Abstract
In plants, the voltage-dependent anion-selective channel (VDAC) is a major component of a pathway involved in transfer RNA (tRNA) translocation through the mitochondrial outer membrane. However, the way in which VDAC proteins interact with tRNAs is still unknown. Potato mitochondria contain two major mitochondrial VDAC proteins, VDAC34 and VDAC36. These two proteins, composed of a N-terminal α-helix and of 19 β-strands forming a β-barrel structure, share 75% sequence identity. Here, using both northwestern and gel shift experiments, we report that these two proteins interact differentially with nucleic acids. VDAC34 binds more efficiently with tRNAs or other nucleic acids than VDAC36. To further identify specific features and critical amino acids required for tRNA binding, 21 VDAC34 mutants were constructed and analyzed by northwestern. This allowed us to show that the β-barrel structure of VDAC34 and the first 50 amino acids that contain the α-helix are essential for RNA binding. Altogether the work shows that during evolution, plant mitochondrial VDAC proteins have diverged so as to interact differentially with nucleic acids, and this may reflect their involvement in various specialized biological functions.
INTRODUCTION
Voltage-dependent anion-selective channels (VDACs) are the most abundant proteins in the outer mitochondrial membrane (OMM) of all eukaryotic cells. These proteins adopt β-barrel structures that form pores integrated in the membrane similar to their bacterial porin counterparts. In contrast to bacterial porins that are only formed by a succession of β-barrel strands, eukaryotic VDACs found in mitochondrial membrane contain an α-helical structure at the N-terminal extremity. Recently, Nuclear Magnetic Resonance (NMR) and crystal structures of the human and mouse VDAC1 confirmed the presence of this segment folded inside the β-barrel composed of 19 β-strands (1–3).
VDACs present evolutionary conserved properties of voltage gating and ion selectivity. Subsequently, the major biological function of VDACs is to mediate and regulate the transport of ions and metabolites such as adenosine triphosphate (ATP) or Nicotinamide Adenine Dinucleotide Hydrogen (NADH) between the cytosol and the mitochondria across the OMM. Furthermore, there is increasing evidence that VDACs represent critical factors in initiating apoptosis (for reviews see for instance (4,5)). Another major role of VDACs is their involvement in the transport of both DNA and tRNA molecules into mitochondria. Indeed, in various organisms, such as yeast, plants and animals, isolated mitochondria have been shown to import double stranded DNA and VDACs were recruited for the translocation through the OMM (6–8). In addition, tRNA import into mitochondria is largely proved in most of the eukaryotic lineages (9–11). The import mechanism is still not well understood and seems to be different according to the studied organism. In higher plants, we previously demonstrated that tRNAs can enter into potato isolated mitochondria through VDACs in the absence of any added protein (12).
Plant VDACs belong to a small multigene family larger than that found in fungi (two genes) or in mammals (three genes) and containing up to ten genes in the Populus trichocarpa (poplar) genome (4). Solanum tuberosum (potato), a model organism to study the plant tRNA mitochondrial import machinery, contains four VDAC genes and two major isoforms, VDAC34 and VDAC36, are present in the OMM (13). Here we show that these two VDACs differentially interact with nucleic acids. VDAC34 binds more efficiently both RNA and DNA than VDAC36. In the absence of any recognizable RNA binding motif, we wondered which regions of VDAC proteins were involved in the interaction with tRNAs. In this context, potato VDAC34 and VDAC36 provide us an ideal system to decipher the molecular basis of this interaction. We thus focused our work on the identification of such domain(s) in VDAC34. To do so, 21 VDAC34 mutants were designed and analyzed by northwestern. While no strict RNA recognition motif could be defined, it appears that the whole structure and a limited set of amino acids scattered at the N-terminal extremity are essential for the interaction. Altogether, our data suggest that, during evolution, VDAC34 has acquired a more specialized function in nucleic acids translocation through the OMM.
MATERIALS AND METHODS
Isolation of plant mitochondria and preparation of outer mitochondrial membranes
Mitochondria were isolated from potato tubers (14). Outer membranes were purified according to (12). Mitochondria (50 mg of mitochondrial proteins) were resuspended in 10 ml of swelling buffer (5 mM potassium phosphate pH 7.2) and kept on ice for 10 min. The same volume of swelling buffer was added and mitochondria were ruptured in a potter homogenizer. Outer membranes were separated from mitoplasts (i.e. mitochondria without outer membrane) by centrifugation through a sucrose step gradient (60–32–15% sucrose in 1 mM EDTA, 1 mM PMSF and 10 mM potassium phosphate pH 7.2) at 4°C for 10 min at 125 000 g. They were collected from the 15–30% interphase, diluted 5× in washing buffer (0.3 M Mannitol, 1 mM EDTA, 0.1% BSA and 10 mM potassium phosphate pH 7.2) and then pelleted for 10 min at 170 000 g.
PCR amplification, cloning, mutagenesis and E. coli overexpression of VDAC proteins
The cDNAs encoding potato VDAC34 and VDAC36 (13) were amplified by RT-PCR and cloned into pQE60 vector. Constructs corresponding to mutant VDAC proteins were obtained using classical oligonucleotide-directed mutagenesis (15) or the quickchange site directed mutagenesis kit (Stratagen, La Jolla, CA, USA). The list of mutant sequences is provided as supplemental information (Supplemental Figure S1). All constructs were overexpressed in Escherichia coli with a His-tag at the C-terminal extremity of the proteins. They were purified on column under denaturing conditions in the presence of 8 M urea according to manufacturer's recommendation (Qiagen, Valencia, CA, USA) as described in (12). When necessary, refolding of the protein was done on column before final elution. It consisted of a three-step urea gradient (8, 4 and 0 M) in buffer solution containing 5 mM MgCl2, 50 mM Tris-Maleate pH 8.5 and 1 or 2% (w/v) of octyl-β-D-glucopyranoside. Proteins were eluted using 250 mM imidazole buffer and concentrated by ultrafiltration using Amicon® Ultra (Amicon® Ultra-4 10K) to about 340 μM. Proteins were stored in buffer solution at 4°C until use. Sample homogeneity was verified by dynamic light scattering (Malvern Zetasizer) at 20°C and corresponding molecular sizes were estimated from the diffusion coefficient using the Stockes–Einstein equation.
