Abstract
Trinucleotide repeats are microsatellite sequences that are polymorphic in length. Their expansion in specific genes underlies a number of neurodegenerative disorders. Using ultraviolet-visible, circular dichroism, nuclear magnetic resonance (NMR) spectroscopies and electrospray ionization mass spectrometry, the structural preferences of RNA molecules composed of two and four repeats of AGG, CGG and UGG in the presence of K+, Na+ and NH4+ were analysed. (AGG)2A, (AGG)4A, p(UGG)2U and p(UGG)4U strongly prefer folding into G-quadruplexes, whereas CGG-containing sequences can adopt different types of structure depending on the cation and on the number of repeats. In particular, the two-repeat CGG sequence folds into a G-quadruplex in potassium buffer. We also found that each G-quadruplex fold is different: A:(G:G:G:G)A hexads were found for (AGG)2A, whereas mixed G:C:G:C tetrads and U-tetrads were observed in the NMR spectra of G(CGG)2C and p(UGG)2U, respectively. Finally, our NMR study highlights the influence of the strand sequence on the structure formed, and the influence of the intracellular environment on the folding. Importantly, we highlight that although potassium ions are prevalent in cells, the structures observed in the HeLa cell extract are not always the same as those prevailing in biophysical studies in the presence of K+ ions.
INTRODUCTION
Guanine-rich DNA and RNA sequences can fold into inter- and intramolecular structures called G-quadruplexes. The fundamental unit of the G-quadruplex is a G-tetrad composed of a planar array of four guanine bases. G-quadruplex formation requires the presence of monovalent cations, in particular, K+ or Na+ (1–4). G-quadruplexes reveal remarkable thermodynamic and kinetic stability resulting from stacking interactions, hydrogen bonding, coordination of cations between the adjacent tetrads and hydration (5). The final topology of G-quadruplexes does not depend solely on their primary sequence, but is greatly affected by environmental conditions (6–9). Recently, many G-quadruplex structures have been reported to contain, apart from G-tetrads, other homo-tetrads (A-, T-, C- or U-tetrads) or mixed tetrads (G:C:G:C, G:T:G:T or A:T:A:T) (10). It has been demonstrated that even more than four nucleobases can be associated into a planar array of H-bonds, forming pentads, hexads, heptads or octads (10–14).
G-rich sequences occur in several biologically important DNA regions, such as telomeres, promoter regions of some human oncogenes and some variable number tandem repeats (15). Additionally, recent reports clearly indicate their existence in vivo (16,17). Also, a number of well-annotated transcripts of human genes have already been shown to contain G-rich sequences (18). Many more are still to be identified within the non-coding part of the transcriptome, which has been shown to cover over two-thirds of the human genome (19). Therefore, RNA G-quadruplexes are likely to be involved in various biological processes. For instance, it was shown that the G-quadruplex structures are implicated in specific protein binding and modulation of gene expression by regulating splicing, polyadenylation, translation and transcript turnover (20).
The trinucleotide repeats (TNRs) belong to a subclass of microsatellite sequences also known as short tandem repeats. TNRs represent the most abundant type of simple sequence repeats in eukaryotic genomes and in transcripts (21,22). Some types of TNRs are widespread in exons while others are very rare, suggesting that their occurrence in exons is not random, but undergoes positive or negative selective pressure. These sequences have received special attention, primarily because some are known to undergo pathogenic expansions that cause triplet repeat expansion diseases (TREDs). More than 20 genetic disorders belong to this group, and most are neurodegenerative and neuromuscular disorders (23).
In several TREDs, stable RNA structures formed by triplet repeats present in untranslated regions of the responsible genes are implicated in pathogenesis (24). Of interest to us are RNA sequences composed solely of AGG, CGG and UGG repeats, with a potential to fold into a G-quadruplex structure. It was determined that AGG and CGG repeats are overrepresented in the human exonic sequences, whereas UGG repeats have been found to be underrepresented. TNRs occurring in different mRNA regions may have different functions. It was shown that AGG-rich sequences occur mainly in the open reading frames (ORF), that CGG repeats are most frequent in the 5′-UTR and that UGG TNRs appear mainly in the 3′-UTR (22). The CGG triplet repeats that have been found within the 5′-UTR of the FMR1 gene are polymorphic in length in a normal population and may undergo pathogenic expansions being responsible for both fragile X-associated tremor/ataxia syndrome (25) and fragile X syndrome, which is the most common inherited form of the mental impairment (26).
An interesting insight into the structural diversity of RNA triplet repeats was published recently (24,27). It was demonstrated that oligoribonucleotides composed of 17 AGG or UGG repeats form G-quadruplexes, while CGG repeats have been found to form hairpins with a tendency to adopt several alternative alignments (27). Recently, a crystal structure of molecule composed of two CGG repeats has been determined (28), in which G(CGG)2C forms an A-type duplex with typical C:G Watson–Crick interactions and the non-canonical G·G pairs. A sequence comprising three GGC repeats was studied by nuclear magnetic resonance (NMR) spectroscopy and a duplex structure was proposed with G·G mismatches intercalated between the adjacent G:C pairs (29). In contrast, there is also evidence that CGG repeats can fold into a G-quadruplex (30–33), so the folding rules are yet unclear and may depend on the exact sequence and experimental conditions.
Tetramolecular and bimolecular G-quadruplex structures containing trinucleotide repeats have also interesting applications as biological therapeutics. An RNA aptamer containing four repeats of GGA has been isolated against a recombinant bovine prion protein and its structure was determined by NMR spectroscopy (34). It was found that two (GGA)4 G-quadruplexes form a dimer stabilized through a stacking interaction between A:(G:G:G:G):A hexad planes of the two G-quadruplexes. Although experimental data available in the literature on RNA UGG repeats underline their tendency to form G-quadruplex structures (27,35), to the best of our knowledge there is yet no detailed structural information on RNA sequences containing UGG repeats.
In the present study, we have focused on the structural preferences of RNA molecules composed of two and four repeats of AGG, CGG and UGG in the presence of K+, Na+ and NH4+ ions. We employed ultraviolet (UV) molecular absorption, circular dichroism (CD), NMR spectroscopies and electrospray ionization mass spectrometry (ESI-MS) to obtain comprehensive information on the structural forms existing in different solution conditions. To maintain comparable conditions between different experimental methods, we have recorded the 1H NMR spectra using the same solutions as for the CD and UV experiments. Only the ESI-MS could be carried out only in NH4+. Additionally, to complement our studies, we have tested the effect of intracellular environment on the folding of analysed molecules. In order to mimic the physiologically relevant conditions the 1H NMR spectra were acquired in the HeLa cell extract.
MATERIALS AND METHODS
RNA sample preparation
All oligoribonucleotides: (AGG)2A, (AGG)4A, G(CGG)2C, G(CGG)4C, p(UGG)2U and p(UGG)4U were chemically synthesized on the Applied Biosystems DNA/RNA synthesizer using β-cyanoethyl phosphoramidite chemistry on solid support (36–38). The samples preparation procedure can be found in the Supplementary Data.
Our earlier experience with molecules composed of AGG, CGG and UGG repeats have shown that occasionally their spectroscopic features can be improved adding 5′-phosphate group. The role of 5′-phosphate has been discussed several times in the context of the G-quadruplex structures (39,40). In this study, this type of modification was used only for UGG repeats, because it improved the quality of the 1H NMR spectra.
