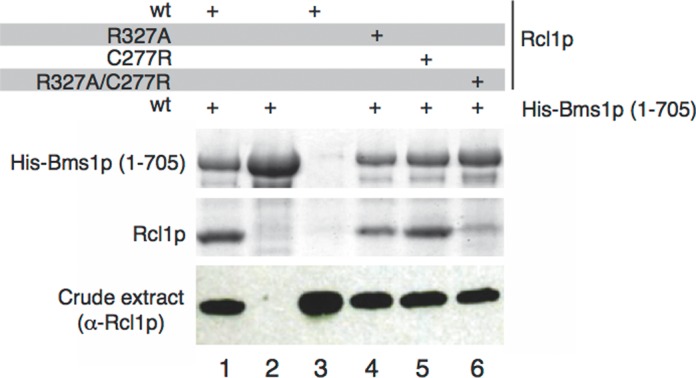
Point mutations altering the Rcl1p–Bms1p interaction in vitro. Point mutations have been introduced in Rcl1p in order to affect protein–protein interactions. Wild-type and mutant Rcl1p were co-expressed in E. coli with N-terminally His-fused Bms1p and pull-down experiments were performed with cobalt-affinity resin. The presence of the proteins was analysed by SDS-PAGE and Coomassie-blue staining. The amount of Rcl1p in crude extracts was controlled using anti-Rcl1p antibodies.
Impairment of Bms1p–Rcl1p interaction affects pre-ribosomal RNA processing in vivo
To assess the relevance of the Bms1p–Rcl1p interaction in yeast cells, we introduced pFL46-derived plasmids encoding the wild-type or C277R, R327A or C277R/R327A altered versions of Rcl1p into a haploid yeast strain bearing a deletion of the chromosomal RCL1 gene by plasmid shuffling. These strains did not display significant growth defects at 30°C on a rich medium, indicating that the altered proteins complement the essential function of Rcl1p in these conditions. To assess the effect of these mutations on pre-rRNA processing (see Supplementary Figure S2 for a detailed description of the pre-rRNA processing pathway in yeast), total RNAs extracted from these strains were analysed by northern blot (Figure 4). Expression of wild-type RCL1 from the pFL46 plasmid (lane 2) results in subtle processing defects as compared to the wild-type BY4741 strain (lane 1) probably due to the overexpression of the protein (Supplementary Figure S3). In comparison, the R327A and C277R/R327A mutants (lanes 4 and 5, respectively) display stronger pre-rRNA processing defects characterized by an accumulation of the 35S, 33S and 23S pre-rRNAs and a decrease in the production of the 27SA2 and 20S pre-rRNAs (Figure 4B). These defects typically result from altered cleavages at sites A0, A1 and A2. However, we also observed a mild accumulation of the combined 21S + 22S pre-rRNAs and of the 22S intermediate detected alone, which may suggest that cleavage defects at sites A1 and A2 are stronger than that at site A0. The C277R substitution (lane 3) reproducibly restores a pre-rRNA accumulation profile comparable to that observed in the BY4741 wild-type cells (lane 1). The corresponding altered Rcl1p protein accumulates at slightly lower levels than the overexpressed wild-type protein (Supplementary Figure S3). We therefore hypothesized that this substitution per se does not affect significantly pre-rRNA processing (consistent with the fact that it does not affect the interaction between Rcl1p and Bms1p in vitro) and that the beneficial effect comes from the decreased expression level.
Figure 4.