In vitro synthesis of radiolabeled RNA transcripts
The construct containing Arabidopsis thaliana cytosolic tRNAAla gene sequence was obtained previously (16). The tRNA gene sequence was directly fused to a T7 RNA polymerase promoter at the 5′ terminus and included a BstNI site at the 3′ terminus. This construct was used as substrate to synthesize in vitro radiolabeled transcript with T7 polymerase using Ribomax™ large scale transcription kit (Promega, Madison WI) as described in (15). BamHI-linearized pBluescript SK+ plasmid (Stratagene) was used as a template to generate a 75 nt long RNA transcript using T7 RNA polymerase as described above.
DNA labeling
To generate a radiolabeled single-stranded DNA substrate, a 76 nt long oligonucleotide corresponding to the sequence of the full-length A. thaliana cytosolic tRNAAla (16) was 32P-labeled with T4 polynucleotide kinase according to classical procedure (17). To obtain a radiolabeled double-stranded DNA substrate, the construct containing the A. thaliana cytosolic tRNAAla gene sequence was used as a template for classical polymerase chain reaction (PCR) amplification in the presence of 25 μCi (3000 Ci/mmole) of 32P-dCTP.
Northwestern, southwestern and gel-shift experiments
Northwestern experiments were performed essentially as described in (12). Briefly, overexpressed proteins were electrophoresed on a sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted on Immobilon-P membrane. Membrane was washed 4× with 0.1 M Tris-HCl pH 7.5, 0.1% NP40, each for 30 min at 4°C with gentle agitation, then saturated with binding buffer containing 5% BSA and 0.01% Triton X-100 for 5 min. The membrane was incubated in presence of 0.1 to 0.2 nM of radiolabeled RNA transcripts. Southwestern experiments were carried out in similar conditions but in the presence of radiolabeled DNA. Gel-shift assays were conducted as described in the Promega (Madison, WI) gel-shift assay system. Briefly, binding reactions contained 50 mM NaCl, 10 mM Tris-HCl pH 7.5, 0.5 mM DTT, 0.5 mM EDTA, 1 mM MgCl2, 2% glycerol, radiolabeled tRNA in excess (1–5 nM) and protein concentrations as indicated. Reactions were incubated for 20 min at 25°C and resolved on 4% native-PAGE gels. For competition assays, 0–30 nM of unlabeled tRNA transcript or ATP was included in the reaction. For each experiment, signals were detected with FLA-7000 phosphor imager (Fujifilm) and quantified using the software ImageGauge (Fujifilm). For quantitative analysis, gel-shift assays were carried out according to (18) in the same conditions as described above but with limiting concentration of radiolabeled tRNA (0.1 nM). Binding curves were graphed and the apparent constant dissociation (Kd) values were calculated with KaleidaGraph 4.0 software as described in (19).
Western blot analysis and antibodies
Western blot analysis was conducted according to standard procedures as described in (17). Antibodies against potato mitochondrial VDAC were kindly provided by H. P. Braun (Hannover University, Hannover, Germany).
Structural analysis
VDAC sequences were aligned with T-coffee (20) and JalView (21) and secondary structure prediction was performed with its JNET plugin (22). Transmembrane segment prediction was done with the online software ProtScale (http://www.expasy.ch/tools/protscale.html) with a window size of 9 amino acids (23). The RNA–protein interaction predictions were performed with BindN software (24). The software Modeler was used for homology modeling of VDAC structures (25) based on the high-resolution crystal structure of mouse VDAC1 (PDB ID: 3EMN). The 3D structures were prepared with PyMOL (Schrödinger—www.pymol.org).
RESULTS
The two major Solanum tuberosum mitochondrial VDAC isoforms differentially interact with tRNA molecules
We previously showed that a 34 kDa potato VDAC can interact with tRNA molecules (12). However, in S. tuberosum two major VDACs are present in the OMM, namely VDAC34 and VDAC36 (13). Indeed, while northwestern experiments performed on total OMM proteins in the presence of radiolabeled plant cytosolic tRNAAla gave only one signal corresponding in size to VDAC34 (Figure 1A), a western blot experiment performed with antibodies recognizing the two VDAC isoforms showed that both proteins were present in equal amount in the OMM (Figure 1A). This suggested that the two VDAC isoforms differentially interact with tRNA. In order to confirm this observation, the two VDACs were overexpressed and His-Tag purified. Northwestern experiments with the purified recombinant proteins confirmed that VDAC34 interacts about 7.5× fold more with labeled tRNAAla transcript than VDAC36 (Figure 1B). This differential interaction, although of less magnitude, was further supported by gel shift assay with labeled tRNAAla transcript and increasing amounts of overexpressed VDAC proteins (Figure 1C), as 2× more tRNA–VDAC complex on an average was formed with VDAC34 than with VDAC36. In order to rule out a defect of refolding and/or of stability of VDAC36, we analyzed VDAC hydrodynamic properties by dynamic light scattering and confirmed that both VDACs are stable and behave as monomers in solution (Supplemental Figure S2). Gel-shift quantitative analyses allowed calculating the apparent constant dissociation (Kd) values for both VDAC proteins (Figure 1D and Supplemental Figure S3). These analyses showed that the Kd values were in the 0.1 μM range and that VDAC36 Kd value was on average 1.4× higher than that of VDAC34. It is worth to note that the Kd for VDAC34 yielded highly reproducible values whereas the VDAC36 Kd was more variable (Figure 1D). Furthermore, the interaction between radiolabeled tRNAAla transcript and VDAC34 remained unchanged when increasing amounts of ATP, one of the major metabolites transported through VDAC, were added to gel shift assays. By contrast, this interaction was almost completely competed out in the presence of an excess of unlabeled tRNA transcript (Figure 1E). This indicates that the binding site(s) of ATP molecules on VDAC34 is likely different to those required for tRNA interaction.
Figure 1.
