Summary
The complex genetic changes underlying metastatic melanoma need to be deciphered to develop new and effective therapeutics. Previously, genome-wide microarray analyses of human melanoma identified two reciprocal gene expression programs, including transcripts regulated by either transforming growth factor, beta 1 (TGFβ1) pathways or microphthalmia-associated transcription factor (MITF)/SRY-box containing gene 10 (SOX10) pathways. We extended this knowledge by discovering that melanoma cell lines with these two expression programs exhibit distinctive microRNA (miRNA) expression patterns. We also demonstrated that hypoxia-inducible factor 1 alpha (HIF1A) is increased in TGFβ1 pathway-expressing melanoma cells and that HIF1A upregulates miR-210, miR-218, miR-224, and miR-452. Reduced expression of these four miRNAs in TGFβ1 pathway-expressing melanoma cells arrests the cell cycle, while their overexpression in mouse melanoma cells increases the expression of the hypoxic response gene Bnip3. Taken together, these data suggest that HIF1A may regulate some of the gene expression and biological behavior of TGFβ1 pathway-expressing melanoma cells, in part via alterations in these four miRNAs.
Keywords: melanoma, miRNA, hypoxia, HIF1, Bnip3
Introduction
Cutaneous melanoma is the most lethal form of skin cancer, demonstrating an increasing incidence and a notable resistance to currently available treatments (Nikolaou and Stratigos, 2014; Siegel et al., 2013). While genetic studies have begun to decipher the diverse alterations that can arise in melanoma as well as the heterogeneity of cells within a melanoma tumor (Hill et al., 2013; Hoek and Goding, 2010), a comprehensive understanding of the molecular pathogenesis that underlies melanoma progression is lacking. Recent genome-wide microarray analyses using collections of melanoma cell lines have identified two reciprocal gene expression programs, and these two programs were shown to include distinctive expression of transcripts regulated by either transforming growth factor, beta 1 (TGFβ1) pathways or microphthalmia-associated transcription factor (MITF)/SRY-box containing gene 10 (SOX10) pathways (Bell et al., 2014; Hoek et al., 2006; Jeffs et al., 2009). Hoek and colleagues defined these two programs as “proliferative” for the MITF/SOX10 pathway-positive (+) melanoma lines and “invasive” for the TGFβ1 pathway+ melanoma lines, based on the corresponding biological behaviors of these cell lines in culture (Hoek et al., 2006). While the increased proliferation rate was subsequently not observed for “proliferative” MITF/SOX10 pathway+ cells by Jeffs et al., the “invasive” TGFβ1 pathway+ cells did exhibit stronger motility by scratch and transwell assays (Jeffs et al., 2009). While it remains to be determined whether “invasive” TGFβ1 pathway+ melanoma cells indeed have increased metastasis propensity in vivo compared with “proliferative” MITF/SOX10 pathway+ melanoma cells, this genetically defined classification provides a useful framework for studying the biological behaviors of melanoma cells that are relevant to their metastasis.
MicroRNAs (miRNAs) are 20–24 nucleotide noncoding RNAs that regulate the stability or translational efficiency of complementary target mRNAs (Mendell and Olson, 2012). MiRNAs are often misexpressed in cancers, playing important roles in tumor formation and progression by acting as oncogenes, tumor suppressors, and metastasis promoters/suppressors (Lujambio and Lowe, 2012; Pencheva and Tavazoie, 2013). Furthermore, increasing evidence suggests miRNAs are involved in melanoma progression and metastasis (Bonazzi et al., 2012; Gaziel-Sovran et al., 2011). Because a single miRNA often regulates multiple targets and because antisense technology exists that allows inhibition of individual miRNAs with high specificity, miRNAs have become an attractive treatment modality for human disease, including cancer (Kasinski and Slack, 2011). A recent report exemplifies how studies of miRNA biological functions present new clinical opportunities to fight melanoma metastasis (Pencheva et al., 2012). From in vivo selected melanoma cell lines, this study identified three miRNAs (miR-1908, miR-199a-5p, and miR- 199a-3p) that cooperatively promoted invasion, angiogenesis and colonization. Inhibition of all three miRNAs strongly suppressed metastasis for a diverse variety of melanoma cells, and furthermore, the individual or aggregate expression level of the three miRNAs predicted metastasis-free survival in melanoma patients.
Hypoxia is a prominent feature of the microenvironment that surrounds tumors, and a well-established effect of hypoxia is to promote metastasis (Sullivan and Graham, 2007). In particular, the role of hypoxia in melanoma metastasis has been emerging (Cheli et al., 2012). Hypoxia-inducible factor 1 alpha (HIF1A) is a master regulator of the cellular hypoxic response (Majmundar et al., 2010), and a direct link between HIF1A and melanoma metastasis was recently reported (Hanna et al., 2013). Hanna et al. found that inactivation of HIF1A greatly reduced metastasis but had no effect on primary tumor formation in a mouse melanoma model (Pten-deficient, Braf-mutant); furthermore, hypoxia induced upregulation of HIF1A in human melanoma cells and promoted invasion in culture, while HIF1A knockdown had the opposite effects. In addition, Widmer et al. recently reported that hypoxia increased invasion of “proliferative” MITF/SOX10+ melanoma cells in vitro but had no effects on invasion of “invasive” TGFβ1+ melanoma cells (Widmer et al., 2013). Taken together, these data raise some interesting questions: is the HIF1A-regulated hypoxic response activated in “invasive” TGFβ1+ melanoma cells even under normoxic conditions, and does it contribute to their heightened invasive potential?