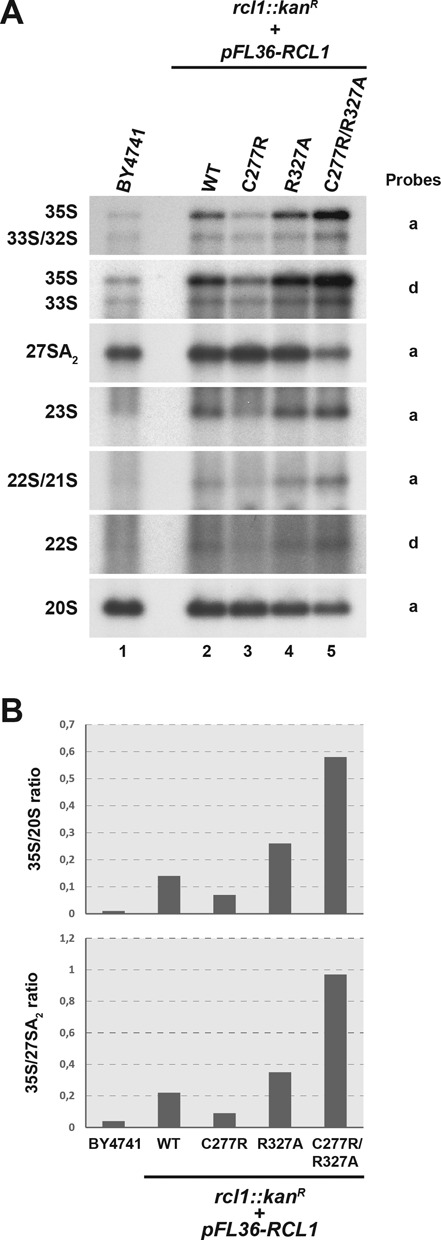
Amino acid substitutions altering the Rcl1p–Bms1p interaction affect early steps of pre-rRNA processing in vivo. (A) Northern blot analysis of pre-rRNA levels in strains expressing altered versions of RCL1. Total RNAs were extracted from yeast strains harbouring a deletion of the chromosomal RCL1 open reading frame (rcl1::kanR) and expressing plasmid-borne copies of wild-type or the indicated mutant versions of RCL1 (RCL1C277R, RCL1R327A, RCL1C277R/R327A). The accumulation levels of the different pre-rRNA species were analysed by northern blot using specific probes (indicated on the right). As a control, RNAs extracted from wild-type cells (BY4741) were analysed in parallel. (B) PhosphorImager quantifications of the radioactive signals obtained in the northern blot experiments presented in panel A. The ratios of the 35S over 20S species intensities and of the 35S over 27SA2 species intensities were calculated after background correction.
Altogether these data indicate that mutations in Rcl1p impairing the interaction with Bms1p in vitro affect pre-rRNA processing at sites A0, A1 and A2 in yeast cells.
Bms1p is required for Rcl1p import into the nucleus
Bms1p is imported in the nucleus via a classical NLS located at its C-terminus and it is required for the incorporation of Rcl1p into pre-ribosomes (13,14). To investigate whether the cellular localization of Rcl1p is affected upon alteration of the Bms1p–Rcl1p interaction, we transformed the BY4741 strain with plasmids expressing the wild-type or C277R/R327A altered version of Rcl1p fused at their N-terminus to the green fluorescent protein (GFP). Fluorescence microscopy analyses (Supplementary Figure S4) showed that, whereas GFP-Rcl1pWT accumulates mainly in the nucleolus consistent with previous results (7), GFP-Rcl1pC277R/R327A is present both in the nucleus and the cytoplasm. The cytoplasmic accumulation of GFP-Rcl1pC277R/R327A suggests that mutations impairing the interaction between Rcl1p and Bms1p in vitro affect the nuclear import of Rcl1p in yeast cells. To confirm this observation, we constructed a strain expressing Bms1p conditionally (GAL::BMS1) and expressing also Rcl1p in fusion to the GFP at the C-terminus. Upon Bms1p depletion, we observed a clear increase in the cytoplasmic accumulation of Rcl1p-GFP (Figure 5, lower rows), in contrast to the strong nucleolar localization in the presence of Bms1p (upper rows). We confirmed these results using independent immunofluorescence experiments (Supplementary Figure S5), which show in addition that the nucleolar localization of Nop1p, another known nucleolar protein, is not affected upon Bms1p depletion, arguing for a specific effect of Bms1p depletion on Rcl1p localization. Altogether these data suggest that Bms1p is required for Rcl1p import into the nucleus. To determine whether the reverse also holds true, we introduced a plasmid encoding GFP-Bms1p into a strain expressing Rcl1p conditionally (GAL::RCL1) and analysed the subcellular localization of GFP-Bms1p upon Rcl1p depletion (Supplementary Figure S6). We observed by fluorescence microscopy that Bms1p remains nucleolar in the absence of Rcl1p, indicating that Rcl1p is not required for the nuclear import of Bms1p and probably not required either for Bms1p recruitment into pre-ribosomes.