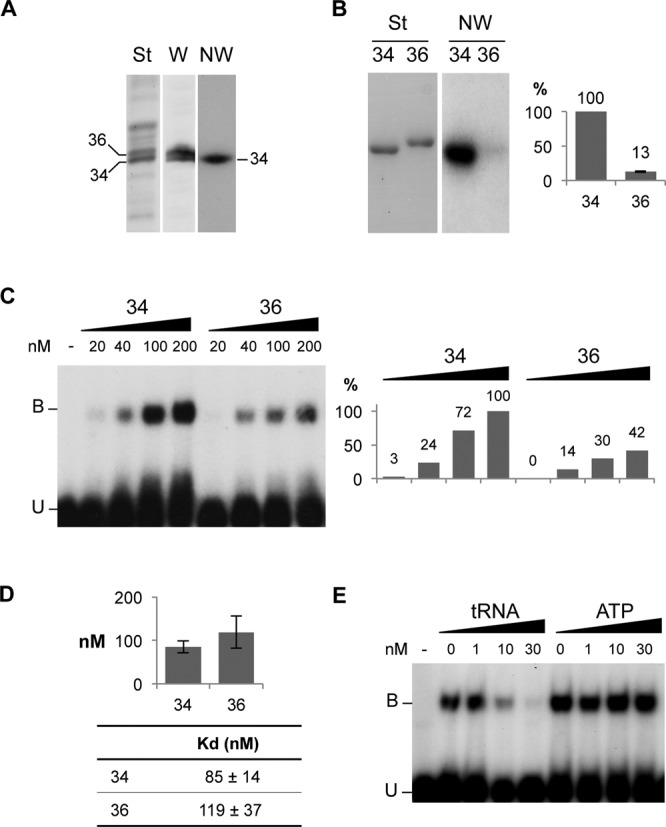
Mitochondrial VDAC proteins from Solanum tuberosum differentially interact with tRNA. (A) VDAC34 interacts with labeled tRNAs. (St) corresponds to the Coomassie blue staining of total mitochondrial outer membrane proteins fractionated on SDS/PAGE and electroblotted onto Immobilon-P membrane. (W) corresponds to the western blot analysis with antibodies raised against S. tuberosum VDAC proteins. The position of the two major VDACs, VDAC34 and VDAC36, is indicated. (NW) corresponds to the northwestern blot analysis. The proteins fixed on the membrane were renatured and incubated with radiolabeled cytosolic Arabidopsis thaliana tRNAAla transcript. (B) Differential interaction was confirmed with the His-tagged purified proteins. (St) corresponds to the Coomassie blue staining of the purified VDAC34 (34) and VDAC36 (36) proteins fractionated by SDS/PAGE and transferred onto nylon membrane. (NW) corresponds to the Northwestern blot analysis with labeled tRNAAla. The histogram shows the percentage of interaction between VDAC36 and tRNA as compared to VDAC34 interaction (100%). It is the average of 11 independent experiments and standard error is indicated for VDAC36. (C) Gel shift assay with 20 to 200 nM of purified VDAC34 or VDAC36 proteins in presence of an excess of labeled tRNAAla (1 nM). Unbound tRNA probe and tRNA probe bound to the VDAC protein are indicated by the letters U and B respectively. The histogram shows the tRNA–VDAC complex signal intensities. The signal intensity observed in presence of 200 nM of VDAC34 was arbitrary taken as 100%. (D) Constant dissociation (Kd) values for VDAC34 and VDAC36. The values are the average of seven and five independent experiments for VDAC34 and VDAC36 respectively and standard errors are indicated. Examples of gel-shift assays and binding curve graphics are shown in supplemental information (Supplemental Figure S3). (E) Gel shift competition assay with 200 nM of VDAC34 and an excess of labeled tRNAAla (1 nM) in presence of increasing amounts of unlabeled tRNAAla or increasing amounts of ATP.
The differential interaction is also observed with other nucleic acid substrates
Plant tRNA mitochondrial import is highly specific, but we previously demonstrated that plant VDACs are not responsible for this selectivity (12). Furthermore, VDACs are involved not only in tRNA import but also in DNA uptake into mitochondria (6). We thus wondered whether the differential interaction observed with tRNA is transposable to other substrates. To answer this question, three radiolabeled substrates including a random RNA transcript, a single and a double stranded DNA fragment were used for northwestern or southwestern experiments. As shown in Figure 2, for each of these nucleic acid substrates, the interaction with VDAC34 increases continuously as a function of protein amounts. By contrast, although an increased signal was observed with VDAC36, it was about 6× weaker on average than with VDAC34 (Figure 2). As a whole, the data indicate that potato VDAC34 and VDAC36 differentially interact with all types of nucleic acids.
Figure 2.
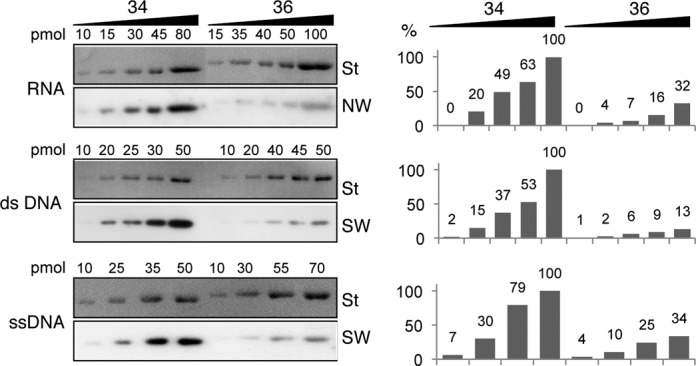
Solanum tuberosum mitochondrial VDACs differentially interact with RNA and DNA in vitro. Coomassie blue-stained profile (St), Northwestern (NW) and Southwestern analyses (SW) of His-tagged purified VDAC34 (34) and VDAC36 (36) proteins. For NW, a radiolabeled random 75 nt long RNA transcript was used. For SW, membranes were incubated with either a radiolabeled double-stranded DNA (dsDNA) substrate corresponding to the Arabidopsis thaliana cytosolic tRNAAla PCR product or with a radiolabeled single-stranded DNA substrate (ssDNA) corresponding to the A. thaliana cytosolic tRNAAla oligonucleotide sequence. NW and SW experiments were performed with increasing amounts of VDAC34 or VDAC36 and in presence of 0.2 nM of radiolabeled nucleic acids. The histograms show the percentages of interaction observed for each VDAC amount. For each nucleic acid substrate the interaction with the highest amount of VDAC34 was arbitrary taken as a reference (100%).
The whole VDAC34 sequence is essential to promote tRNA interaction
In order to determine which regions are important for the interaction with tRNA, different mutant versions of VDAC34 were constructed. To do so, VDAC proteins were arbitrarily divided into three segments: I (position 1–90), II (position 91–171) and III (position 172–276) (Figure 3A). In total, seven deletion mutants (D1–D7) and nine chimeric (C1–C9) were designed (Figure 4). Northwestern analysis on deletion mutants showed that none of the VDAC34 segments with the exception of the D3 mutant are able to interact efficiently with tRNAs (Figure 4A). In addition, analysis on chimeric mutants indicated that the gradual replacement of VDAC34 sequence by VDAC36 sequence from the C-terminal side (C1-C2-C3-C4) or from the N-terminal side (C5-C6-C7-C8-C9) leads to decreased interaction with tRNA (Figure 4B). These two observations are in agreement with the fact that the VDAC34–tRNA interaction does not involve a specific RNA binding domain but rather a set of amino acids distributed all along the sequence (Figure 3B). Furthermore, analyzing deletion mutants highlighted the importance of preserving a complete VDAC34 sequence since short deletions of 25 amino acids at the N-terminus (D2) or 30 amino acids at the C-terminus (D4) induced an important decrease of interaction up to 70%. Even the deletion of 10 amino acids (D3) decreased the interaction level by 10%. Indeed, the comparison of deleted and corresponding chimeric structures such as D5 with C2, D6 with C1 and D2 with C6, showed that deleted proteins could not interact as efficiently as full-length proteins.