In this study, we investigated miRNA expression patterns in both “proliferative” MITF/SOX10 pathway+ and “invasive” TGFβ1 pathway+ human melanoma cell lines. We identified a set of miRNAs that exhibited differential expression between these two expression profile-defined subtypes of melanoma cells. We then demonstrated HIF1A expression was increased in TGFβ1 pathway+ melanoma cells and contributed to the increased expression of miR-210, miR-218, miR-224, and miR-452. We also demonstrated that reduced expression of this set of four miRNAs in TGFβ1 pathway+ melanoma cells caused cell cycle arrest, suggesting these miRNAs may contribute to cell cycle progression. Overexpression of miR-210, miR-218, miR-224, and miR-452 in mouse melanoma cells increased the expression of the hypoxic response gene BCL2/adenovirus E1B interacting protein 3 (Bnip3) as well as the stress response gene activating transcription factor 3 (Atf3). Our results expand the reciprocal gene expression programs in melanoma cell lines to include miRNAs, and they also provide evidence that HIF1A might function in regulating the gene expression program and biological behavior of “invasive” TGFβ1 pathway+ melanoma cells.
Results
Expression of specific miRNAs correlates with the MITF/SOX10- and TGFβ1-regulated mRNA expression programs in metastatic melanoma cell lines
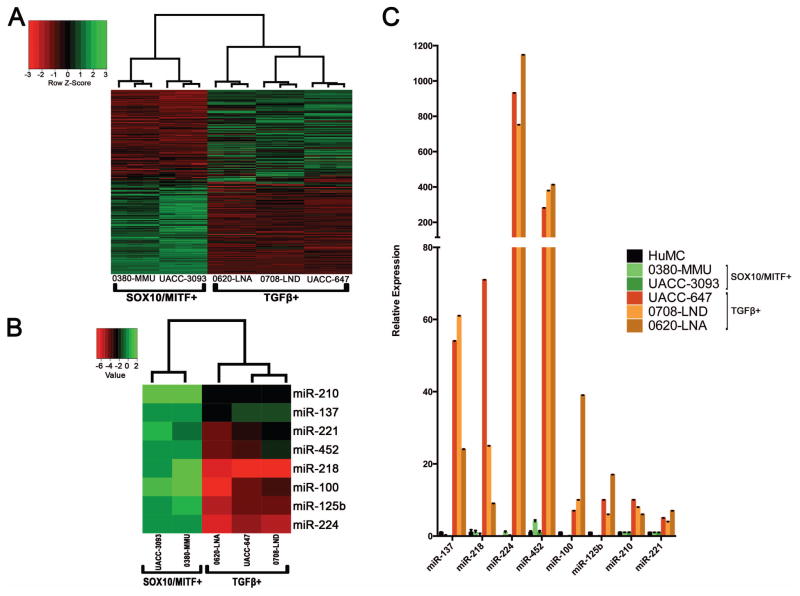
We first performed genome-wide mRNA microarray analysis on a panel of five human melanoma cell lines: 0380-MMU, UACC-3093, 0620-LNA, 0708-LND, and UACC-647. Hierarchical clustering analysis using all transcripts separated these lines into two groups, one containing 0380-MMU and UACC-3093, and the other containing 0620-LNA, 0708-LND, and UACC-647 (data not shown). Hoek and colleagues previously reported a 223-gene molecular signature that classifies metastatic melanoma cell lines into the two phenotypically distinct cohorts of “proliferative,” showing expression of MITF/SOX10-regulated pathways (hereafter MITF/SOX10+), and “invasive,” showing expression of TGFβ1-regulated pathways (hereafter TGFβ1+) (Hoek et al., 2006). Application of this 223-gene molecular signature to our melanoma cell line panel clustered the melanoma cell lines into the same two groups as genome-wide analysis, and showed that the 0380-MMU and UACC-3093 lines exhibited an MITF/SOX10+ gene expression pattern, while a TGFβ1+ gene expression pattern was present in the 0620-LNA, 0708-LND, and UACC-647 lines (Figure 1A).
Figure 1.
Eight miRNAs are differentially expressed between two expression profile-defined groups of human melanoma cell lines. A. Hierarchical cluster analysis with the 223-gene signature divides the melanoma cell lines into two groups: 0380-MMU and UACC-3093, which exhibit the “proliferative” SOX10/MITF+ gene expression profile, and 0620-LNA, 0708-LND, and UACC-647, which exhibit the “invasive” TGFβ1+ expression profile. B. Hierarchical cluster analysis using expression of 377 miRNAs separates these melanoma cell lines into the same two SOX10/MITF+ and TGFβ1+ groups as in A; this heatmap shows the 8 miRNAs that are differentially expressed between the two groups. Values = ΔΔCt. C. Quantitative RT-PCR confirms differential expression of the 8 miRNAs between the two groups of melanoma cell lines. HuMC, primary human melanocytes. Except for miR-224, all expression levels are normalized to primary human melanocytes. For miR-224, which was not detectable in primary human melanocytes, expression level was normalized to that of the SOX10/MITF+ cell line 0380-MMU.
We next assayed miRNA expression patterns in these five melanoma cell lines using TaqMan Low Density Array (TLDA). To identify miRNA expression changes specific to melanomagenesis, we also performed TLDA analysis in primary human melanocytes (HuMC; data in Table S1). Interestingly, hierarchical clustering based on miRNA expression profiles classified the five cell lines into the same two groups identified in the mRNA analysis: 0380-MMU with UACC-3093 (MITF/SOX10+), and 0620-LNA with 0708-LND and UACC-647 (TGFβ1+; Supporting Information Figure S1). TLDA analysis identified eight miRNAs (miR-137, miR-218, miR-224, miR-452, miR-100, miR-125b, miR-210 and miR-221) that were expressed at similar levels in HuMC and the MITF/SOX10+ group but were upregulated in the TGFβ1+ group (Figure 1B). In contrast, the expression level of miR-211, a known suppressor for melanoma invasion that is silenced during melanoma progression (Levy et al., 2010), was similar between HuMC and MITF/SOX10+ cells, but almost undetectable in the TGFβ1+ cells (data not shown). The increased expression of the eight miRNAs in the TGFβ1+ cell lines identified in TLDA analysis was subsequently validated by TaqMan qPCR (Figure 1C). Taken together, these data suggested specific upregulation of these 8 miRNAs occurred alongside the TGFβ1+ gene expression program.