Bms1p and Rcl1p are both recruited and released simultaneously in the pre-ribosome maturation pathway
Since Bms1p is required for the import of Rcl1p into the nucleus, we next investigated the chronology of recruitment and release of each protein in the maturation pathway. To get an overview of the pre-ribosomal particles containing Bms1p and Rcl1p, we purified Rcl1p-TAP or Bms1p-3HA from total cellular extracts and analysed the co-precipitated pre-rRNAs by northern blot. As shown in Figure 6A, the early 35S, 33S/32S and 23S precursors are precipitated above background levels with both Bms1p and Rcl1p. The 22S and 21S precursors are barely detectable in the total extracts, suggesting that these intermediates display a short half-life at steady state as compared to other species. Surprisingly, however, these species are best co-immunoprecipitated with both Rcl1p and Bms1p. Very low levels of 27SA2 intermediate are co-precipitated with Bms1p and Rcl1p suggesting that these proteins are not incorporated into early pre-60S particles following A2 cleavage. In contrast, significant amounts of the 20S precursor are associated with Bms1p and Rcl1p, indicating that both proteins are not immediately released from the particles following A2 cleavage but remain associated to some extent with pre-40S pre-ribosomal particles.
Figure 6.
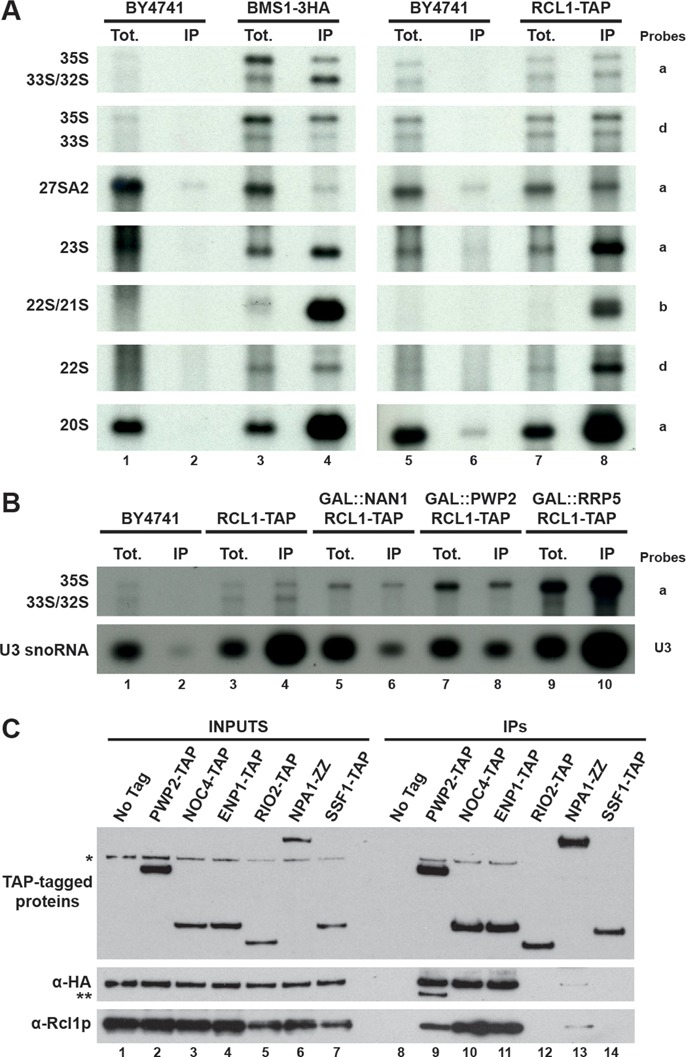
Rcl1p and Bms1p are both recruited and released at similar stages of the pre-ribosome maturation pathway. (A) Pre-rRNAs associated with Rcl1p-TAP or Bms1p-3HA. The pre-ribosomal particles containing Rcl1p-TAP or Bms1p-3HA were immunoprecipitated by affinity chromatography. The associated RNAs were purified and analysed by northern blot using specific probes (indicated on the right) allowing detection of the indicated pre-rRNAs. (B) Association of Rcl1p-TAP with early pre-ribosomal particles in presence or absence of Nan1p (UTP-A module), Pwp2p (UTP-B module) or Rrp5p. Strains GAL::NAN1, GAL::PWP2 and GAL::RRP5 expressing Rcl1p-TAP were shifted from a galactose- to a glucose-containing medium to deplete the corresponding proteins. A strain expressing Rcl1p-TAP but otherwise wild type (WT) was used as a positive control and strain BY4741 (no tag) was used to reveal background immunoprecipitation signals. Total extracts were prepared from these different strains and the pre-ribosomal particles containing Rcl1p-TAP were isolated. The co-immunoprecipitated RNAs were analysed by northern blot using specific probes (mentioned on the right) allowing detection of the indicated pre-rRNAs. (C) Association of Bms1p and Rcl1p with a series of pre-ribosomal particles corresponding to different stages of the maturation pathway. Total extracts were prepared from strains expressing Bms1p-3HA and either Enp1p-TAP, Pwp2p-TAP, Noc4p-TAP, Rio2p-TAP, Npa1p-ZZ, Ssf1p-TAP or otherwise WT and the pre-ribosomal particles containing the TAP-tagged proteins were isolated. The co-immunoprecipitated proteins were analysed by western blot using anti-Rcl1p and anti-HA antibodies to detect endogenous Rcl1p and Bms1p-3HA, respectively. *, Bms1p-3HA signal remaining from the previous incubation with anti-HA antibodies. **, Pwp2p-TAP detected with the anti-HA antibodies.