Electrospray mass spectrometry
ESI-MS experiments were carried out with a Synapt HDMS (Waters, Manchester, UK) with electrospray ionization in the negative ion mode (direct infusion at 4 μl/min). Sodium and potassium ions were not used in ESI-MS experiments because they condense around negatively charged nucleic acid backbones in solution and during solvent droplet evaporation, without a possibility of clean-up by evaporation once in the gas phase. To overcome this problem, we used NH4+ ions, which can stabilize G-quadruplexes by coordination in-between G-tetrads in a similar way as K+ ions (41), but can evaporate as ammonia when bound to external phosphate sites (42–44). The stock solution was prepared from 500 μM RNA (single strand concentration) in 150 mM NH4OAc. All the samples were annealed by heating at 90°C for 5 min and then slowly cooled down to the room temperature and stored at 4°C at least one day before the experiments. The final injection concentration of RNA (single strand) was 18 μM (prepared from 500 μM) in 150 mM NH4OAc. About 1 h before measurement methanol was added to the final concentration 10% v/v. A fixed concentration of dT6 molecule (as an intensity reference) was also present in the sample. The part where ions are most activated is upon entering the IMS cell, because a bias voltage is required for the ions to enter the higher-pressure region of the cell. Therefore, the mass spectra were systematically recorded at different bias voltages, from 15 to 25 V, and the number of ammonium ions remaining bound to the RNA multistranded structures was monitored. Typically, ammonium ions persisting at up to 25 V bias voltage indicate very tight binding sites in-between G-tetrads. For more experimental details see the Supplementary Data.
UV thermal denaturation curves
Thermal denaturation curves were obtained at 295 nm with JASCO V-650 spectrophotometer using quartz optical cuvettes of 0.5 and 1 cm path lengths with the sample volumes of 150 and 300 μl, respectively. The samples were protected against evaporation by the silicone oil. Before measurements the cuvettes filled with samples were spun at 5000 revolutions per minute by 3 min in order to avoid air bubbles formation during measurements. The temperature range was 5–93°C, using the scan rate 0.5°C min−1.
Thermal difference spectra (TDS)
The TDS were obtained by subtracting the UV spectra recorded above and below the melting temperature (90°C and 20°C, respectively). TDS were recorded in the range between 220 and 340 nm on JASCO V-650 spectrophotometer using quartz optical cuvettes of 0.5 and 1 cm path lengths with the sample volumes of 150 and 300 μl, respectively.
Circular dichroism
The JASCO J815 spectropolarimeter equipped with a Peltier temperature controller was used to collect CD spectra. A quartz cuvettes with a path length 0.5 and 1 cm were used with the sample volumes of 1300 and 2500 μl, respectively. Spectra were collected in the range between 220 and 340 nm at 25°C, from three scans and a buffer baseline was subtracted from each spectrum. CD was expressed as the difference in the molar absorption of the right- and left-handed circularly polarized light. Δϵ, in units of cm2 mmol−1, was normalized to the number of nucleoside residues in the RNA samples (45).
Nuclear magnetic resonance
The 1D 1H NMR spectra as well as 2D homo- and hetero-nuclear spectra were collected on a 700 MHz Bruker AVANCE III spectrometer, equipped with a QCI CryoProbe. The 5 mm thin wall tubes were used with the final sample volume of 600 μl (90% H2O/10% D2O, v/v). The 1H NMR spectra were collected with water suppression using excitation sculpting scheme with gradients (46), typically from 20 480 scans with 1.0 s relaxation delay. Experiments were performed at 25°C for AGG and UGG, whereas for G(CGG)2C and G(CGG)4C NMR spectra were recorded at 10°C in order to observe imino proton resonances from less thermodynamically stable forms, which were not observed at 25°C. The 1H-1H NOESY and 1H-15N HSQC spectra were collected for selected samples. Experimental details are given in the Supplementary Data. Spectra were processed and prepared with TopSpin 3.0 Bruker Software.
NMR studies in HeLa cell extract
HeLa cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), 1× RPMI-1640 vitamin solution and 1× antibiotic antimycotic in 5% CO2 for 48 h. In the next step, cells were trypsinized and subsequently washed three times with phosphate buffered saline to remove any remaining trypsin and medium. Cell extract was prepared by sonication of 1.6 × 107 cells in 1 ml deionized water with subsequent centrifugation at 20 000 × g, 5 min. For the NMR experiments 0.15–0.5 mM RNA samples were dissolved in 180 μl HeLa cell extract and 20 μl D2O. The 3 mm thin wall sample tubes were used with the final sample volume of 200 μl. At those RNA concentrations, 256–2048 scans were usually sufficient to obtain a good signal-to-noise ratio. All spectra were recorded immediately after dissolving in HeLa extract and after 24 h. In all cases, the spectra remained unchanged, indicating that RNA samples were stable in HeLa cell extract.
RESULTS
(AGG)2A and (AGG)4A fold into G-quadruplex structures
The melting profiles of (AGG)2A and (AGG)4A obtained at 295 nm in the presence of 50 mM K+, 150 mM NH4+ or 150 mM Na+ were typical of G-quadruplexes, with a decrease of absorbance upon melting (Figure 1A, Supplementary Figure S1A and B) (47). The TDS of (AGG)2A and (AGG)4A shown in Supplementary Figure S2A and B, respectively, also display a negative band at 295 nm in all conditions and support G-quadruplex formation (48). All these profiles are similar, with the exception of the TDS of (AGG)2A obtained in the presence of sodium ions for which a slight shift of the band at ∼250 nm toward a higher wavelength was observed (Figure 1B).
Figure 1.
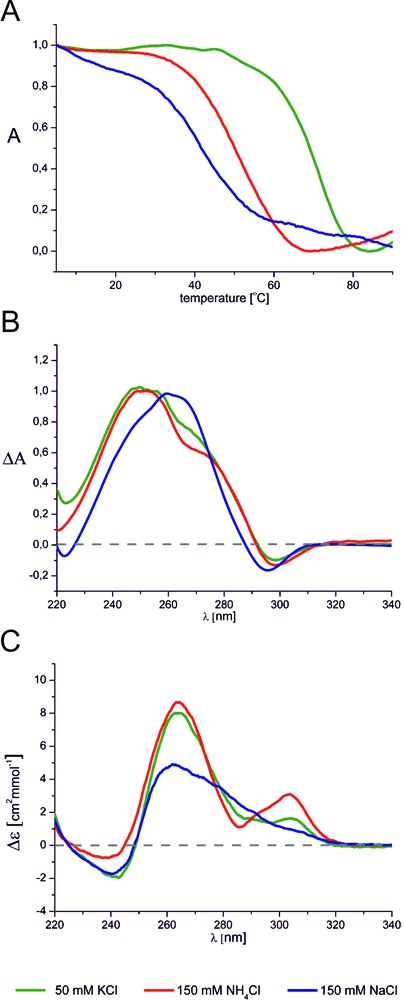
(A) Normalized UV melting profiles of (AGG)4A at 295 nm, (B) normalized thermal differential spectra of (AGG)2A and (C) CD spectra of (AGG)2A at 25°C. All the spectra were recorded in the presence of 50 mM KCl, 10 mM potassium phosphate and 0.1 mM EDTA, pH 6.8 (green), 150 mM NH4Cl, 10 mM Tris/HCl, 0.1 mM EDTA, pH 6.8 (red) and 150 mM NaCl, 10 mM sodium phosphate, 0.1 mM EDTA, pH 6.8 (blue).