HIF1A is overexpressed and regulates miR-210, miR-218, miR-224 and miR-452 in TGFβ1 melanoma cell lines
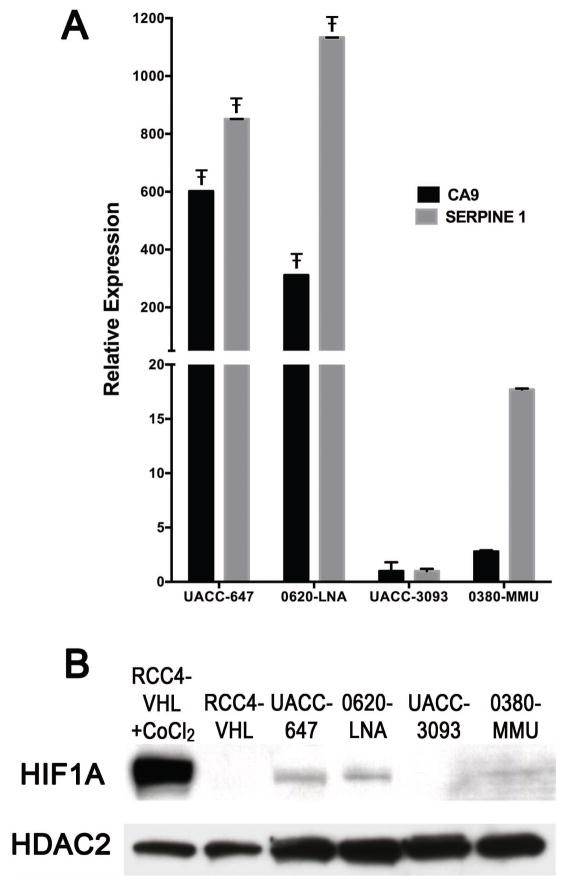
One of the eight miRNAs, miR-210, is a well-known target of HIF1A and a marker for hypoxia (Huang et al., 2010). The overexpression of miR-210 in the TGFβ1+ melanoma cell lines suggested these cells might have increased HIF1A activities under normoxic conditions. In support of this, the mRNA expression levels of two known HIF1A targets, CA9 and SERPINE1, were significantly greater in TGFβ1+ cell lines compared to MITF/SOX10+ cell lines (Figure 2A). Furthermore, examination of the expression of HIF1A protein in the TGFβ1+ and MITF/SOX10+ melanoma cell lines under normoxic conditions showed HIF1A expression in the TGFβ1+ cell lines, and either absent or weak expression in the MITF/SOX10+ cell lines (Figure 2B), consistent with the mRNA expression results. High levels of HIF1A transcriptional activity in the TGFβ1+ UACC-647 and 0620-LNA cell lines were confirmed using a luciferase reporter containing HIF-responsive elements (HREs) in its promoter along with a control reporter containing mutated HREs that prevent HIF1A binding (Supporting Information Figure S2). These experiments demonstrated that HIF1A is overexpressed in these TGFβ1+ melanoma cell lines under normoxic conditions, suggesting it may play a role in their increased metastatic potential.
Figure 2.
HIF1A is expressed in TGFβ1+ melanoma cell lines under normoxic conditions. A. qRT-PCR demonstrates significantly higher expression under normoxic conditions of the HIF1A targets CA9 and SERPINE1 in the TGFβ1+ melanoma cell lines UACC-647 and 0620-LNA as compared to the SOX10/MITF+ melanoma cell lines 0380-MMU and UACC-3093 (Ŧ indicates multiplicity-adjusted p value < 0.0001, One-way ANOVA with Tukey’s post-test, comparing each TGFβ1+ melanoma cell line with each SOX10/MITF+ melanoma cell line). Expression levels are normalized to 18S rRNA. B. Immunoblot analysis demonstrates the presence of HIF1A under normoxic conditions in in the TGFβ1+ melanoma cell lines UACC-647 and 0620-LNA and absent/faint expression in the SOX10/MITF+ cell lines UACC-3093 and 0380-MMU, respectively. The faint HIF1A expression in 0380-MMU cells correlates with the modest expression of SERPINE1 in (A). RCC4-VHL cells treated with Cobalt Chloride CoCl2 to induce HIF1A expression and RCC4-VHL cells serve as positive and negative controls, respectively. HDAC2 is the loading control.
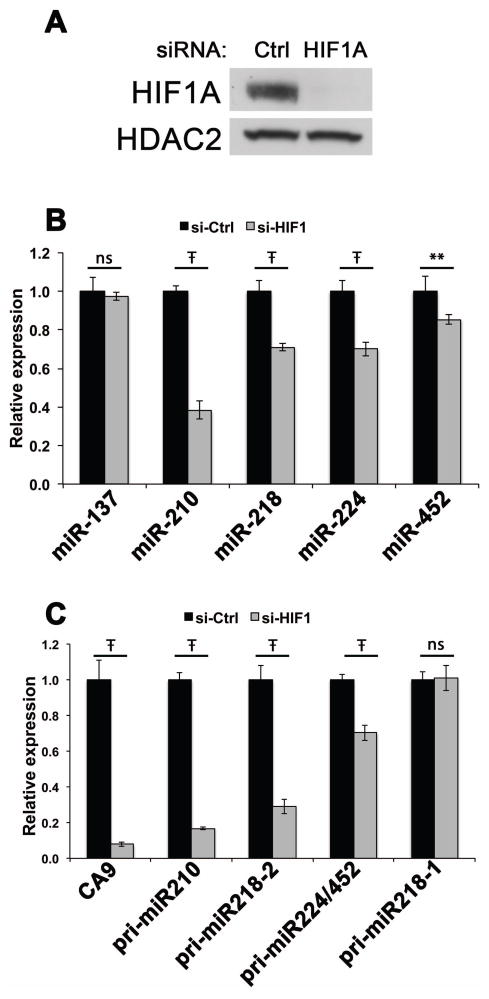
Among the eight miRNAs upregulated in the TGFβ1+ lines, four of them (miR-137, miR-218, miR-224 and miR-452) were undetectable in HuMC and either undetectable or expressed at very low levels in the MITF/SOX10+ group. We hypothesized that the absence of expression of these four miRNAs in HuMC might indicate their selection during melanoma progression, and thus we focused on these four miRNAs for further analysis. Since none of these miRNAs were previously associated with hypoxia, we asked whether HIF1A plays a role in the upregulation of these miRNAs in TGFβ1+ human melanoma cells, and included miR-210 as a positive control. After verifying that complete HIF1A knockdown was achieved by siRNA transfection (Figure 3A), miRNA levels were assayed by qRT-PCR in both HIF1A siRNA- and control siRNA-transfected TGFβ1+ melanoma cells. As expected, HIF1A knockdown resulted in more than 50% reduction in miR-210 expression (Figure 3B).