We next determined whether the prerequisites governing Rcl1p recruitment into pre-ribosomes are the same as those previously described for Bms1p (27). We TAP-tagged Rcl1p in yeast strains expressing Nan1p (UTP-A module), Pwp2p (UTP-B module) or Rrp5p under the control of a galactose promoter and we assessed whether Rcl1p-TAP is physically associated with the early pre-rRNAs upon depletion of these factors (Figure 6B). Depletion of Nan1p (lane 6) or Pwp2p (lane 8) resulted in a significant decrease (by 3-fold) in the immunoprecipitation efficiency of the 35S pre-rRNA with Rcl1p-TAP as compared to the wild-type control (lane 4) and a strong decrease in the co-precipitation of U3 snoRNA. In contrast, no drastic difference in the physical association between Rcl1p and U3 snoRNA is observed upon Rrp5p depletion (lane 10). The association with the 35S pre-rRNA is even increased, suggesting that Rcl1p is recruited into the pre-ribosomal particles in the absence of Rrp5p and remains trapped into the aberrant particles formed due to the defect in Rrp5p function. These data show that recruitment of both Bms1p and Rcl1p into the pre-ribosomal particles requires prior incorporation of the UTP-A and UTP-B modules but occurs independently from Rrp5 recruitment, suggesting that the two proteins are recruited into the particles at a similar stage of the maturation pathway, probably as a complex.
To delineate when Rcl1p and Bms1p are released from the pre-ribosomal particles in the maturation pathway, we purified pre-ribosomal particles at different stages of the maturation pathway by Tandem Affinity Purification (TAP) purifications using different baits and we determined by western blot whether Bms1p and Rcl1p are present within these particles (Figure 6C). For the small subunit biogenesis pathway, the selected baits were Pwp2p-TAP and Noc4p-TAP associated with particles ranging from early 90S pre-ribosomes to early pre-40S particles; Enp1p-TAP recruited within early nucleolar 90S pre-ribosomes and which remains associated with late cytoplasmic pre-40S particles; Rio2p-TAP, present in intermediate pre-40S particles but not in early 90S pre-ribosomes. For the large subunit maturation pathway, we selected Npa1p-ZZ as a component of very early pre-60S particles containing the 27SA2 pre-rRNA and Ssf1p-TAP allowing purification of intermediate pre-60S particles. Bms1p was 3HA-tagged into the strains expressing the different TAP-tagged baits to allow its detection in the final western blot experiments. As shown in Figure 6C, both Rcl1p and Bms1p-3HA are found associated with Pwp2p-TAP (lane 9), Noc4p-TAP (lane 10) and Enp1p-TAP (lane 11), which are all present in early 90S pre-ribosomes and early pre-40S particles. In contrast, neither Rcl1p nor Bms1p-3HA are detected in the intermediate pre-40S particles co-purifying with Rio2p-TAP (lane 12), suggesting that both proteins have been released from the pre-ribosomal particles at that stage of the maturation pathway. Bms1p and Rcl1p are not present within intermediate pre-60S particles containing Ssf1p-TAP (lane 14) and only trace amounts are detected in the very early pre-60S particles co-purifying with Npa1p-ZZ (lane 13). These results indicate that neither Bms1p nor Rcl1p is incorporated into pre-60S particles following A2 cleavage. The weak association of these proteins with Npa1p-ZZ, consistent with the weak co-immunoprecipitation of the 27SA2 intermediate (Figure 6A), may reflect the presence of these proteins in very transient 90S-like particles in which A2 cleavage has just occurred but that did not yet split into pre-60S and pre-40S particles.
To summarize, these data indicate that Bms1p and Rcl1p are both incorporated into the very early pre-ribosomal particles and are both present in the particles containing the pre-rRNA substrates of A2 cleavage. Following A2 cleavage, both proteins are incorporated into pre-40S particles and are subsequently released before the incorporation of Rio2p. Importantly, all the observations made for Rcl1p also hold true for Bms1p suggesting that the proteins behave like a complex all along the maturation pathway.