Figure 3.
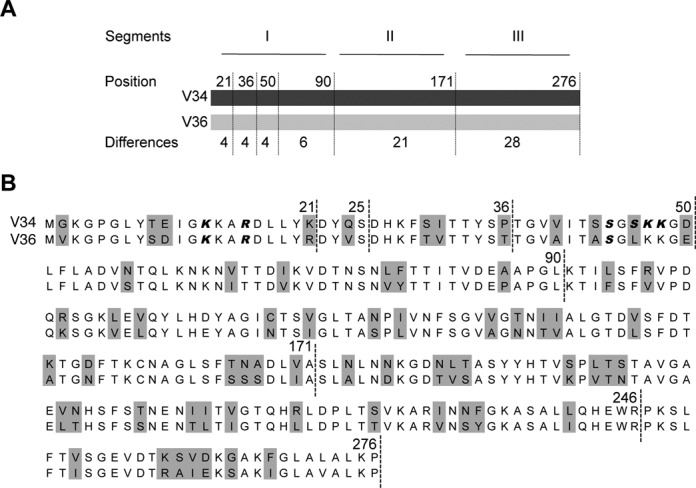
VDAC34 and VDAC36 sequences. (A) Schematic representation of VDAC34 (dark gray) and of VDAC36 (light gray). The three segments set up for the design of the different mutants are indicated. Subsequently, for segment I, two more divisions were done. The number of amino acids that differ between the two VDAC is indicated for each segment. (B) Sequence alignment of VDAC34 and VDAC36. Amino acids that differ between the two VDAC sequences are depicted on a gray background. Positions delimiting the different segments used for design of the different VDAC34 mutants are indicated. Amino acids of the first 50 residues predicted to potentially interact with RNA by BindN software are indicated in italics bold.
Figure 4.
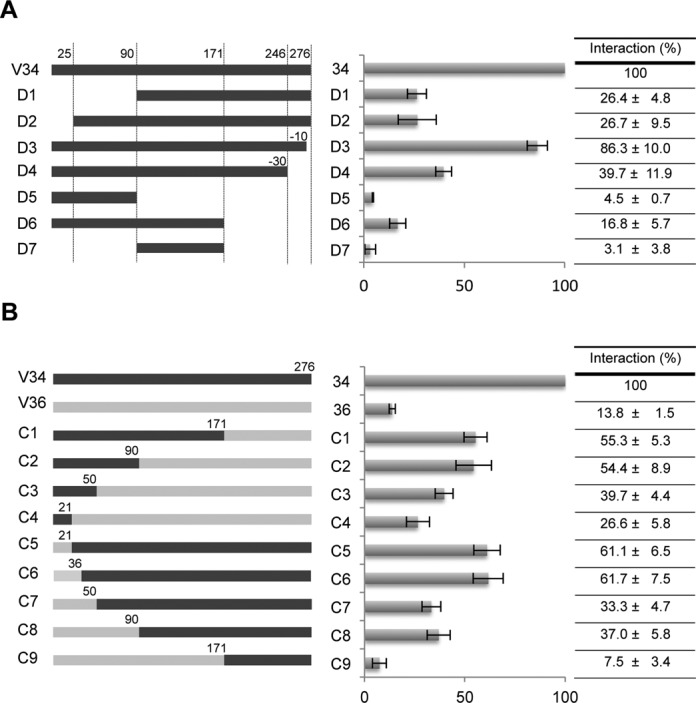
The N-terminal part of VDAC34 is essential for an efficient interaction with tRNA. Two kinds of mutants were designed: (A) deleted versions of VDAC34 (D1–D7) and (B) chimeric proteins (C1–C9) in which segments of VDAC34 were replaced by segments of VDAC36. Numbers indicate position within VDAC sequences. The variants overexpressed in Escherichia coli were purified and used to perform NW experiments in the presence of Arabidopsis thaliana cytosolic tRNAAla in order to quantify percentage of interaction between mutant proteins and tRNA. Obtained values are represented on a diagram and are given in a summary table. They are the average of 2–9 independent experiments and correspond to the percentage of interaction of the mutant protein with tRNA compared to the wild-type VDAC34 interaction. Standard error is indicated for each value. Examples of NW experiments allowing the quantification of interactions are given as supplemental information (Supplemental Figure S4).
As a whole, these data support the idea that the full sequence is required for proper and efficient interaction. Moreover, the strong RNA binding property observed for VDAC34 must be linked to part of the 67 amino acids over the 276 that are different between the two VDACs and scattered all along the sequence.
The N-terminal region plays a major role on tRNA interaction
Comparing interaction of VDAC34 with mutants C1 and C2, we noticed that segment I corresponding to the first 90 amino acids of VDAC34 was enough to maintain 50% of interaction with tRNA (Figure 4B). Furthermore, when segment I of VDAC34 was replaced by VDAC36 homologous region (mutant C8), the interaction was decreased by 60%. This suggested that the N-terminal part of VDAC34 played a major role for tRNA interaction. Analysis with mutants C3 to C7 confirmed this result and allowed to propose that the first 50 amino acids of VDAC34 were crucial. For instance, in mutant C3, the first 50 amino acids of VDAC34 in a VDAC36 context was sufficient to restore the interaction to a level of 40%, as compared to 14% for VDAC36. Finally, the comparison of mutants C5, C6 and C7 permitted us to delimit two main regions for interaction within the first 50 amino acids corresponding to the regions from positions 1 to 21 and from position 37 to 50 (Figure 4B).