Figure 3.
HIF1A regulates the expression of miR-210, miR-218, miR-224 and miR-452 in TGFβ1+ melanoma cells. A. Immunoblot analysis shows expression of HIF1A in the TGFβ1+ melanoma cell line 0620-LNA (Ctrl, left) and the complete knockdown of HIF1A in the TGFβ1+ melanoma cell line 0620-LNA by HIF1A siRNA transfection (HIF1A, right). HDAC2 serves as a loading control. Ctrl, non-targeting control siRNA. B. qRT-PCR demonstrates significantly decreased expression of miR-210, miR-218, miR-224 and miR-452 as a result of HIF1A knockdown in TGFβ1+ melanoma cells. C. qRT-PCR demonstrates significantly decreased expression of pri-miR-210, pri-miR-218-2, and pri-miR-224/452. CA9 serves as a positive control. For panels B and C, the expression level for each miRNA and pri-miRNA is normalized to control siRNA-transfected cells (si-Ctrl); ** and Ŧ indicate multiplicity adjusted p values of <0.005 and p<0.0001, respectively, One-way ANOVA with Sidak’s multiple comparisons test; ns indicates no significant difference.
While the expression of miR-137 was unaffected, the abundance of miR-218, miR-224 and miR-452 was significantly decreased by HIF1A knockdown. Because mature miRNAs in general have longer half-lives compared with mRNAs (Kim et al., 2009), we also measured the abundance of the miRNA primary transcripts (pri-miRNAs) that are precursors to miR-210, miR-218, miR-224 and miR-452 to provide a more sensitive readout of HIF1A knockdown. In the human genome, miR-224 and miR-452 are processed from the same primary transcript while miR-218 is encoded by two loci, pri-miR-218-1 and pri-miR-218-2. We found that pri-miR-210 expression was significantly reduced by HIF1A knockdown and the expression levels of pri-miR-218-2 and pri-miR-224/452 were also significantly decreased by 71% and 30%, respectively (Figure 3C). The transcript pri-miR-218-1 was unaffected, demonstrating specificity for the pri-miR-218-2 locus. Taken together, the pri-miRNA and miRNA results demonstrated that miR-210, miR-218, miR-224 and miR-452 expression is positively regulated by HIF1A in TGFβ1+ human melanoma cell lines.
Inhibition of miR-210, miR-218, miR-224 and miR-452 causes cell cycle arrest
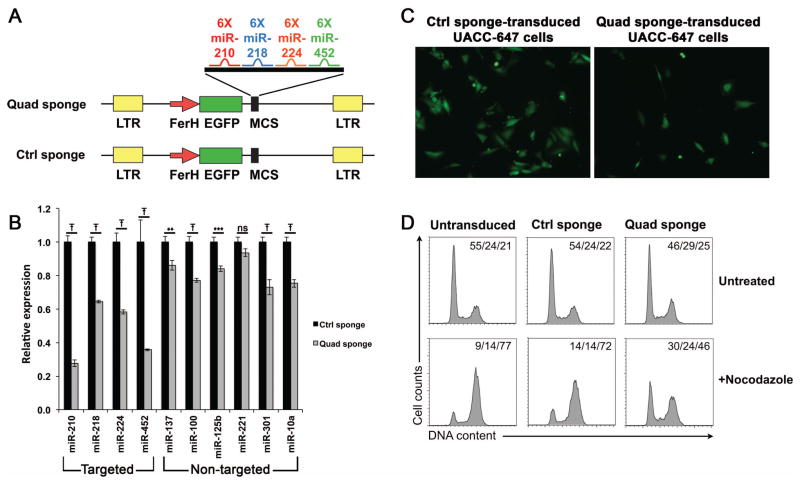
Previous studies suggest multiple miRNAs cooperate to promote cancer progression and metastasis (Garofalo et al., 2012; Pencheva et al., 2012). To investigate the collective biological function of the four HIF1A-regulated miRNAs we identified in TGFβ1+ human melanoma cell lines, we constructed a lentiviral miRNA sponge that contained 6 binding sites each for miR-210, miR-218, miR-224 and miR-452, thus permitting their simultaneous inhibition (Quad sponge, Figure 4A). Stable inhibitory cell lines were established by transducing the TGFβ1+ cell lines 0620-LNA and UACC-647 with the Quad sponge or with an empty sponge as a control (Ctrl sponge), and the GFP-positve transduced cells were subsequently selected by fluorescence-activated cell sorting. Since the competitive inhibition mediated by miRNA sponges results in a decrease in miRNA expression (Ebert and Sharp, 2010), we assayed miRNA expression in Quad sponge- and Ctrl sponge-transduced cells as a surrogate indicator for miRNA inhibition. Of note, both the targeted miRNAs and the non-targeted controls, with the exception of miR-221, showed significant decreases in relative expression levels in Quad sponge-transduced cells (Figure 4B). However, the targeted miRNAs had 2–3 fold greater reductions in expression levels (0.35—0.72) than those seen in the non-targeted miRNAs (0.14—0.27), which was consistent with successful targeting of the four miRNAs of interest in Quad sponge-transduced cells. Interestingly, following flow sorting and plating at equal densities, Quad sponge-transduced cells soon stopped dividing, in stark contrast to the Ctrl sponge-transduced cells, which continued to proliferate (Figure 4C and data not shown from multiple experiments in both TGFβ1+ cell lines). This result suggested one or more of these four miRNAs are responsible for proper cell-cycle progression in TGFβ1+ cells. To further analyze this blockage, cell-cycle analysis was performed by flow cytometry in UACC-647 cells. Comparison of untransduced, Ctrl sponge-transduced, and Quad sponge-transduced cells showed no significant alterations in cell cycle profiles (Figure 4D, top 3 panels). However, it has been shown that cell cycle profiling at steady state may not be sensitive enough to detect G1-phase arrest (Lanni and Jacks, 1998). Therefore, we also performed cell cycle analysis using the nocodazole trap assay, a commonly used method to examine G1-phase arrest in which nocadozole holds cycling cells in the G2/M phase but does not affect cells that are arrested before the G2/M checkpoint (Gerald et al., 2002). Nocodazole treatment trapped the majority of untransduced and Ctrl sponge-transduced cells at the G2/M phase (77% and 72%, respectively), and only small percentages of cells remained at G1 (9% and 14%, respectively). In contrast, less than 50% of Quad sponge-transduced cells were trapped by nocodazole while 30% of cells remained in G1 (Figure 4D, bottom right panel), suggesting that the inhibition of these four HIF1A-regulated miRNAs in TGFβ1+ melanoma cells caused cell-cycle arrest prior to G2/M.