GTP binding by Bms1p is required for pre-rRNA processing but not for Rcl1p incorporation into pre-ribosomal particles
To investigate the importance of GTP binding to Bms1p, we took advantage of the K82A amino acid substitution in Bms1p that alters the P-loop motif of the nucleotide-binding site (13). This mutation is lethal in vivo and the altered protein shows a reduced ability to bind GTP. Plasmids expressing Bms1pWT, Bms1pK82A or the empty vector as a control were transformed into the strain bearing the chromosomal GAL::BMS1 construct. We studied the localization of Rcl1p-3HA in the resulting strains following transfer to a glucose-containing medium to deplete endogenous Bms1p (Figure 7A). Rcl1p-3HA localizes as expected in the nucleolus in cells expressing Bms1pWT and is partially retained in the cytoplasm in the absence of Bms1p (empty vector) as described above. Surprisingly, Rcl1p-3HA is present in the nucleolus in cells expressing Bms1pK82A. This observation suggests that in vivo, GTP binding to Bms1p is not required for the import of Rcl1p into the nucleus. We then analysed the sedimentation profile of Rcl1p on sucrose gradient (Figure 7B). In cells expressing Bms1pWT, Rcl1p-3HA co-sediments with a variety of pre-ribosomal particles containing the 35S, 23S and 20S pre-rRNAs in fractions 7–18. In the absence of Bms1p (empty vector), a significant fraction of Rcl1p-3HA is detected in the very top fractions of the gradient containing free proteins or small particles. This population of Rcl1p probably corresponds to the pool of proteins that accumulates in the cytoplasm due to defective nuclear import, as observed by microscopy (Figure 7A). In cells expressing Bms1pK82A, the sedimentation profile of Rcl1p-3HA is very comparable to that observed in the wild-type control. Notably, the protein is not detected in the top fractions of the gradient and exclusively co-sediments with the early pre-ribosomal particles. These results strongly suggest that Rcl1p is normally incorporated into the pre-ribosomal particles in cells expressing Bms1pK82A. Using immunoprecipitation experiments (Figure 7C) we observed a clear enrichment of the 35S and 23S pre-rRNAs as well as the U3 snoRNAs in the material co-immunoprecipitated with Rcl1p-3HA using extracts from cell expressing Bms1pK82A (lane 6) as compared to cells lacking Bms1p (empty vector, lane 5). The slightly lower amount of pre-rRNAs and U3 snoRNA co-purifying with Rcl1p3HA from cells expressing Bms1pK82A relatively to the wild-type control (lane 4) is correlated with a lower amount of precipitated Rcl1p-3HA. This does not, therefore, reflect a decreased association with the pre-rRNAs and U3 snoRNA. Interestingly, analysis of the pre-rRNA accumulation profile in cells expressing Bms1pK82A shows a clear defect in the accumulation of the 20S pre-rRNA, indicating that early cleavages of the pre-rRNA are affected in these cells. We propose that GTP binding to Bms1p is not required for the incorporation of the Bms1p–Rcl1p complex into pre-ribosomes but rather for a later stage of the maturation pathway important for pre-rRNA cleavages (see Discussion).
DISCUSSION
Amino acid substitutions in Rcl1p altering the interaction with Bms1p in vitro affect early pre-rRNA cleavages in yeast cells
We report in this study the crystal structure of S. cerevisiae Rcl1p protein bound to a central region of the GTPase Bms1p encompassing residues 547–636. On the basis of this structure, we identified residues of Rcl1p, in particular, C277 and R327, establishing direct contacts with Bms1p. Amino acid substitutions altering R327 (R327A) and C277 (C277R) impair the interaction with Bms1p in vitro. Although we do not have a complete structural picture of the Bms1p–Rcl1p complex due to Bms1p proteolysis, these results suggest that the Bms1p (547–636) domain probably constitutes the only Rcl1p-interacting surface of Bms1p, consistent with the fact that neither the GTPase nor the GAP domains of Bms1p interact on their own with Rcl1p in vitro. To assess the importance of the Rcl1p–Bms1p interaction in vivo, we expressed in yeast cells the C277R, R327A and C277R/R327A altered versions of Rcl1p and we analysed pre-rRNA processing in the resulting mutant cells. We observed that early cleavages at sites A0, A1 and A2 are affected in cells expressing Rcl1p R327A and Rcl1p C277R/R327A and the severity of these phenotypes is consistent with the extent of the interaction defects observed in vitro. Importantly, accumulation of the 33S precursor (resulting from cleavage of the 35S pre-rRNA at site A0 but not at sites A1 and A2) and the 22S pre-rRNA (resulting from 23S precursor cleavage at site A0 but not at sites A1 and A2) in the mutant cells may suggest that not only cleavage at site A2, but also cleavage at site A1, are strongly affected in these mutant strains. This result is consistent with the observation that depletion of Rcl1p induces accumulation of 33S and 22S pre-rRNAs (7). As mentioned in the Introduction, cleavage at site A2 is dispensable for synthesis of the mature 18S rRNA and can be bypassed through cleavages at other sites. Since Rcl1p is essential for production of the mature 18S rRNA, it must have another function in the rRNA processing pathway in addition to its nuclease activity at site A2. Our data and previous results (7) show that Rcl1p is also required for productive A1 cleavage but it is unclear whether this requirement implies a catalytic or a structural function of the protein.