Three amino acids at the N-terminal region are particularly critical for tRNA interaction
Between positions 1 to 21, four amino acids differ between the two VDACs (Figure 3). Comparison of the physicochemical properties showed that one out of the four amino acids was not equivalent: at position 2 there is a Glycine residue (G2) in VDAC34 whereas in VDAC36 there is a Valine residue (Figure 3B). Consequently, two mutants were designed: mutant M1 corresponding to VDAC34 with a Valine instead of a Glycine at position 2 and mutant M2 introducing a Glycine at position 2 in VDAC36. Northwestern analysis showed that in mutant M1, the interaction was reduced 2-folds whereas reciprocally in mutant M2, the interaction was increased 2-folds compared to VDAC36 (Figure 5). This is in accordance with what was observed for mutants C4 and C5 (Figure 4B) and demonstrates the importance of the Glycine residue at position 2 for the interaction with tRNAs.
Figure 5.
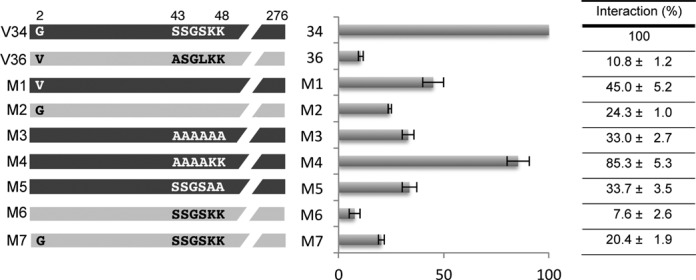
Two Lysines and one Glycine present in the N-terminal region of VDAC34 are essential for the interaction with tRNA. Mutants with point mutations (M1–M7) are schematically represented. Amino acids are indicated by the one letter code. Quantification of interaction between mutant proteins and tRNA were done by NW experiments in presence of Arabidopsis thaliana cytosolic tRNAAla. Obtained values are represented on a diagram and are given in a summary table. They are the average of 2–10 independent experiments and correspond to the percentage of interaction of the mutant protein with tRNA compared to the wild-type VDAC34 interaction. Standard error is indicated for each value. Examples of NW experiments allowing the quantification of interactions are given as supplemental information (Supplemental Figure S4).
Between positions 37 to 50, there are also four amino acids differing between the two VDAC sequences. The physicochemical properties change for only two of them, those at positions 43 and 46. Moreover, the analysis with BindN software that calculates the potentiality of a protein to interact with RNA pointed out three amino acids on VDAC34, namely S46K47K48, which might interact with RNAs whereas the amino acids at equivalent positions on VDAC36 might not (Figure 3B and Supplemental Figure S5). Therefore, we further analyzed the sequence S43SGSKK48 present in VDAC34 and five more mutants, M3–M7, were constructed (Figure 5). When the six amino acids were mutated into Alanine (mutant M3), a strong decrease (67%) of interaction was observed, thus confirming that this region was indeed important for interaction. However, if only the first four amino acids between positions 43 and 46 were changed into Alanine (mutant M4), the interaction remained almost unchanged as compared to wild-type VDAC34. On the contrary, if only the four residues S43SGS46 out of the six were kept (mutant M5 versus mutant M3) on VDAC34 or if they were in the VDAC36 environment (mutant M6 where the A43SGL46 sequence of VDAC36 was replaced by the S43SGS46 sequence from VDAC34), there was no improvement of the interaction with tRNAs. This demonstrates that the S43SGS46 sequence do not participate in the interaction with tRNAs. Furthermore, when only the K47 and K48 of VDAC34 were mutated into Alanine residues (mutant M5), we observed a strong reduction of interaction similar to the one obtained with mutant M3. By contrast, a strong interaction (85%) remained in mutant M4 where K47 and K48 were not replaced. As a whole, this shows that among the six residues only the two Lysines appear essential. Finally, when both a Glycine at position 2 and the S43SGSKK48 sequence are introduced in VDAC36 (mutant M7), only a 2-fold increase of the interaction similar to what was observed for mutant M2 was obtained. This indicates that the G2 alone can improve the interaction and that the two Lysines residues need the VDAC34 context to efficiently interact with tRNA. Altogether, these analyses show the importance of the G2, K47 and K48 residues at the N-terminal region of VDAC34 but also suggest that the G2 and the 2 Lysines act independently.
Solanum tuberosum VDAC architecture and tRNA binding
In order to gain an insight into the organization of tRNA binding sites in VDACs we built 3D models of S. tuberosum VDACs and of their mutants (Supplemental Figure S6) based on the crystal structure of the mouse VDAC1 (3). Sequence alignment with mammal proteins unambiguously showed that VDAC34 and VDAC36 adopt a β-barrel structure composed of 19 antiparallel β-strands (Figure 6A), in agreement with ProtScale prediction of 19 hydrophobic segments separated by hydrophilic loops (not shown). The N-terminal extremity contains a short α-helix that is positioned inside the β-barrel close to strands 10–15 (Figure 6B). The amphiphilic nature of the α-helix (Supplemental Figure S6) allows its anchoring to the β-barrel wall via hydrophobic contacts and exposes polar and charged side chains inside the pore. As observed in mammal VDAC1, this N-terminal helix is positioned at the midpoint of the pore and restricts its inner diameter to about 15 Å, whereas both entries of the barrel have a diameter of at least 25 Å (Figure 6B). Hydrophilic residues are mainly found in the loops connecting the β-strands or inside the pore alternating with hydrophobic residues that point towards the membrane. This organization provides pores with an inner channel that is mainly positively charged to guide anions through the OMM (Supplemental Figure S6).
Figure 6.