The use of CD spectroscopy to determine the topology of DNA G-quadruplexes is well established (49) and was applied here. Note, however, that its use in studies of RNA G-quadruplexes can be problematic because CD spectra of the DNA and RNA parallel G-quadruplexes are very similar to those of A-form duplexes. The CD spectra recorded for (AGG)2A and (AGG)4A in the presence of K+, Na+ and NH4+ ions have positive bands at ∼265 nm and negative bands at 240 nm (Supplementary Figure S3A and B), a pattern characteristic of a parallel G-quadruplex. In a solution containing K+ or NH4+ ions, an additional positive band of small intensity was detected around 305 nm. This band was previously observed in CD spectra of other RNA G-quadruplexes and was attributed to the formation of a dimer of G-quadruplexes stabilized by hexad–hexad stacking (50,51). The shape of the CD spectrum of (AGG)2A in solution containing Na+ ions was different (Figure 1C). That observation, together with the observed shift of the maximum TDS band, suggest the formation of a G-quadruplex with a different topology.
We then established the molecularity of the G-quadruplexes as well as the number of the adducted ammonium ions preserved in the structure using negative ion mode ESI-MS. For parallel-stranded structures, ammonium ion preservation in the gas phase is easier than for antiparallel structures (42–44). As the structures formed here are parallel-stranded both in ammonium and in potassium, we are in a favourable case where the ammonium ion count by ESI-MS allows us to deduce the number of adjacent stacked tetrads. The ESI-MS spectrum of (AGG)2A shows two significant peaks (Supplementary Figure S4A). The peak at m/z 1151.86 corresponds to a monomer at charge state 2- without adducted ammonium ions, whereas the peak at m/z 1859.57 corresponds to the tetramolecular G-quadruplex structure at charge state 5- with three, four or five trapped ammonium ions. The distribution of ammonium ion adducts is presented in the insert of Supplementary Figure S4A. The ESI-MS spectrum of (AGG)4A is presented in Figure 2. Only one peak at m/z 1747.82 and charge state 5- was observed, corresponding to the bimolecular G-quadruplex structure stabilized by three ammonium ions, suggesting the contiguous stacking of four tetrads.
Figure 2.
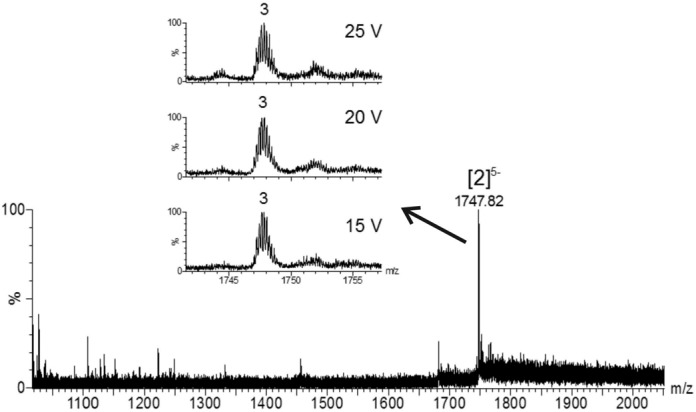
Electrospray mass spectrum of (AGG)4A in the presence 150 mM ammonium acetate. The final single strand RNA concentration was 18 μM. 10% v/v of methanol was added 1 h before measurement. The peak annotation [n]z- indicates the number of strands (n) and the total charge (z). The distribution of the number of ammonium ions preserved in the structure is shown in the inset. The distribution is presented at three bias voltages.
Another method that we used to assess G-quadruplex formation was NMR spectroscopy. We have used the same RNA solutions for NMR measurements as for CD and UV experiments. Consequently, the very low concentration of NMR samples (0.01 mM) entailed long acquisition times, typically about 14 h, in order to obtain a satisfactory signal-to-noise (S/N) ratio. Additionally, we have recorded 1H NMR spectra at higher concentrations for each RNA sequence, in order to determine the effect of strand concentration on RNA structure. A comparison of 1H NMR spectra of (AGG)2A recorded in solutions containing K+, Na+ or NH4+ ions is depicted in Figure 3A (a–c). A spectrum recorded in K+ solution revealed the presence of two major and one minor forms. However, when the sample concentration was increased (1.14 mM) only one of these two major species was observed (Figure 3Ad). In NMR spectra of both concentrated and diluted samples, a characteristic signal appeared around 10 ppm. Signals in this region were previously observed in the NMR spectra of oligoribonucleotides containing an A:(G:G:G:G):A hexad motif (Figure 4A) and were assigned to guanosine amino protons involved in hydrogen bonding with adenosine (34,51,52). We recorded the 1H-15N HSQC spectrum for concentrated samples, in order to confirm that this high-field signal is due to amino proton (Supplementary Figure S5). The imino and amino protons can be easily distinguished based on the analysis of 15N chemical shifts in 1H-15N HSQC spectra (53). Indeed, the chemical shift of the nitrogen atom corresponding to the proton at 10.14 ppm allowed us to unambiguously attribute this signal to the guanosine amino group. Finally, taking into account the results from the ESI-MS, we assumed that (AGG)2A folds into a dimer of bimolecular G-quadruplexes (Figure 4E), similar to that previously published (51).
Figure 3.
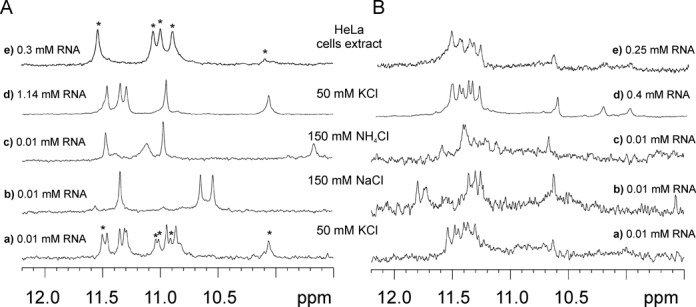
Imino region of the 1H NMR spectra of (A) (AGG)2A and (B) (AGG)4A at 25°C in the presence of 50 mM KCl, 10 mM potassium phosphate and 0.1 mM EDTA, pH 6.8 (Aa, Ba, Ad, Bd), 150 mM NaCl, 10 mM sodium phosphate, 0.1 mM EDTA, pH 6.8 (Ab, Bb), 150 mM NH4Cl, 10 mM Tris/HCl, 0.1 mM EDTA, pH 6.8 (Ac, Bc) and the HeLa cell extract (Ae, Be). The poor S/N ratio observed for (AGG)4A is due to polymorphism or/and the presence of higher order structures.
Figure 4.
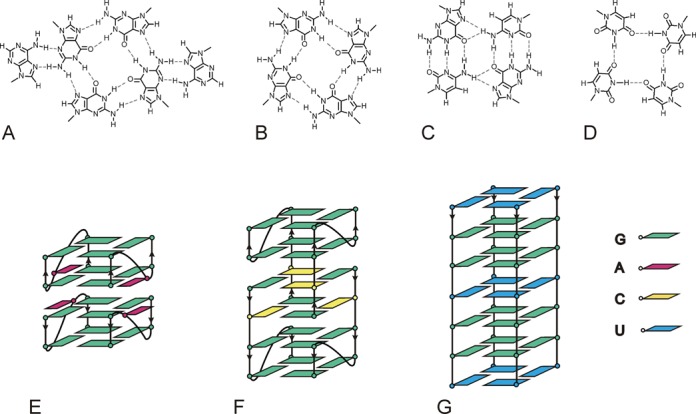
Schematic outline of (A) A:(G:G:G:G):A hexad, (B) G-tetrad, (C) G:C:G:C tetrad, (D) U-tetrad motifs and fold of (E) (AGG)2A, (F) G(CGG)2C and (G) p(UGG)2U.