Recruitment and release of Bms1p and Rcl1p during the pre-rRNA maturation pathway
Interfering with the interaction between Rcl1p and Bms1p impairs pre-rRNA processing in vivo. In the case of cells expressing Rcl1pC277A/R327A we observed that the processing defects are correlated with a significant defect in the nuclear import of the altered Rcl1p protein (Supplementary Figure S4) and probably as consequence, with a reduction in its incorporation into the pre-ribosomal particles purified with Pwp2p-TAP, Enp1p-TAP or Noc4p-TAP (Supplementary Figure S7). We showed in addition that upon Bms1p depletion, Rcl1p import into the nucleus is impaired, precluding its recruitment into pre-ribosomes. Therefore, Rcl1p likely associates with Bms1p in the cytoplasm and the two proteins may travel together as a complex through the nuclear pores, although further experiments will be needed to formally demonstrate this co-import. Alternatively, we cannot formally exclude that Bms1p is simply required for the nuclear retention of Rcl1p. The two proteins are then incorporated into the earliest nascent pre-ribosomal particles as attested by their physical association with the 35S, 33S/32S and 23S species. Furthermore, our data combined with previous results (27) indicate that incorporation of both Bms1p and Rcl1p requires prior loading of the tUTP/UTP-A and UTP-B modules but occurs independently of Rrp5p recruitment. Bms1p and Rcl1p are therefore recruited at a similar stage of pre-ribosome assembly, suggesting that they are loaded as a complex into the nascent pre-ribosomal particles.
Interestingly, both Bms1p and Rcl1p co-immunoprecipitate with best efficiencies the 22S and 21S pre-rRNAs. Other components of the SSU processome, such as Noc4p (28) or Utp10p and Utp20p (29), have also been reported to be strongly associated with the 21S and 22S intermediates. The 21S intermediate has been detected at low levels in some conditions in wild-type strains (30). In addition, the 21S pre-rRNA that accumulates in several mutant strains in which A2 cleavage is strongly inhibited (11–12,31) is normally processed at site D to generate the mature 18S rRNA. These results suggest that these intermediates are natural precursors to the mature 18S rRNA in yeast, resulting from minor processing pathways. They probably derive from the 23S pre-rRNA, another minor natural intermediate in yeast cells (30,32) which most likely results either from nascent transcript cleavage (33) at site A3, and/or from post-transcriptional cleavage of the released 35S pre-rRNA (33) at site A3. The strong association of SSU processome components with the 21S and 22S pre-rRNAs as compared to other early precursors, such as the 35S, 32S and 23S intermediates, suggests that these proteins are either more stably associated with the particles containing the 22S or 21S intermediates or more accessible to the affinity matrix within these particles during the immunoprecipitation, or both. These associations could also reflect the presence of SSU processome components within 21S and 22S pre-rRNA-containing particles possibly undergoing some proofreading steps. It is also possible that the 21S and 22S species represent dead-end processing intermediates that transiently accumulate before being degraded by the nucleolar surveillance pathway.
Our results indicate that both Rcl1p and Bms1p are present within the earliest pre-ribosomal particles and remain associated with particles containing the 20S precursor following A2 cleavage. Therefore, not only Rcl1p but also Bms1p are present within pre-ribosomal particles when A2 cleavage occurs, and neither of them is released from the particles immediately after A2 cleavage. It is still unclear how the GTPase Bms1p regulates Rcl1p function (see below) but our results indicate that both proteins seem to function as a complex all the way through during the maturation pathway. We cannot exclude, however, that in some pre-ribosomal particles the two proteins are present but not directly associated.