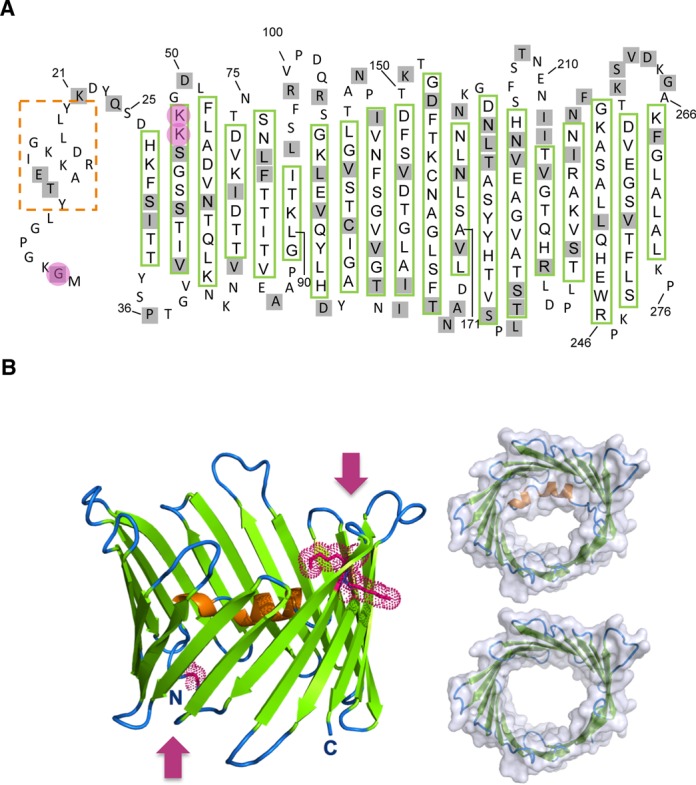
Plant VDAC architecture and tRNA binding regions. When performing modeling, VDAC 34 and 36 architecture models show no significant differences due to their high sequence identity (75%). Thus, this figure focuses on VDAC 34 features. (A) Topological arrangement of VDAC34. The N-terminal α-helix is indicated by an orange dashed box and the 19 β-strands of the β-barrel are depicted by open boxes (green frames). Amino acid residues different in VDAC36 are under gray background. G2, K47 and K48 residues are under pink background. (B) 3D model of VDAC34 generated using Modeler (25) and the mouse mitochondrial VDAC1 crystal structure as a template (PDB ID: 3EMN). It displays a characteristic VDAC architecture with the N-terminal helix in orange docked along the inner channel wall at the midpoint with the β-strands in green. Key residues for tRNA binding, G2 at one entry and K47K48 at the opposite entry, are highlighted by dotted volumes and are indicated by pink arrows. The N-terminus including the helix partially closes the pore and leaves a channel of about 25 × 15 Å (top right). The pore diameter is 25 Å large at the extremities and could be open more widely by moving the N-terminal region out of the channel (bottom right).
Comparative binding assays with a series of chimers and deletion mutants (Figure 4, Supplemental Figure S7) indicated that nucleic acid binding sites in VDAC34, although they may be discretely present all along the sequence, are mainly located in the N-terminal part of the protein including the helix and the first two strands (Figure 6A). Deletions in this region are detrimental to the binding (mutants D1–D2) and, conversely, its partial transplantation in VDAC36 can increase the efficiency of binding (mutants C1–C4). The β-barrel structure is also required since the binding property is lost as soon as more than 10 residues are deleted (mutants D3–D4), probably hampering the closure and, with larger deletions, the formation of a pore. The N-terminal region alone shows no binding activity (mutants D5–D7).
Key residues revealed by point mutagenesis, G2, K47 and K48, are located at both entries of the pore and probably do not act synergistically (Figures 5 and 6B). From the behavior of chimeric and point mutants it is obvious that the role of these residues is influenced by the local structural context. The protein orientation is not elucidated yet in plants, but the position of the three amino acids in the protein let us suppose that they may provide entry or sorting contact points with tRNA. They may be involved in primary recognition events assisted by the overall electropositive environment of the pore, which is favorable for interactions with polyanions. Alternatively, they could also maintain an optimal structure allowing critical contact points to be accessible.
DISCUSSION
The VDAC proteins of the OMM assume a crucial role in mitochondrial function and in cell life and death. Primarily known as responsible for the exchange of metabolites between the cytosol and mitochondria, they are also essential key players in aging, diseases or apoptosis. In addition, more recent studies have demonstrated that in higher plants, VDAC proteins are recruited for translocation of tRNA through the OMM. Understanding how VDAC proteins can perform such a variety of functions requires a better knowledge on structure-function relations.
Here, by two different methods, i.e. northwestern hybridizations and gel shift assays, we provide strong evidence that the two major S. tuberosum VDACs, VDAC34 and VDAC36, do not behave the same regarding their interaction with tRNA molecules: VDAC34 interacts more efficiently with tRNAs than VDAC36. The differential binding capacity of the two proteins are not restricted to tRNAs and was also seen with all types of nucleic acids i.e. single-stranded or double-stranded molecules DNA or RNA. This is likely linked to the fact that, as reported previously, both RNA and DNA can enter mitochondria isolated from plant cells and in an unspecific manner (11,26).
The analysis by northwestern of a collection of mutant proteins overexpressed in E. coli allowed us to show that the efficient interaction between VDAC34 and tRNA does not rely on a precise RNA-binding motif but rather on amino acids distributed all along the VDAC structure. The shape of the protein is likely to be crucial as well. Compared to the Kd value of human VDAC1 for nucleotides which is in the 1 mM range (27), the Kd value of about 0.1 μM obtained for both VDAC34 and 36 shows that these proteins present a relatively strong interaction with tRNAs. This appears in disagreement with the fact that the affinity between tRNA and VDAC is expected to be tenuous since it represents a transitory event where tRNA needs to rapidly cross the OMM via the pore of VDAC. However, we should take into account that the VDAC–tRNA interaction also depends on competition or synergy between various factors present in vivo. Indeed, different studies indicate that associated proteins can modulate VDAC permeability (28). Furthermore, tRNAs are usually not naked within the cell but rather strongly associated with proteins such as aminoacyl-tRNA synthetases (29). If tRNAs enter alone into mitochondria through VDAC, the interaction between tRNA and VDAC must be strong enough to allow the dissociation between tRNA and the cognate aminoacyl-tRNA synthetase with a Kd value in the range of 0.1–0.5 μM (30). Moreover, plant membranes contain a large variety of sterol types, and the type of sterol and its abundance in the mitochondrial membrane modulate VDAC properties (31). Thus, it is possible that the tRNA–VDAC interaction and/or tRNA translocation is also modulated by the lipid membrane content.
Our studies showed that the N-terminal region and few residues on this region, namely G2, K48 and K49, seem to be essential for the functional interaction. The importance of the N-terminal region of VDAC34 for tRNA interaction is reminiscent to what was observed in other VDAC biological functions. For instance, in human VDAC1, this region is a key element in regulating apoptosis and the target of anti-apoptotic proteins such as members of the Bcl-2 protein family (32). Furthermore, a study on rat VDAC1 demonstrates the presence of nucleotide-binding sites in the protein including one localized in the N-terminal part (33). This site corresponds to a sequence similar to the highly conserved ATP-binding motif A, G(X/G)XGXGKT (X denote any amino acid) described by Walker et al. (34). It is important to point out that this ATP-binding motif A is evocative to the G2KGPGLYT9 sequence in the N-terminal extremity of VDAC34 and that, interestingly, the VDAC34 G2 was shown to be an important residue for tRNA interaction. Moreover, a GLK triplet was also identified as a putative nucleotide-binding site in a porin from the pathogenic Neisseria species (35). The two other important identified residues are K48 and K49. These residues are present on both VDAC34 and VDAC36 proteins. Thus, they cannot confer by their own, the capacity to fix efficiently nucleic acids and they rather represent key elements of an overall environment of the pore favorable for tRNA interaction. It is worth to note that these three residues, G2, K48 and K49, are not present in VDAC proteins from other eukaryotic lineages. However, yeast and mammals VDAC proteins have also the capacity to interact with nucleic acids (7,8). In the light of these results, it is clear that analysis of VDAC sequences does not allow predicting their capacity to interact efficiently with tRNAs.