Figure 4.
Inhibition of HIF1A-regulated miRNAs in TGFβ1+ melanoma cells results in cell cycle arrest. A. Schema showing the design of the miRsponge lentiviral vectors. For each miRNA, six bulged binding sites were inserted using the MCS of the Quad sponge vector. The Ctrl sponge vector consisted of the vector backbone with no insertions. LTR, long-terminal repeat; FerH, hybrid SV40 enhancer-human ferritin promoter; EGFP, enhanced green fluorescent protein; MCS, multiple cloning site. B. qRT-PCR on the targeted miRNAs miR-210, miR-218, miR-224, and miR-452 as well as the non-targeted controls miR-137, miR-100, miR-125b, miR-221, miR-301, and miR10a in Quad sponge- and Ctrl sponge-transduced TGFβ1+ UACC-647 melanoma cells. The results demonstrate significant reduction in miRNA expression levels in Quad sponge-transduced cells for 9 out of the 10 miRNAs examined, however the four miRNAs targeted by the Quad sponge show 2–3 times greater reduction in expression levels. **, ***, and Ŧ indicate multiplicity adjusted p values p<0.005, p<0.001, and p<0.0001, respectively, One-way ANOVA with Sidak’s multiple comparisons test; ns indicates no significant difference. Expression levels are normalized to Ctrl sponge-transduced cells. C. Fluorescent microscopy images show the decrease in proliferation in Quad sponge-transduced cells (right) compared to Ctrl sponge-transduced cells (left). Images were taken 24 hours after FACS sorting to isolate EGFP+ cells followed by plating at equivalent densities. D. Cell cycle analysis using a nocodazole trap assay demonstrated cell-cycle arrest in Quad sponge-transduced cells (bottom 3 panels). Cell cycle analysis of untreated cells showed no significant difference (top 3 panels). Numbers in the upper right corners of each histogram indicate the percentages of cells in G1/S/G2-M phases, respectively.
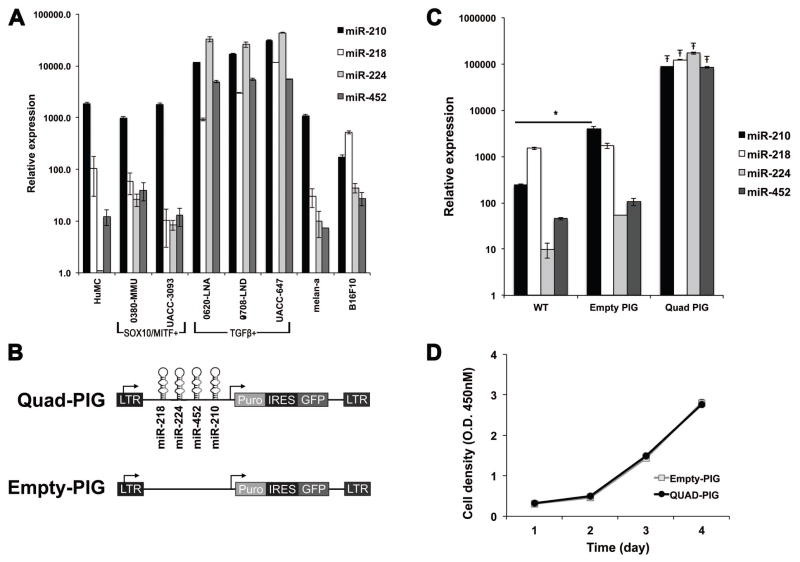
Overexpression of miR-210, miR-218, miR-224 and miR-452 causes increased Bnip3 and Atf3 expression in murine melanoma cells
As an alternative strategy to investigate the potential collective function of the four HIF1A-regulated miRNAs (miR-210, miR-218, miR-224 and miR-452), we overexpressed all four miRNAs in B16F10 mouse melanoma cells. B16F10 cells were chosen because they showed endogenous expression of these four miRNAs at similar levels to those of HuMC, immortalized mouse melanocytes (melan-a cells), and SOX10/MITF+ melanoma cell lines, but at lower levels than the those of TGFβ1+ human melanoma cells (Figure 5A). B16F10 cells were transduced with a retroviral construct containing all four miRNAs (Quad-PIG) or an empty construct (Empty-PIG) to generate stable cell lines (Figure 5B). Overexpression of all four miRNAs was confirmed by qPCR (Figure 5C) and had no effects on growth in culture (Figure 5D).