Role of GTP binding to Bms1p on the function of the protein in vivo
It was hypothesized on the basis of quantitative biochemical analyses that Bms1p functions in the deposition of Rcl1p into pre-ribosomes in a GTP-controlled process (15). Our in vitro results indicate that the interaction between Bms1p (1–705) and Rcl1p does not require GTP addition. This observation has also been made by Karbstein et al., who showed that although GTP increases the affinity of Bms1p for Rcl1p, the two proteins do interact efficiently in vitro without GTP addition (15). However, these in vitro studies were performed with a fragment of Bms1p lacking the C-terminal domain encompassing a predicted GAP domain and proposed to promote GTP hydrolysis of the N-terminal GTPase domain. The influence of GTP on the interaction between Bms1p and Rcl1p in vitro could be different in the presence of the C-terminal domain. We showed here that the putative GAP domain of Bms1p interacts in trans with the isolated GTPase domain in agreement with structure predictions highlighting the clear similarity between Bms1p and SelB.
In mutant cells expressing the GTPase defective version of Bms1p (Bms1pK82A), Rcl1p accumulates in the nucleolus and is found associated with early pre-rRNAs. These results strongly suggest that GTP binding to Bms1p is not required for the interaction with Rcl1p nor for the incorporation of the complex into pre-ribosomes in vivo. The accumulation levels of the 20S pre-rRNA are reduced in these mutant cells indicating that early processing of the pre-rRNA is affected. It was predicted that residues R327/D328/K330 of Rcl1p may be involved in the binding of the RNA substrate 3′ to the cleavage site (10). Here, we show that residue R327 of Rcl1p is directly involved in the interaction with Bms1p, prompting the question of whether Bms1p occludes Rcl1p substrate binding site, preventing premature cleavage at site A2. In the light of our in vivo data suggesting that GTP binding to Bms1p is important for pre-rRNA cleavage, it is tempting to speculate that GTP binding to Bms1p and/or GTP hydrolysis induces a conformational rearrangement within the Bms1p–Rcl1p complex that releases the RNA substrate binding site of Rcl1p thereby allowing pre-rRNA cleavage. Therefore, the role of Bms1p may not be limited to a chaperon activity driving and addressing Rcl1p to pre-ribosomal particles, it may also extend to the regulation of the cleavage event itself.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited to the Protein Data Bank under the accession number 4clq.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and we would like to thank staff members for assistance in using beamline ID14–1 and ID29. We also acknowledge the Synchrotron SOLEIL and Dr Pierre Legrand for providing access to Proxima-1 beamline. We gratefully thank Hélène Rogniaux and the staff of the BIBS platform (INRA, UR1268 Biopolymers Interactions Assemblies F-44316 NANTES) for conducting the mass spectrometry analyses with excellent expertise. We are grateful to the members of the H/H, Gleizes and Gadal groups for advice and helpful discussions.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be considered as Joint First Authors.
FUNDING
ANR Blanc 2010 [RIBOPRE40S to Y.H. and S.F.]; Ligue Nationale Contre le Cancer [to Y.H., équipe labellisée]; INSERM and CNRS. Postdoctoral fellowship from the Association pour la Recherche contre le Cancer (ARC) {to A.D.]. Ph.D. fellowship from the Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche and from the ARC [to Y.A.K.]. Funding for open access charge: INSERM, CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Dragon F., Gallagher J.E., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schafer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 3.Schafer T., Strauss D., Petfalski E., Tollervey D., Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henras A.K., Soudet J., Gerus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woolford J.L., Jr, Baserga S.J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195:643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strunk B.S., Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Billy E., Wegierski T., Nasr F., Filipowicz W. Rcl1p, the yeast protein similar to the RNA 3’-phosphate cyclase, associates with U3 snoRNP and is required for 18S rRNA biogenesis. EMBO J. 2000;19:2115–2126. doi: 10.1093/emboj/19.9.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palm G.J., Billy E., Filipowicz W., Wlodawer A. Crystal structure of RNA 3’-terminal phosphate cyclase, a ubiquitous enzyme with unusual topology. Structure. 2000;8:13–23. doi: 10.1016/s0969-2126(00)00076-9. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka N., Smith P., Shuman S. Crystal structure of Rcl1, an essential component of the eukaryal pre-rRNA processosome implicated in 18s rRNA biogenesis. RNA. 2011;17:595–602. doi: 10.1261/rna.2571811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horn D.M., Mason S.L., Karbstein K. Rcl1 protein, a novel nuclease for 18 S ribosomal RNA production. J. Biol. Chem. 2011;286:34082–34087. doi: 10.1074/jbc.M111.268649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torchet C., Hermann-Le Denmat S. Bypassing the rRNA processing endonucleolytic cleavage at site A2 in Saccharomyces cerevisiae. RNA. 2000;6:1498–1508. doi: 10.1017/s1355838200000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vos H.R., Faber A.W., de Gier M.D., Vos J.C., Raue H.A. Deletion of the three distal S1 motifs of Saccharomyces cerevisiae Rrp5p abolishes pre-rRNA processing at site A(2) without reducing the production of functional 40S subunits. Eukaryotic Cell. 2004;3:1504–1512. doi: 10.1128/EC.3.6.1504-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelperin D., Horton L., Beckman J., Hensold J., Lemmon S.K. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegierski T., Billy E., Nasr F., Filipowicz W. Bms1p, a G-domain-containing protein, associates with Rcl1p and is required for 18S rRNA biogenesis in yeast. RNA. 2001;7:1254–1267. doi: 10.1017/s1355838201012079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karbstein K., Jonas S., Doudna J.A. An essential GTPase promotes assembly of preribosomal RNA processing complexes. Mol. Cell. 2005;20:633–643. doi: 10.1016/j.molcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D Biol. Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 18.Vonrhein C., Blanc E., Roversi P., Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 19.Langer G., Cohen S.X., Lamzin V.S., Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bricogne G., Blanc E., Brandl M., Flensburg C., Keller P., Paciorek W., Roversi P., Smart O.S., Vonrhein C., Womack T. BUSTER, version 2.8.0. Cambridge, United Kingdom: Global Phasing Ltd.; 2009. [Google Scholar]
- 21.Bonnart C., Gerus M., Hoareau-Aveilla C., Kiss T., Caizergues-Ferrer M., Henry Y., Henras A.K. Mammalian HCA66 protein is required for both ribosome synthesis and centriole duplication. Nucleic Acids Res. 2012;40:6270–6289. doi: 10.1093/nar/gks234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerus M., Bonnart C., Caizergues-Ferrer M., Henry Y., Henras A.K. Evolutionarily conserved function of RRP36 in early cleavages of the pre-rRNA and production of the 40S ribosomal subunit. Mol. Cell. Biol. 2010;30:1130–1144. doi: 10.1128/MCB.00999-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leibundgut M., Frick C., Thanbichler M., Bock A., Ban N. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marneros A.G. BMS1 is mutated in aplasia cutis congenita. PLoS Genet. 2013;9:e1003573. doi: 10.1371/journal.pgen.1003573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka N., Smith P., Shuman S. Structure of the RNA 3’-phosphate cyclase-adenylate intermediate illuminates nucleotide specificity and covalent nucleotidyl transfer. Structure. 2010;18:449–457. doi: 10.1016/j.str.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez-Fernandez J., Martin-Marcos P., Dosil M. Elucidation of the assembly events required for the recruitment of Utp20, Imp4 and Bms1 onto nascent pre-ribosomes. Nucleic Acids Res. 2011;39:8105–8121. doi: 10.1093/nar/gkr508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn H., Hierlmeier T., Merl J., Jakob S., Aguissa-Toure A.H., Milkereit P., Tschochner H. The Noc-domain containing C-terminus of Noc4p mediates both formation of the Noc4p-Nop14p submodule and its incorporation into the SSU processome. PLoS ONE. 2009;4:e8370. doi: 10.1371/journal.pone.0008370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dez C., Dlakic M., Tollervey D. Roles of the HEAT repeat proteins Utp10 and Utp20 in 40S ribosome maturation. RNA. 2007;13:1516–1527. doi: 10.1261/rna.609807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallagher J.E., Baserga S.J. Two-hybrid Mpp10p interaction-defective Imp4 proteins are not interaction defective in vivo but do confer specific pre-rRNA processing defects in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:1404–1413. doi: 10.1093/nar/gkh318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torchet C., Jacq C., Hermann-Le Denmat S. Two mutant forms of the S1/TPR-containing protein Rrp5p affect the 18S rRNA synthesis in Saccharomyces cerevisiae. RNA. 1998;4:1636–1652. doi: 10.1017/s1355838298981511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunbar D.A., Wormsley S., Agentis T.M., Baserga S.J. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol. Cell. Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kos M., Tollervey D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell. 2010;37:809–820. doi: 10.1016/j.molcel.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]