The mechanism of tRNA translocation is not clear yet. Our previous work demonstrated that the primary anchoring site of the tRNA to the OMM is not achieved by the VDAC protein itself but rather by a membrane receptor (12). Furthermore, as mentioned before, we do not exclude the recruitment of other proteins to promote the transport through VDAC. The diameter of the pore (25 Å) could be compatible with the docking of a DNA/RNA molecule and even with the passage of a nucleic acid hairpin if the N-terminal helix is displaced out of the β-barrel. Recent data however suggest that such movements are not required for VDAC transport activity since mVDAC1 with the helix covalently linked to the β-barrel displays unaltered anion transport properties (36). Other contributions advocate for a rather flexible structure of the β-barrel pore (37). This idea is supported by crystal structure of the β-barrel protein FimD from E. coli, where large conformational changes involving pore shape and size modifications were observed (38).
Phylogenetic analysis (4,39) shows that gene duplication is at the origin of potato VDAC34 and VDAC36. We can therefore hypothesize that upon gene duplication, the two major S. tuberosum VDAC isoforms acquired specialized function, and VDAC34 (or other plant homologues) has evolved to develop a dedicated gate for tRNA import in plants from the time where the loss of mitochondrial genetic material also led to the disappearance of tRNA genes from plant mitochondrial genomes. Multiple putative roles of plant VDACs have been proposed, e.g. example in pathogen response, apoptosis and abiotic stress (40–44). It has also been shown that plant VDACs are differentially dual targeted to both mitochondria and plasma membrane (45). Here, the comparison between potato VDAC34 and VDAC36 brings additional arguments in favor of the acquisition, within this small protein family, of specific functions besides their primary role in the exchange of metabolites.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We wish to thank J.M. Grienenberger and P. Giegé for fruitful discussion. This work has been also published under the framework of the LABEX (ANR-11-LABX-0057_MITOCROSS) and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program.
ACCESSION NUMBER
PDB ID: 3EMN
FUNDING
Centre National de la Recherche Scientifique (CNRS) in association with the University of Strasbourg, by the Agence Nationale pour la Recherche (ANR) [ANR-09-BLAN-0240-01, ANR-09-BLAN-0091-03]; ANR [ANR-09-BLAN-0240-01 to T.S.]; LabEx consortium «MitoCross» [ANR-11-LABX-0057_MITOCROSS to T.S.]; French Ministère de l'Education et de la Recherche [to S.E.-A.]. Funding for open access charge: Centre National de la Recherche Scientifique (CNRS) in association with the University of Strasbourg, by the Agence Nationale pour la Recherche (ANR) [ANR-09-BLAN-0240-01, ANR-09-BLAN-0091-03]; ANR [ANR-09-BLAN-0240-01 to T.S.]; LabEx consortium «MitoCross» [ANR-11-LABX-0057_MITOCROSS to T.S.]; French Ministère de l'Education et de la Recherche [to S.E.-A.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Bayrhuber M., Meins T., Habeck M., Becker S., Giller K., Villinger S., Vonrhein C., Griesinger C., Zweckstetter M., Zeth K. Structure of the human voltage-dependent anion channel. Proc. Natl. Acad. Sci. U.S.A. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiller S., Garces R.G., Malia T.J., Orekhov V.Y., Colombini M., Wagner G. Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science. 2008;321:1206–1210. doi: 10.1126/science.1161302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ujwal R., Cascio D., Colletier J.P., Faham S., Zhang J., Toro L., Ping P., Abramson J. The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17742–17747. doi: 10.1073/pnas.0809634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homblé F., Krammer E.M., Prevost M. Plant VDAC: facts and speculations. Biochim. Biophys. Acta. 2012;1818:1486–1501. doi: 10.1016/j.bbamem.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 5.Shoshan-Barmatz V., Ben-Hail D. VDAC, a multi-functional mitochondrial protein as a pharmacological target. Mitochondrion. 2012;12:24–34. doi: 10.1016/j.mito.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Koulintchenko M., Konstantinov Y., Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koulintchenko M., Temperley R.J., Mason P.A., Dietrich A., Lightowlers R.N. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum. Mol. Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 8.Weber-Lotfi F., Ibrahim N., Boesch P., Cosset A., Konstantinov Y.N., Lightowlers R.N., Dietrich A. Biochim. Biophys. Acta. 2009;1787:320–327. doi: 10.1016/j.bbabio.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salinas T., Duchêne A.M., Maréchal-Drouard L. Recent advances in tRNA mitochondrial import. Trends Biochem. Sci. 2008;33:320–329. doi: 10.1016/j.tibs.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 10.Alfonzo J.D., Söll D. Mitochondrial tRNA import - the challenge to understand has just begun. Biol. Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sieber F., Duchêne A.M., Maréchal-Drouard L. Mitochondrial RNA import from diversity of natural mechanisms to potential applications. Int. Rev. Cell Mol. Biol. 2011;287:145–190. doi: 10.1016/B978-0-12-386043-9.00004-9. [DOI] [PubMed] [Google Scholar]
- 12.Salinas T., Duchêne A.M., Delage L., Nilsson S., Glaser E., Zaepfel M., Maréchal-Drouard L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18362–18367. doi: 10.1073/pnas.0606449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heins L., Mentzel H., Schmid A., Benz R., Schmitz U.K. Biochemical, molecular, and functional characterization of porin isoforms from potato mitochondria. J. Biol. Chem. 1994;269:26402–26410. [PubMed] [Google Scholar]
- 14.Duchêne A.M., Giritch A., Hoffmann B., Cognat V., Lancelin D., Peeters N.M., Zaepfel M., Maréchal-Drouard L., Small I.D. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Placido A., Gagliardi D., Gallerani R., Grienenberger J.M., Maréchal-Drouard L. Fate of a larch unedited tRNA precursor expressed in potato mitochondria. J. Biol. Chem. 2005;280:33573–33579. doi: 10.1074/jbc.M505269200. [DOI] [PubMed] [Google Scholar]