In the presence of NH4+ ions the spectrum indicative of a single G-quadruplex form was acquired (Figure 3Ac). The high-field resonance characteristic of the hexad motif also occurred, suggesting that (AGG)2A adopts a similar fold to that obtained in the potassium buffer. In the NMR spectrum of (AGG)2A in a buffer containing Na+ ions (Figure 3Ab), only three major imino resonances were visible, two of which were shifted high-field in respect to the spectra recorded in K+ and NH4+ solutions. The absence of characteristic guanosine amino resonance around 10 ppm, together with the observed shift of a positive band in the TDS spectrum and the different shape of the CD spectrum, suggested that (AGG)2A folds differently in the presence of Na+ ions. Next, we have performed a study in the HeLa cell extract, to find out how the intracellular environment could influence the (AGG)2A folding. Since the 1H NMR spectrum of such an extract does not contain any signals in the region above 9.5 ppm, therefore, this model system is very convenient to monitor RNA structures. The 1H NMR spectrum recorded in the HeLa cell extract is presented in Figure 3Ae. The number of proton resonances indicated the formation of a single G-quadruplex form. However, the spectrum was different from those recorded in other conditions. The chemical shifts of the observed imino protons were similar to that of the minor form (marked by *) present in the 1H NMR spectrum recorded in the potassium buffer at a low concentration of RNA (Figure 3Aa,e).
The 1H NMR spectra of diluted and concentrated (AGG)4A samples obtained in the presence of K+ ions are similar (Figure 3Ba,d) and resemble that previously published for (GGA)4 (34), suggesting the formation of the dimer of unimolecular subunits including the A:(G:G:G:G):A hexad (Figure 4E). It is difficult to interpret the NMR spectrum of (AGG)4A obtained in solution containing NH4+ ions (Figure 3Bc), because of the lack of spectral resolution and the extensive line broadening leading to a low signal-to-noise ratio. Inspection of Figure 3Bb implies that the major structure of (AGG)4A in buffer containing sodium ions can be different from that existing in potassium and ammonium buffers, which together with results obtained for (AGG)2A suggests that the presence of sodium ions can induce a different fold in the case of AGG-rich sequences. Finally, the spectra of (AGG)4A obtained in the HeLa cell extract and potassium buffer were virtually identical, regardless of the RNA concentration (Figure 3Be).
Structural preferences of G(CGG)2C and G(CGG)4C
The same set of experimental methods was applied to the other sequences. Supplementary Figure S1C and D show the UV melting curves obtained at 295 nm for G(CGG)2C and G(CGG)4C, respectively, in the presence of K+, NH4+ and Na+ ions. Here, we observed melting profiles typical of G-quadruplexes only in the presence of K+ ions. In the melting curve of G(CGG)4C, besides the decrease of absorbance visible at higher temperatures, an additional, low temperature transition with absorbance increase occurred in K+ solution. This observation suggested that other types of structure could co-exist with the G-quadruplex. No features characteristic of the G-quadruplexes were detected in the melting curves of G(CGG)2C and G(CGG)4C obtained in sodium and ammonium-containing buffers.
To get more insight into the type of structures which were formed in the sodium and ammonium buffers, we analysed the melting curves at 260 nm (Supplementary Figure S6). The concentration dependence of melting temperature (Tm) of G(CGG)2C was characteristic of melting profile of bimolecular form, such as a duplex (Supplementary Figure S7). The sigmoidal shape of the profiles at 260 nm obtained for G(CGG)4C in the ammonium and sodium buffers suggested that this molecule can fold into a duplex or hairpin structure. TDS of G(CGG)2C and G(CGG)4C are shown in Supplementary Figure S2C and D, respectively. A negative band at 295 nm was found only when potassium ions were present in solution. The shapes of the CD spectra of both molecules were similar, except for the spectrum of G(CGG)2C recorded in potassium buffer (Supplementary Figure S3C and D).
The ESI-MS spectrum of G(CGG)2C shown in Supplementary Figure S4C revealed the presence of bimolecular and tetramolecular structures. The major peak (100%) at m/z 1288.89 and minor peak at m/z 1718.85 correspond to a bimolecular structure at charge states 4- and 3-, respectively. The absence of specifically bound ammonium ion adducts suggests the absence of stacked G-tetrads, and hence that this bimolecular structure may be stabilized by Watson–Crick base pairs instead of G-tetrads. Another minor peak at m/z 1724.54 corresponding to the tetramolecular structure at charge state 6- appeared, in this case with adducted ammonium ions. The ammonium ion distribution was monitored with the increase of the bias voltage and is shown in the insert of Supplementary Figure S4C, and the biased distribution toward two coordinated ammonium ions suggests that this tetramolecular structure, in contrast to the bimolecular one, contains stacked G-tetrads. In the ESI-MS spectrum of G(CGG)4C (Supplementary Figure S4D) the main peak observed at m/z 1141.49 and the minor peak at m/z 1522.42 correspond to a monomolecular form without adducted ammonium ions at charge state 5- and 3-, respectively. An additional, less intense peak corresponding to a bimolecular structure was observed at m/z 1844.32 (charge 5-). The distribution of ammonium adducts in this structure at low voltages is broad as a result of the mild desolvation conditions and makes it difficult to conclude whether or not different structures (G-quadruplex with labile ammonium ions (43) and non-G-quadruplex) coexist.
Figure 5Aa represents an 1H NMR spectrum of G(CGG)2C in the presence of K+ ions. In these conditions, four imino resonances appear in the region distinctive of G-quadruplex structures and one guanosine imino proton resonates at 12.44 ppm, a region characteristic of Watson–Crick G·C base pairs. It has been previously demonstrated that the presence of 5′-GC ends can lead to the association of two G-quadruplex moieties through G·C base pairs and the formation of two G:C:G:C-tetrads (54). Overall, the observation of the inverted UV melting profile at 295 nm and a negative band at this wavelength in the TDS spectrum, together with the analysis of the ESI-MS and 1H NMR spectra indicated the formation of a highly symmetric tetramolecular G-quadruplex structure with G:G:G:G and mixed G:C:G:C tetrads (Figure 4B, C and F) in the K+-containing solution.
Figure 5.
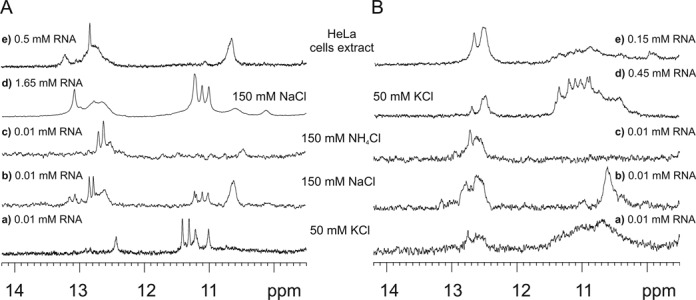
Imino regions of the 1H NMR spectra of (A) G(CGG)2C and (B) G(CGG)4C at 10°C in the presence of (Aa, Ba, Ad, Bd) 50 mM KCl, 10 mM potassium phosphate and 0.1 mM EDTA, pH 6.8, (Ab, Bb) 150 mM NaCl, 10 mM sodium phosphate, 0.1 mM EDTA, pH 6.8, (Ac, Bc) 150 mM NH4Cl, 10 mM Tris/HCl, 0.1 mM EDTA, pH 6.8 and (Ae, Be) HeLa cell extract. The poor S/N ratio observed for G(CGG)4C is due to polymorphism or/and the presence of higher order structures.