Figure 5.
Overexpression of miR-210, miR-218, miR-224, and miR-452 in B16F10 melanoma cells. A. Endogenous expression levels of the 4 miRNAs in B16F10 mouse melanoma cells (B16F10, far right) are similar to those seen in normal human melanocytes (HuMC), immortalized mouse Melan-Ink4a-Arf1 melanocytes (melan-a), and SOX10/MITF+ human melanoma cell lines, but are lower than those seen in TGFβ1+ human melanoma cell lines. Expression level data are presented in terms of 2−Ct × 1012. B. Schema showing the design of the Quad-PIG and Empty-PIG vectors. C. qRT-PCR demonstrates the significantly increased expression of miR-210, miR-218, miR-224 and miR-452 in B16F10 cells stably transduced with the Quad-PIG vector compared to both untransduced cells (WT) and to cells transduced with the Empty-PIG vector. Ŧ indicates multiplicity adjusted p value < 0.0001 for pair-wise comparisons of each miR with both WT and Empty-PIG, One-way ANOVA with Tukey’s post-test. Comparison of WT and Empty-PIG demonstrates no significant differences with the exception of a modest increase in miR-210 (* indicates multiplicity adjusted p value = 0.035, One-way ANOVA with Tukey’s post-test). Expression level data are presented in terms of 2−Ct × 1012. D. Proliferation assay showing similar growth curves between Empty-PIG- and Quad-PIG-transduced cells.
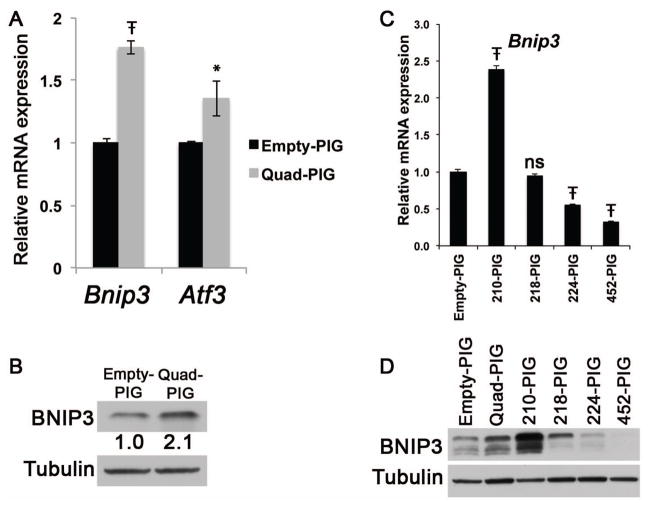
To identify gene expression changes resulting from overexpression of the four HIF1A-regulated miRNAs, we performed microarray gene expression profiling of both Quad-PIG and Empty-PIG B16F10 cells. Generally modest changes were observed: 39 genes were downregulated and 3 genes were upregulated in Quad-PIG cells (Table 1). None of the 39 genes downregulated in Quad-PIG cells was a predicted target of any of the four miRNAs. Interestingly, two of the three upregulated genes, Bnip3 and Atf3, are involved in cellular responses to low oxygen levels (Ameri et al., 2007; Mellor and Harris, 2007). The upregulation of Bnip3 and Atf3 identified by microarray in Quad-PIG B16F10 cells was confirmed by qRT-PCR (Figure 6A) and increased BNIP3 protein expression in Quad-PIG B16F10 cells was also confirmed by immunoblot (Figure 6B). To determine which miRNA(s) are responsible for the upregulation of Bnip3, four additional B16F10 stable cell lines (210-PIG, 218-PIG, 224-PIG, and 452-PIG) were generated by retroviral transduction of each individual miRNA, and the expression of Bnip3 mRNA and protein in these cell lines was measured by qRT-PCR and immunoblot, respectively (Figure 6C, D). Interestingly, miR-210 overexpression resulted in higher expression of Bnip3 mRNA and protein, while overexpression of miR-224 or miR-452 actually decreased Bnip3 expression at both the mRNA and protein level. MiR-218 overexpression had no effects on Bnip3 mRNA, but modestly increased BNIP3 protein expression. Taken together, these results suggest that in B16F10 mouse melanoma cells, miR-210 overexpression alone was sufficient to increase expression of BNIP3. Additionally, analysis of Atf3 mRNA expression in these cell lines showed increased Atf3 expression in both 210-PIG and 218-PIG lines, and decreased Atf3 expression in 224-PIG and 452-PIG lines (data not shown).
Table 1.
Differentially expressed genes with a fold-change ≥1.3 identified in microarray analysis of Quad-PIG B16F10 cells and Empty-PIG B16F10 cells.