- 16.Carneiro V.T., Dietrich A., Maréchal-Drouard L., Cosset A., Pelletier G., Small I. Characterization of some major identity elements in plant alanine and phenylalanine transfer RNAs. Plant Mol. Biol. 1994;26:1843–1853. doi: 10.1007/BF00019497. [DOI] [PubMed] [Google Scholar]
- 17.Delage L., Duchêne A.M., Zaepfel M., Maréchal-Drouard L. The anticodon and the D-domain sequences are essential determinants for plant cytosolic tRNA(Val) import into mitochondria. Plant J. 2003;34:623–633. doi: 10.1046/j.1365-313x.2003.01752.x. [DOI] [PubMed] [Google Scholar]
- 18.Ryder S.P., Recht M.I., Williamson J.R. Quantitative analysis of protein-RNA interactions by gel mobility shift. Methods Mol. Biol. 2008;488:99–115. doi: 10.1007/978-1-60327-475-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warf M.B., Diegel J.V., von Hippel P.H., Berglund J.A. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Notredame C., Higgins D.G., Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 21.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole C., Barber J.D., Barton G.J. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 24.Wang L., Brown S.J. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 2006;34:243–248. doi: 10.1093/nar/gkl298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fiser A., Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 26.Ibrahim N., Handa H., Cosset A., Koulintchenko M., Konstantinov Y., Lightowlers R.N., Dietrich A., Weber-Lotfi F. DNA delivery to mitochondria: sequence specificity and energy enhancement. Pharm. Res. 2011;28:2871–2882. doi: 10.1007/s11095-011-0516-4. [DOI] [PubMed] [Google Scholar]
- 27.Villinger S., Giller K., Bayrhuber M., Lange A., Griesinger C., Becker S., Zweckstetter M. Nucleotide interactions of the human voltage-dependent anion channel. J. Biol. Chem. 2014;289:13397–13406. doi: 10.1074/jbc.M113.524173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shoshan-Barmatz V., De Pinto V., Zweckstetter M., Raviv Z., Keinan N., Arbel N. VDAC, a multi-functional mitochondrial protein regulating cell life and death. Mol. Aspects Med. 2010;31:227–285. doi: 10.1016/j.mam.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Negrutskii B.S., Deutscher M.P. Channeling of aminoacyl-tRNA for protein synthesis in vivo. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4991–4995. doi: 10.1073/pnas.88.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giegé R., Puglisi J.D., Florentz C. RNA structure and aminoacylation efficiency. Prog. Nucleic Acid Res. Mol. Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- 31.Mlayeh L., Chatkaew S., Leonetti M., Homblé F. Modulation of plant mitochondrial VDAC by phytosterols. Biophys. J. 2010;99:2097–2106. doi: 10.1016/j.bpj.2010.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abu-Hamad S., Arbel N., Calo D., Arzoine L., Israelson A., Keinan N., Ben-Romano R., Friedman O., Shoshan-Barmatz V. The VDAC1 N-terminus is essential both for apoptosis and the protective effect of anti-apoptotic proteins. J. Cell Sci. 2009;122:1906–1916. doi: 10.1242/jcs.040188. [DOI] [PubMed] [Google Scholar]
- 33.Yehezkel G., Hadad N., Zaid H., Sivan S., Shoshan-Barmatz V. Nucleotide-binding sites in the voltage-dependent anion channel: characterization and localization. J. Biol. Chem. 2006;281:5938–5946. doi: 10.1074/jbc.M510104200. [DOI] [PubMed] [Google Scholar]
- 34.Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudel T., Schmid A., Benz R., Kolb H.A., Lang F., Meyer T.F. Modulation of Neisseria porin (PorB) by cytosolic ATP/GTP of target cells: parallels between pathogen accommodation and mitochondrial endosymbiosis. Cell. 1996;85:391–402. doi: 10.1016/s0092-8674(00)81117-4. [DOI] [PubMed] [Google Scholar]
- 36.Teijido O., Ujwal R., Hillerdal C.O., Kullman L., Rostovtseva T.K., Abramson J. Affixing N-terminal alpha-helix to the wall of the voltage-dependent anion channel does not prevent its voltage gating. J. Biol. Chem. 2012;287:11437–11445. doi: 10.1074/jbc.M111.314229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rostovtseva T.K., Kazemi N., Weinrich M., Bezrukov S.M. Voltage gating of VDAC is regulated by nonlamellar lipids of mitochondrial membranes. J. Biol. Chem. 2006;281:37496–37506. doi: 10.1074/jbc.M602548200. [DOI] [PubMed] [Google Scholar]
- 38.Phan G., Remaut H., Wang T., Allen W.J., Pirker K.F., Lebedev A., Henderson N.S., Geibel S., Volkan E., Yan J., et al. Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate. Nature. 2011;474:49–53. doi: 10.1038/nature10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young M.J., Bay D.C., Hausner G., Court D.A. The evolutionary history of mitochondrial porins. BMC Evol. Biol. 2007;7:31–51. doi: 10.1186/1471-2148-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai M.K., Mishra R.N., Verma D., Nair S., Sopory S.K., Reddy M.K. Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum) Plant Physiol. Biochem. 2006;44:483–493. doi: 10.1016/j.plaphy.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Jones A.M., Thomas V., Bennett M.H., Mansfield J., Grant M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006;142:1603–1620. doi: 10.1104/pp.106.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kusano T., Tateda C., Berberich T., Takahashi Y. Voltage-dependent anion channels: their roles in plant defense and cell death. Plant Cell Rep. 2009;28:1301–1308. doi: 10.1007/s00299-009-0741-z. [DOI] [PubMed] [Google Scholar]
- 43.Lacomme C., Roby D. Identification of new early markers of the hypersensitive response in Arabidopsis thaliana. FEBS Lett. 1999;459:149–153. doi: 10.1016/s0014-5793(99)01233-8. [DOI] [PubMed] [Google Scholar]
- 44.Lee A.C., Xu X., Blachly-Dyson E., Forte M., Colombini M. The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J. Membr. Biol. 1998;161:173–181. doi: 10.1007/s002329900324. [DOI] [PubMed] [Google Scholar]
- 45.Robert N., d'Erfurth I., Marmagne A., Erhardt M., Allot M., Boivin K., Gissot L., Monachello D., Michaud M., Duchêne A.M., et al. Voltage-dependent-anion-channels (VDACs) in Arabidopsis have a dual localization in the cell but show a distinct role in mitochondria. Plant Mol. Biol. 2012;78:431–446. doi: 10.1007/s11103-012-9874-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