The 1H NMR spectrum of G(CGG)2C obtained in Na+ solution revealed the presence of a mixture of conformers (Figure 5Ab). Although neither the UV melting analysis at 295 nm nor the TDS spectrum indicated the formation of a G-quadruplex under the Na+ environment, the appearance of four sharp signals in the 11.01–11.22 ppm region of the 1H NMR spectrum revealed that a minor population of the G-quadruplex form also exists in these conditions. We attributed resonances of the major form in the 12.5–13.2 ppm region to the G:C base pairs, and the broad signal at 10.61 ppm to a G·G mismatch (Figure 5Ab). Subsequently, we have studied the effect of oligoribonucleotide concentration on the G-quadruplex-duplex equilibrium in the sodium buffer. When the RNA concentration was increased, signals corresponding to the duplex diminished notably, and the G-quadruplex emerged as the dominant form (Figure 5Ad). The observed down-field shift of G:C imino resonance relative to that in the presence of K+ could be related to the deshielding effect of the ion, or could reflect different metal binding sites within the G:C:G:C tetrad (55). In the ammonium buffer, only two sharp imino resonances at 12.63 and 12.71 ppm were visible and no signals corresponding to the G-quadruplex form could be determined (Figure 5Ac).
The dependence of the 1H NMR spectra of G(CGG)4C on the solution conditions is presented in Figure 5B. Severe broadening of imino resonances was observed in a buffer containing potassium ions, indicating the coexistence of several species. We recorded 1H NMR spectra as a function of the temperature in order to verify whether these species differ in thermal stability. Because the 1H NMR spectra of diluted (0.01 mM) and concentrated (0.45 mM) samples were similar (Figure 5Ba,d), we performed this experiment on the most concentrated sample (Supplementary Figure S8). Imino resonances at about 12.7 ppm and a broad signal at 10.65 ppm diminished upon heating and vanished at 50ºC. On the other hand, resonances around 11.4 ppm sharpened upon heating and remained very intense even at 50ºC, a behaviour that is typical of G-quadruplexes. When the 1H NMR spectrum of G(CGG)4C was recorded in the presence of Na+ ions, only signals representing a hairpin (or duplex) form were detected (Figure 5Bb). No resonances corresponding to a G-quadruplex form were observed in buffer containing NH4+ ions either (Figure 5Bc). The ESI-MS spectrum of G(CGG)4C corresponds to the monomolecular form without adducted ammonium ions (Supplementary Figure S4D). This stoichiometry suggests that a hairpin, and not a duplex, is the preferred form of G(CGG)4C.
In the NMR spectrum of G(CGG)2C recorded in the HeLa cell extract, only signals characteristic of Watson–Crick base pairs appeared, demonstrating that the duplex is the dominant structure (Figure 5Ae). In the NMR spectrum of G(CGG)4C, the hairpin is the major form, even though a small population of G-quadruplex could also be detected (Figure 5Be).
G-quadruplex is the preferred form of p(UGG)2U and p(UGG)4U
Inverted UV melting profiles observed at 295 nm for p(UGG)2U and p(UGG)4U in the presence of K+, Na+ and NH4+ indicated the formation of G-quadruplexes (Supplementary Figure S1E and F). The tendency of these molecules to form G-quadruplexes was further confirmed by the presence of the negative band at 295 nm observed in the TDS (Supplementary Figure S2E and F). All CD profiles of p(UGG)2U were similar (Supplementary Figure S3E), including the presence of a small negative band at ∼290 nm. The CD shape of p(UGG)4U obtained in ammonium buffer (Supplementary Figure S3F, red) differed from that recorded in potassium and sodium buffers (Supplementary Figure S3F, green and blue curves, respectively). In addition to the positive band at ∼264 nm, a relatively intense positive band at ∼290 nm and the absence of the negative band at 240 nm are clearly notable.
We have observed two types of peaks in ESI-MS spectra of p(UGG)2U. The peak at m/z 1157.86 corresponding to a single strand at charge state 2-, and two peaks at m/z 1555.31 and 1870.34 corresponding to charge states 6- and 5- of a tetramolecular structure with four or five ammonium ion adducts (Supplementary Figure S4E). The evolution of the ammonium ion distribution upon increasing bias voltage for both peaks (shown in the insert of Supplementary Figure S4E) revealed that the structure stabilized by four ions were the most stable. In the ESI-MS spectrum of p(UGG)4U (Supplementary Figure S4F) only one peak at m/z 1733.50 was observed, corresponding to a bimolecular structure at charge state 5-, stabilized by three ammonium ions.
The 1H NMR spectra of p(UGG)2U obtained in solutions containing K+, Na+ and NH4+ (Figure 6A) provided additional evidence for a G-quadruplex formation. In the 1H NMR spectrum of p(UGG)2U recorded in the presence of K+ (Supplementary Figure S9), several resonances sharpened with the increase in temperature and were visible even at 80°C. Additionally, new resonances appeared at 3°C. To elucidate their origin and, in particular, to distinguish guanosine from uridine imino protons, we acquired a 1H –15N HSQC spectrum at 3°C. Four guanosine and three uridine imino protons were identified based on the chemical shifts of the attached nitrogen (Supplementary Figure S10). The appearance of uridine imino signals suggested the formation of U-tetrads (56). To verify whether the uridine residues constitute part of the G-quadruplex architecture, we assigned all guanosine and uridine imino protons based on the analysis of a 2D NOESY spectrum (data not shown). Each of the three observed uridine imino resonances exhibited a strong NOE to H5 and much weaker one to H6 protons (Supplementary Figure S11), a pattern that is distinctive of a U-tetrad motif (55). The 3′-end uridine imino proton at 11.08 ppm and that at 9.85 ppm, assigned to the internal uridine residue, sharpened with increasing temperature and were still observed at a high temperature, revealing the formation of two stable U-tetrads. Although there is no NMR structure known with an internal U-tetrad, a similar shift ∼9.5 ppm was observed for imino protons of the thymidine tetrad accommodated in the centre of the DNA G-quadruplex (13,57,58). The 5′-end uridine imino proton at 10.74 ppm was observed only below 25°C (Supplementary Figure S9). These results, together with ESI-MS data (four very stable cation binding sites and one additional, more labile binding site in the tetramer), indicate that p(UGG)2U folds into a symmetrical tetramolecular parallel G-quadruplex with four G-tetrads and three U-tetrads (Figure 4B, D and G).
Figure 6.
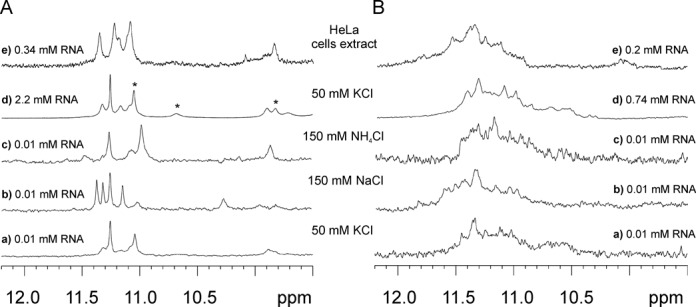
Imino region of the 1H NMR spectra of (A) p(UGG)2U and (B) p(UGG)4U at 25°C in the presence of (Aa, Ba, Ad, Bd) 50 mM KCl, 10 mM potassium phosphate and 0.1 mM EDTA, pH 6.8, (Ab, Bb) 150 mM NaCl, 10 mM sodium phosphate, 0.1 mM EDTA, pH 6.8, (Ac, Bc) 150 mM NH4Cl, 10 mM Tris/HCl, 0.1 mM EDTA, pH 6.8 and (Ae, Be) HeLa cell extract. Asterisks indicate imino proton resonances of uridines. The poor S/N ratio observed for p(UGG)4U is due to polymorphism or/and the presence of higher order structures.