| Upregulated in Quad-PIG B16F10 cells | ||
|---|---|---|
| Gene Symbol | Fold-Change | p-value |
| Bnip3 | 1.39 | 4.10E-08 |
| Pycr1 | 1.34 | 1.13E-06 |
| Atf3 | 1.32 | 3.96E-06 |
| Downregulated in Quad-PIG B16F10 cells | ||
|---|---|---|
| Gene Symbol | Fold-Change | p-value |
| Ttr | −1.30 | 6.8f5E-06 |
| Clock | −1.31 | 1.01E-06 |
| Mug1 | −1.32 | 4.56E-05 |
| Mat1a | −1.32 | 3.54E-05 |
| Sgpl1 | −1.32 | 4.36E-07 |
| Snora20 | −1.32 | 1.96E-04 |
| Pzp | −1.33 | 6.30E-05 |
| Eid3 | −1.34 | 5.74E-05 |
| Slc27a2 | −1.34 | 1.80E-07 |
| Hmgcs2 | −1.34 | 1.18E-05 |
| Gas5 | −1.34 | 4.82E-02 |
| Atp13a3 | −1.34 | 6.56E-03 |
| Rbp4 | −1.35 | 3.28E-05 |
| Serpina1b | −1.35 | 3.15E-03 |
| C3 | −1.35 | 4.17E-08 |
| Angptl3 | −1.36 | 1.50E-04 |
| Ahsg | −1.37 | 1.82E-06 |
| Myo9a | −1.37 | 2.53E-02 |
| Mup2 | −1.37 | 3.43E-07 |
| Serpina1e | −1.38 | 6.16E-07 |
| Fabp1 | −1.40 | 3.10E-05 |
| Serpina1a | −1.40 | 4.75E-06 |
| Mup11 | −1.40 | 1.97E-07 |
| Mup7 | −1.41 | 9.57E-08 |
| Fgb | −1.42 | 5.52E-06 |
| Bhmt | −1.44 | 9.18E-07 |
| Cyp3a11 | −1.45 | 6.45E-05 |
| Cyp4a10 | −1.46 | 5.81E-04 |
| Fgg | −1.47 | 6.10E-07 |
| Cyp2e1 | −1.50 | 5.21E-08 |
| Cyp4a32 | −1.54 | 1.16E-06 |
| Mup1 | −1.56 | 8.29E-05 |
| Cyp4a31 | −1.58 | 5.82E-05 |
| Alb | −1.62 | 9.30E-11 |
| Gc | −1.62 | 1.42E-06 |
| Mup3 | −1.68 | 8.59E-06 |
| Serpina1c | −1.70 | 4.41E-04 |
| Cyp4a14 | −1.71 | 1.28E-07 |
| Mup20 | −1.84 | 2.34E-07 |
Figure 6.
Overexpression of miR-210, miR-218, miR-224, and miR-452 increased Bnip3 and Atf3 expression in B16F10 cells. A. qRT-PCR showing significantly higher Bnip3 and Atf3 mRNA expression in Quad-PIG cells compared to Empty-PIG cells (Ŧ indicates multiplicity adjusted p value < 0.0001; * indicates multiplicity adjusted p value = 0.0125, Unpaired t-test). Expression levels are normalized to Empty-PIG cells. B. Immunoblot demonstrating a 2.1-fold increase in BNIP3 protein abundance in Quad-PIG cells. C. qRT-PCR showing Bnip3 mRNA expression in B16F10 cells overexpressing each of the four HIF1A-regulated miRNAs. 210-PIG, 218-PIG, 224-PIG, and 452-PIG indicates cells with individual overexpression of miR-210, miR-218, miR-224, and miR-452, respectively. Significant differences relative to Empty-PIG expression levels were seen in 210-PIG, 224-PIG, and 452-PIG; Ŧ indicates multiplicity adjusted p value < 0.0001; One-way ANOVA with Dunnett’s multiple comparison test. Expression levels are normalized to Empty-PIG cells. D. Immunoblot demonstrating changes in BNIP3 protein abundance from combined or individual overexpression of the four HIF1A-regulated miRNAs in B16F10 cells (labels as in (C). Tubulin serves as a loading control.
Discussion
Previous reports have shown that two reciprocal gene expression programs exist in metastatic melanoma cell lines, defined in part by the expression of either MITF/SOX10-regulated pathways or TGFβ1-regulated pathways (Bell et al., 2014; Hoek et al., 2006). Our microarray profiling studies extended these results to include 5 melanoma lines that had no previous characterization of their gene expression patterns. Furthermore, our studies have expanded these two gene expression programs to include miRNAs, by demonstrating that a subset of miRNAs exhibited differential expression in parallel with the mRNA expression patterns. The distinction of these two melanoma subsets by miRNA expression is consistent with a previous report that miRNAs can provide accurate classification of human cancer samples (Lu et al., 2005). It is not clear how these two expression programs are regulated in melanoma cells, but one potential regulatory candidate is HIF1A. In support of this, the 223-gene signature identified by Hoek et al. that distinguishes these two gene expression programs includes the well-known HIF1A targets CTGF, LOX, and SERPINE1 (Benita et al., 2009). By combining qRT-PCR, reporter assays, and immunoblot analyses, we now provide strong evidence that TGFβ1+ melanoma cells have sustained expression of HIF1A under normoxic conditions. Hypoxia is known to increase the tumorigenicity and metastatic potential of melanoma through HIF1A (Cheli et al., 2012; Hanna et al., 2013), therefore it will be of interest to determine the functional roles of HIF1A in TGFβ1+ melanoma cells in future studies.
We have identified four miRNAs (miR-210, miR-218, miR-224 and miR-452) that show overexpression and are regulated by HIF1A in TGFβ1+ melanoma cells. With the exception of miR-210, these were not detectable in primary human melanocytes by TLDA array, suggesting that their expression might be selected to perform biological functions in TGFβ1+ melanoma cells. In fact, simultaneous inhibition of all four miRNAs resulted in cell-cycle arrest, indicating that at least one of these miRNAs contributes to cell-cycle progression in these cells. Another possible function of these miRNAs might be promoting the TGFβ1+ gene expression program. In support of this, miR-218 and miR-452 were identified from an independent panel of melanoma cell lines as important suppressors of the MITF/SOX10+ gene expression program (Bell et al., 2014). Interestingly, this study also identified miR-211 as a repressor the TGFβ1+ gene expression program, consistent with our finding that miR-211 expression is lost in TGFβ1+ melanoma cells. Future studies to identify targets of these miRNAs in melanoma cells should provide important insights into the regulation of MITF/SOX10+ and TGFβ1+ gene expression programs in melanoma cells.