The imino proton spectrum obtained in ammonium buffer was similar to that recorded in the presence of K+ (Figure 6Aa,c). The main change observed in the spectrum recorded in Na+ solution (Figure 6Ab) was the sharpening of two resonances and the down-field shift of one of the uridine resonances (10.27 ppm). This could be explained by the different binding of sodium ions within U-tetrads, due to the smaller ionic radii of Na+ relative to K+ and NH4+. We have also observed sharp imino resonances in the NMR spectrum of p(UGG)2U in the HeLa cell extract (Figure 6Ae). The similarity between the spectra obtained in the cell extract and in potassium buffer strongly suggest that also in cellular conditions, the p(UGG)2U adopts a G-quadruplex structure with at least two stable U-tetrads.
As for the four-repeat sequence p(UGG)4U, independently of the conditions used including cell extract, all 1H NMR spectra were typical of G-quadruplexes. However, a strong tendency to form aggregates and multiple conformations was observed (Figure 6B). Due to the broadness of imino resonances we were not able, using the methods of NMR spectroscopy, to determine the role of uridine residues in a G-quadruplex fold of p(UGG)4U, as we had succeeded in doing for p(UGG)2U.
DISCUSSION
We examined here the structural differences between molecules composed of two or four AGG, CGG and UGG repeats. In general, all these molecules can fold into G-quadruplex structures, but for sequences comprising CGG repeats, the G-quadruplex structure competes with duplex or hairpin structures. We have identified G-quadruplex formation by combining different experimental techniques: UV, CD and NMR spectroscopies along with ESI-MS. The observation of an inverted profile of melting curve at 295 nm demonstrated, with a high degree of confidence, the existence of a G-quadruplex structure. The use of CD spectroscopy in the study of RNA G-quadruplexes has some limitations arising from the fact that spectra of RNA G-quadruplexes are very similar to those of A-form duplexes. Nevertheless, careful analysis of the shape of the CD curve may provide supporting arguments for the presence of specific structural motifs. For example, a small negative band at ∼290 nm in the CD spectrum of p(UGG)2U was attributed to the U-tetrad formation. A similar negative band had also been observed, but not discussed, in the CD spectra of other RNA sequences containing U-tetrads (55,59).
Using ESI-MS in NH4+, we determined that all three two-repeat sequences form tetramolecular G-quadruplexes, while (AGG)4A and p(UGG)4U fold into bimolecular G-quadruplexes. The number of ammoniuim ions remaining coordinated as detected by ESI-MS, however, differed depending on the sequence, suggesting different structural arrangements. The interpretation of the ESI-MS results obtained for G(CGG)4C was ambiguous: the presence of peaks with no ammonium ions remaining bound can be attributed to non-G-quadruplex structures (as confirmed with the other technique) but ESI-MS alone would be inconclusive because the absence of bound ammonium ions could also be due particularly to high lability in the gas phase (44).
The presence of structural motifs, such as A:(G:G:G:G):A hexads, mixed G:C:G:C tetrads and U-tetrads, was confirmed mainly through the analysis of 1D and 2D NMR spectra. NMR spectroscopy helped not only in the identification of the various structural motifs, but also allowed us to observe the formation of various alternate structures in different environments. DNA and RNA sequences containing the GGAGG motif are known to preferentially fold into a dimer of G-quadruplex subunits composed of the canonical G:G:G:G tetrad and A:(G:G:G:G):A hexad planes (34,50,51,60–62). The formation of the hexads increases the stacking contact between two subunits and facilitates their association. Both (AGG)2A and (AGG)4A, contain such a motif and, indeed, adopt similar dimeric G-quadruplex structures involving A:(G:G:G:G):A hexads (Figure 4E). Furthermore, we have acquired the NMR spectra of all molecules in the HeLa cell extract to determine which in vitro observed structure exists in a native-like environment. Interestingly, although the potassium cations are predominant in the cellular environment, the NMR spectrum obtained in the K+ solution was not always the same as that recorded in the cellular extract. Accordingly, for G(CGG)2C in K+ solution a G-quadruplex was found to be the major form, but in the HeLa cell extract a duplex was formed almost exclusively.
The structural preferences of molecules composed of CGG repeats have been a matter of debate (28,31,32,63,64). Our study revealed that G(CGG)2C and G(CGG)4C are indeed polymorphic, and that their structures depend on both the length of sequence and type of cation. We have shown that G(CGG)2C fold exclusively into G-quadruplex in K+ solution but a G-quadruplex/duplex equilibrium was observed in the presence of Na+ ions. Under all conditions, G(CGG)4C showed a greater tendency to fold into a hairpin than into a G-quadruplex structure. Our data have shown that it is possible to switch between a duplex and G-quadruplex structures of G(CGG)2C by changing the type of monovalent cation or the concentration of the RNA strand. In a recent study on DNA G-wire formation, the dimerization of G-quadruplex structures through formation of G:C:G:C tetrads based on Watson−Crick G:C base pairing was tested (65). It was found that the presence of 5′GC-ends, but not 3′GC-ends, promotes self-assembly via Watson−Crick base pairing. Although for the RNA CGG TNRs the presence of G-quadruplex was postulated, no structure was proposed (30–33). We have demonstrated that G(CGG)2C folds into a symmetrical, tetramolecular parallel G-quadruplex structure with G:G:G:G and mixed G:C:G:C tetrads (Figure 4F). Both the presence of complementary 5′GC-ends in the sequence and G:C:G:C tetrads strongly suggest that G(CGG)2C adopts the same architecture as that determined by Webba da Silva for DNA (54).
We also established that p(UGG)2U folds into tetramolecular parallel G-quadruplex with two terminal and one internal uridine tetrads (Figure 4G). Until now, there have been no reports on RNA G-quadruplexes with a uridine tetrad intercalated into the G-quadruplex core. Analysis of the available X-ray structures of RNA G-quadruplexes indicates that internal uridine residues prefer to adopt the bulged-out conformation (66,67). Our data demonstrate for the first time that uridine residues can form U-tetrad inside G-tetrads. The stacking of a single bulge in or out of a G-quadruplex resembles the structures of DNA and RNA duplexes with a base being bulged out or stacked into a helix. In general, in crystal structures single bulges are more likely to be looped out, while in NMR structures the bases are usually found stacked into the helix (68–73). Following our finding that this holds true for short, tetramolecular G-quadruplexes, more thorough structural investigations on larger systems are needed to generalize these conclusions to longer sequences.
CONCLUSIONS
In summary, using UV molecular absorption, CD, NMR spectroscopies and ESI-MS, we have demonstrated that all oligoribonucleotides comprising two TNRs fold preferentially into G-quadruplexes in solution containing potassium ions. In their structure, these G-quadruplexes have distinct structural motifs characteristic of the type of repeats. Additionally, spectral features indicative of A:(G:G:G:G):A hexads have been found for (AGG)2A, whereas signals characteristic of mixed G:C:G:C tetrads and U-tetrads have been observed in NMR spectra of G(CGG)2C and p(UGG)2U, respectively. The knowledge of the structures formed by CGG repeats is crucial for understanding the pathogenesis of the family of fragile X-associated disorders, where the structure of RNA molecules play important role. Then, detailed understanding of the structural preferences of AGG and UGG trinucleotide tandem repeats may help in the recognition of the biological functions of these sequences.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be considered as Joint First Authors.
FUNDING
National Science Center [N N301 707040 to Z.G.; UMO-2013/08/A/ST5/00295, UMO-2011/03/B/NZ1/00576 and UMO-2011/03/B/ST5/011098 to R.K.]; Ministry of Science and Higher Education [N N301 255536 to Z.G.]; Ministry of Science and Higher Education; European Fund for Regional Development [UDA-POIG.02.01.00–30–182/09 to Z.G.]; EU COST action MP0802 [STSM n°11227 to M.M.]; Fonds de la Recherche Scientifique-FNRS [research associate position and FRFC grant 2.4528.11 to V.G.]. Source of open access funding: National Science Center [UMO-2013/08/A/ST5/00295].
Conflict of interest statement. None declared.
REFERENCES
- 1.Simonsson T. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 2001;382:621–628. doi: 10.1515/BC.2001.073. [DOI] [PubMed] [Google Scholar]
- 2.Davis J.T. G-quartets 40 years later: from 5’-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 3.Phan A.T., Kuryavyi V., Patel D.J. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane A.N., Chaires J.B., Gray R.D., Trent J.O. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyoshi D., Nakao A., Sugimoto N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002;41:15017–15024. doi: 10.1021/bi020412f. [DOI] [PubMed] [Google Scholar]
- 7.Ambrus A., Chen D., Dai J., Bialis T., Jones R.a., Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heddi B., Phan A.T. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 9.Marchand A., Ferreira R., Tateishi-Karimata H., Miyoshi D., Sugimoto N., Gabelica V. Sequence and solvent effects on telomeric DNA bimolecular G-quadruplex folding kinetics. J. Phys. Chem. B. 2013;117:12391–12401. doi: 10.1021/jp406857s. [DOI] [PubMed] [Google Scholar]
- 10.Tran P.L.T., De Cian A., Gros J., Moriyama R., Mergny J.-L. Tetramolecular quadruplex stability and assembly. Top. Curr. Chem. 2013;330:243–273. doi: 10.1007/128_2012_334. [DOI] [PubMed] [Google Scholar]
- 11.Matsugami A., Ouhashi K., Kanagawa M., Liu H., Kanagawa S., Uesugi S., Katahira M. An intramolecular quadruplex of (GGA)4 triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 12.Phan A.T., Kuryavyi V., Ma J.-B., Faure A., Andréola M.-L., Patel D.J. An interlocked dimeric parallel-stranded DNA quadruplex: a potent inhibitor of HIV-1 integrase. Proc. Natl. Acad. Sci. U.S.A. 2005;102:634–639. doi: 10.1073/pnas.0406278102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borbone N., Amato J., Oliviero G., D'Atri V., Gabelica V., De Pauw E., Piccialli G., Mayol L. d(CGGTGGT) forms an octameric parallel G-quadruplex via stacking of unusual G(:C):G(:C):G(:C):G(:C) octads. Nucleic Acids Res. 2011;39:7848–7857. doi: 10.1093/nar/gkr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Islam B., Sgobba M., Laughton C., Orozco M., Sponer J., Neidle S., Haider S. Conformational dynamics of the human propeller telomeric DNA quadruplex on a microsecond time scale. Nucleic Acids Res. 2013;41:2723–2735. doi: 10.1093/nar/gks1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maizels N., Gray L.T. The G4 genome. PLoS Genet. 2013;9:e1003468. doi: 10.1371/journal.pgen.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipps H.J., Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Biffi G., Tannahill D., McCafferty J., Balasubramanian S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013;5:182–186. doi: 10.1038/nchem.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert J.L., Bugaut A., Kumari S., Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djebali S., Davis C.a., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millevoi S., Moine H., Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip. Rev. RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 21.Tóth G., Gáspári Z., Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozlowski P., de Mezer M., Krzyzosiak W.J. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 24.Galka-Marciniak P., Urbanek M.O., Krzyzosiak W.J. Triplet repeats in transcripts: structural insights into RNA toxicity. Biol. Chem. 2012;393:1299–1315. doi: 10.1515/hsz-2012-0218. [DOI] [PubMed] [Google Scholar]
- 25.Jacquemont S., Hagerman R.J., Leehey M., Grigsby J., Zhang L., Brunberg J.a., Greco C., Des Portes V., Jardini T., Levine R., et al. Fragile X premutation tremor/ataxia syndrome: molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glass I.A. X-linked mental retardation. J. Med. Genet. 1991;28:361–371. doi: 10.1136/jmg.28.6.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobczak K., Michlewski G., de Mezer M., Kierzek E., Krol J., Olejniczak M., Kierzek R., Krzyzosiak W.J. Structural diversity of triplet repeat RNAs. J. Biol. Chem. 2010;285:12755–12764. doi: 10.1074/jbc.M109.078790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiliszek A., Kierzek R., Krzyzosiak W.J., Rypniewski W. Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 2011;39:7308–7315. doi: 10.1093/nar/gkr368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zumwalt M., Ludwig A., Hagerman P.J., Dieckmann T. Secondary structure and dynamics of the r(CGG) repeat in the mRNA of the fragile X mental retardation 1 (FMR1) gene. RNA Biol. 2007;4:93–100. doi: 10.4161/rna.4.2.5039. [DOI] [PubMed] [Google Scholar]
- 30.Handa V., Saha T., Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khateb S., Weisman-Shomer P., Hershco I., Loeb L.A., Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khateb S., Weisman-Shomer P., Hershco-Shani I., Ludwig A.L., Fry M. The tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance the in vivo translation of fragile X premutation mRNA. Nucleic Acids Res. 2007;35:5775–5788. doi: 10.1093/nar/gkm636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ofer N., Weisman-Shomer P., Shklover J., Fry M. The quadruplex r(CGG)n destabilizing cationic porphyrin TMPyP4 cooperates with hnRNPs to increase the translation efficiency of fragile X premutation mRNA. Nucleic Acids Res. 2009;37:2712–2722. doi: 10.1093/nar/gkp130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mashima T., Matsugami A., Nishikawa F., Nishikawa S., Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joachimi A., Benz A., Hartig J.S. A comparison of DNA and RNA quadruplex structures and stabilities. Bioorg. Med. Chem. 2009;17:6811–6815. doi: 10.1016/j.bmc.2009.08.043. [DOI] [PubMed] [Google Scholar]
- 36.Beaucage S.L., Caruthers M.H. Deoxynucleoside phosphoramidites—a new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981;22:1859–1862. [Google Scholar]
- 37.Xia T., SantaLucia J., Burkard M.E., Kierzek R., Schroeder S.J., Jiao X., Cox C., Turner D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 38.Kierzek E., Kierzek R. The synthesis of oligoribonucleotides containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines via post-synthetic modification of precursor oligomers. Nucleic Acids Res. 2003;31:4461–4471. doi: 10.1093/nar/gkg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uddin M.K., Kato Y., Takagi Y., Mikuma T., Taira K. Phosphorylation at 5’ end of guanosine stretches inhibits dimerization of G-quadruplexes and formation of a G-quadruplex interferes with the enzymatic activities of DNA enzymes. Nucleic Acids Res. 2004;32:4618–4629. doi: 10.1093/nar/gkh766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen E.B., Nielsen J.T., Nielsen C., Filichev V.V. Enhanced anti-HIV-1 activity of G-quadruplexes comprising locked nucleic acids and intercalating nucleic acids. Nucleic Acids Res. 2011;39:2470–2481. doi: 10.1093/nar/gkq1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hud N.V., Schultze P., Feigon J. Ammonium ion as an NMR probe for monovalent cation coordination sites of DNA quadruplexes. 1998;120:6403–6404. [Google Scholar]
- 42.Gros J., Rosu F., Amrane S., De Cian A., Gabelica V., Lacroix L., Mergny J.-L. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collie G.W., Parkinson G.N., Neidle S., Rosu F., De Pauw E., Gabelica V. Electrospray mass spectrometry of telomeric RNA (TERRA) reveals the formation of stable multimeric G-quadruplex structures. J. Am. Chem. Soc. 2010;132:9328–9334. doi: 10.1021/ja100345z. [DOI] [PubMed] [Google Scholar]
- 44.Balthasart F., Plavec J., Gabelica V. Ammonium ion binding to DNA G-quadruplexes: do electrospray mass spectra faithfully reflect the solution-phase species. J. Am. Soc. Mass Spectrom. 2013;24:1–8. doi: 10.1007/s13361-013-0649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sosnick T.R., Fang X., Shelton V.M. Application of circular dichroism to study RNA folding transitions. Methods Enzymol. 2000;317:393–409. doi: 10.1016/s0076-6879(00)17026-0. [DOI] [PubMed] [Google Scholar]
- 46.Hwang T.-L., Shaka A.J. Water suppression that works. Excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Magn. Reson. Ser. A. 1995;112:275–279. [Google Scholar]
- 47.Mergny J.-L., Lacroix L. UV melting of G-quadruplexes. Curr. Protoc. Nucleic Acid Chem. 2009;37:17.1–17.15. doi: 10.1002/0471142700.nc1701s37. [DOI] [PubMed] [Google Scholar]
- 48.Mergny J.-L., Li J., Lacroix L., Amrane S., Chaires J.B. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Randazzo A., Spada G.P., Webba da Silva M. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013;330:67–86. doi: 10.1007/128_2012_331. [DOI] [PubMed] [Google Scholar]
- 50.Nishikawa F., Murakami K., Matsugami A., Katahira M., Nishikawa S. Structural studies of an RNA aptamer containing GGA repeats under ionic conditions using microchip electrophoresis, circular dichroism, and 1D-NMR. Oligonucleotides. 2009;19:179–190. doi: 10.1089/oli.2008.0167. [DOI] [PubMed] [Google Scholar]
- 51.Lipay J.M., Mihailescu M.-R. NMR spectroscopy and kinetic studies of the quadruplex forming RNA r(UGGAGGU) Mol. Biosyst. 2009;5:1347–1355. doi: 10.1039/b911555b. [DOI] [PubMed] [Google Scholar]
- 52.Liu H., Kugimiya A., Matsugami A., Katahira M., Uesugi S. Quadruplex structures of RNA 14-mer, r(GGAGGUUUUGGAGG) and DNA 14-mer, d(GGAGGTTTTGGAGG) Nucleic Acids Res. 2002;2:177–178. doi: 10.1093/nass/2.1.177. [DOI] [PubMed] [Google Scholar]
- 53.Fürtig B., Richter C., Wöhnert J., Schwalbe H. NMR spectroscopy of RNA. Chembiochem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 54.Webba da Silva M. Association of DNA quadruplexes through G:C:G:C tetrads. Solution structure of d(GCGGTGGAT) Biochemistry. 2003;42:14356–14365. doi: 10.1021/bi0355185. [DOI] [PubMed] [Google Scholar]
- 55.Xu Y., Ishizuka T., Kimura T., Komiyama M. A U-tetrad stabilizes human telomeric RNA G-quadruplex structure. J. Am. Chem. Soc. 2010;132:7231–7233. doi: 10.1021/ja909708a. [DOI] [PubMed] [Google Scholar]
- 56.Cheong C., Moore P.B. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structured. Biochemistry. 1992;31:8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 57.Patel P.K., Hosur R.V. NMR observation of T-tetrads in a parallel stranded DNA quadruplex formed by Saccharomyces cerevisiae telomere repeats. Nucleic Acids Res. 1999;27:2457–2464. doi: 10.1093/nar/27.12.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oliviero G., Amato J., Borbone N., Galeone A., Varra M., Piccialli G., Mayol L. Synthesis and characterization of DNA quadruplexes containing T-tetrads formed by bunch-oligonucleotides. Biopolymers. 2006;81:194–201. doi: 10.1002/bip.20399. [DOI] [PubMed] [Google Scholar]
- 59.Randazzo A., Esposito V., Ohlenschläger O., Ramachandran R., Mayola L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004;32:3083–3092. doi: 10.1093/nar/gkh629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kettani A., Gorin A., Majumdar A., Hermann T., Skripkin E., Zhao H., Jones R., Patel D.J. A dimeric DNA interface stabilized by stacked A · (G · G · G · G) · A hexads and coordinated monovalent cations. J. Mol. Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]
- 61.Liu H., Matsugami A., Katahira M., Uesugi S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J. Mol. Biol. 2002;322:955–970. doi: 10.1016/s0022-2836(02)00876-8. [DOI] [PubMed] [Google Scholar]
- 62.Mergny J.-L., De Cian A., Amrane S., Webba da Silva M. Kinetics of double-chain reversals bridging contiguous quartets in tetramolecular quadruplexes. Nucleic Acids Res. 2006;34:2386–2397. doi: 10.1093/nar/gkl098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sobczak K., de Mezer M., Michlewski G., Krol J., Krzyzosiak W.J. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Broda M., Kierzek E., Gdaniec Z., Kulinski T., Kierzek R. Thermodynamic stability of RNA structures formed by CNG trinucleotide repeats. Implication for prediction of RNA structure. Biochemistry. 2005;44:10873–82. doi: 10.1021/bi0502339. [DOI] [PubMed] [Google Scholar]
- 65.Ilc T., Šket P., Plavec J., Webba da Silva M., Drevenšek-Olenik I., Spindler L. Formation of G-wires: the role of G:C-base pairing and G-quartet stacking. J. Phys. Chem. C. 2013;117:23208–23215. [Google Scholar]
- 66.Pan B., Xiong Y., Shi K., Sundaralingam M. Crystal structure of a bulged RNA tetraplex at 1.1 Å resolution. Structure. 2003;11:1423–1430. doi: 10.1016/j.str.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 67.Pan B., Shi K., Sundaralingam M. Base-tetrad swapping results in dimerization of RNA quadruplexes: implications for formation of the i-motif RNA octaplex. Proc. Natl. Acad. Sci. U.S.A. 2006;103:3130–3134. doi: 10.1073/pnas.0507730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Popenda L., Adamiak R.W., Gdaniec Z. Bulged adenosine influence on the RNA duplex conformation in solution. Biochemistry. 2008;47:5059–5067. doi: 10.1021/bi7024904. [DOI] [PubMed] [Google Scholar]
- 69.Popenda L., Bielecki L., Gdaniec Z., Adamiak R. Structure and dynamics of adenosine bulged RNA duplex reveals formation of the dinucleotide platform in the C:G-A triple. Arkivoc. 2009;2009:130–144. [Google Scholar]
- 70.Hermann T., Patel D. RNA bulges as architectural and recognition motifs. Structure. 2000;8:47–54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 71.Sudarsanakumar C., Xiong Y., Sundaralingam M. Crystal structure of an adenine bulge in the RNA chain of a DNA·RNA hybrid, d(CTCCTCTTC)·r(gaagagagag) J. Mol. Biol. 2000;299:103–112. doi: 10.1006/jmbi.2000.3730. [DOI] [PubMed] [Google Scholar]
- 72.Thiviyanathan V., Guliaev A.B., Leontis N.B., Gorenstein D.G. Solution conformation of a bulged adenosine base in an RNA duplex by relaxation matrix refinement. J. Mol. Biol. 2000;300:1143–1154. doi: 10.1006/jmbi.2000.3931. [DOI] [PubMed] [Google Scholar]
- 73.Xiong Y., Deng J., Sudarsanakumar C., Sundaralingam M. Crystal structure of an RNA duplex r(gugucgcac)2 with uridine bulges. J. Mol. Biol. 2001;313:573–582. doi: 10.1006/jmbi.2001.5045. [DOI] [PubMed] [Google Scholar]