Accumulating evidence suggests that miRNAs are part of the cellular hypoxic response and that miR-210 plays a critical role in coordinating the gene expression program in response to hypoxia (Huang et al., 2010). MiR-210 is a direct HIF1A target, and is almost invariably induced by hypoxia in cells irrespective of their tissue origins (Chan and Loscalzo, 2010). In contrast, the association between hypoxia and other miRNAs is more context-dependent, as the lists of hypoxia-induced miRNAs from different experimental systems often have little overlap other than miR-210 (Huang et al., 2010; Kulshreshtha et al., 2007). We have provided evidence that in the context of TGFβ1+ melanoma cells, miR-218, miR-224 and miR-452 expression is positively regulated by HIF1A. Intronic miRNAs are derived from common transcripts with their host genes and their expression is usually coordinated with that of their host gene mRNAs (Baskerville and Bartel, 2005). Although miR-218 has not been associated with hypoxia or HIF1A, the expression levels of Slit2 and Slit3, the host genes of miR-218, are increased by hypoxia in choriocarcinoma cells and human umbilical vein endothelial cells, respectively (Liao et al., 2012). Compared with miR-210 and miR-218, we found that knockdown of HIF1A has less effect on the expression of miR-224 and miR-452 in TGFβ1+ cells, especially at the primary transcript level. This suggests that additional regulators may be responsible for their expression. Interestingly, miR-452 was shown to be highly enriched in the neural crest and was required for neural crest cell differentiation during mouse development (Sheehy et al., 2010). Since melanocytes are derived from the neural crest, the tissue-restricted expression pattern of miR-452 might explain why its association with hypoxia remained undiscovered until now, and genes that control neural crest development might provide clues to miR-452 regulation in melanoma cells.
The overexpression of miR-210, miR-218, miR-224 and miR-452 together in B16F10 cells resulted in modest effects on overall gene expression at the mRNA level and did not result in repression of any predicted targets of the four miRNAs. Previous studies demonstrated a stronger effect from miRNA perturbation at the protein level compared with the transcript level (Baek et al., 2008; Selbach et al., 2008), thus it remains possible that changes in protein abundance did occur but were not detected. Intriguingly, overexpression of miR-210 alone augmented the expression of two genes that are induced by hypoxia, Bnip3 and Atf3. ATF3 is a transcription factor that activates an adaptive program to help cells cope with stress, while BNIP3 plays an important role in hypoxia-induced autophagy (Azad and Gibson, 2010; Thompson et al., 2009). Bnip3 is a direct HIF1A target (Benita et al., 2009), and overexpression of miR-210 stabilizes HIF1A and increases the expression of HIF1A targets (Kelly et al., 2011). Thus it is possible that overexpression of miR-210 increased Bnip3 expression through HIF1A stabilization. On the contrary, the upregulation of Atf3 expression by miR-210 and miR-218 is unclear and most likely occurs through other intermediates. Since ATF3 induction under low oxygen is independent of HIF1A (Ameri et al., 2007), miR-210 may further amplify the hypoxic response by activating additional gene networks in addition to its stabilization of HIF1A. It will be interesting to examine the relationship between HIF1A, ATF3, BNIP3 and endogenous miR-210/miR-218 in melanoma and other types of cancer.
In summary, we demonstrated two distinct gene expression programs in melanoma cell lines that include both mRNAs and miRNAs. We discovered a role for HIF1A in regulating the gene expression program of TGFβ1+ melanoma cell lines and also identified four HIF1A-regulated miRNAs in these lines. Cell cycle arrest resulted from the inhibition of these 4 miRNAs, suggesting they are necessary for cell cycle progression in TGFβ1+ melanoma cells. Overexpression studies indicated that two of these HIF1A-regulated miRNAs might amplify the hypoxic response by activating additional effector genes. Future studies to elucidate the functional roles of HIF1A and HIF1A-regulated miRNAs in “invasive” TGFβ1+ melanoma cells will provide important insights into the biological behaviors of melanoma and potentially facilitate the development of therapeutics for this devastating disease.
Methods
Cell culture
The human melanoma cell lines used were generated from patients following standard surgical consent and under IRB approval. UACC-647 and UACC-3093 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM). 0708-LND, 0620-LNA and 0380-MMU cells were grown in Iscove’s Modified Dulbecco’s Medium. B16F10 mouse melanoma cells were obtained from ATCC and were grown in DMEM. Melan-INK4a-ARF1 (INK4a/ARF double null) cells were grown in RPMI +200 nM 12-0-tetradecanoyl phorbol acetate and 200 pM cholera toxin. All media for these cell lines was supplemented with 10% FBS. Normal human melanocytes were obtained from the William Gahl lab (NHGRI, NIH) and were maintained as previously described (Westbroek et al., 2012). For HIF1A analyses, the following cell lines were used as controls: RCC4, a renal carcinoma cell line that constitutively expresses HIF1A, RCC4-VHL, a derivative of RCC4 that only expresses HIF1A in hypoxia, and RCC4-VHL treated with CoCl2 to induce HIF1A expression. For CoCl2 treatment of RCC4-VHL cells, cells were cultured in media + 50μM CoCl2 for 24 hours prior to harvest and subcellular fractionization. Cell proliferation assays were performed using Cell Counting Kit-8 (Dojindo). For HIF1A knockdown, SMARTpool siRNA against HIF1A or control siRNA (Dharmacon) was transfected into 0620-LNA and UACC-647 cells using DharmaFECT (Dharmacon). Seventy-two hours after transfection, total RNA and protein lysate were harvested for analysis.
Microarray analysis
Microarrays were performed using Affymetrix GeneChip Human Genome U133A 2.0 Arrays for human melanoma cell lines in three replicates, and Affymetrix GeneChip Mouse Gene 1.0 ST Arrays for B16F10 mouse melanoma cell lines in five replicates (Affymetrix, Inc., Santa Clara, CA, USA). Microarray experiments were performed using standard Affymetrix recommended protocols. Arrays were scanned using the Affymetrix Gene Chip Scanner 3000 and gene expression intensities were calculated using the Affymetrix GeneChip Command Console software (AGCC). Affymetrix .CEL files were normalized using the RMA (Robust Multi-Array Analysis) algorithm within Partek Genomics Suite software (Partek Inc., St. Louis, MO). Analysis of variance (ANOVA) and linear contrasts were used to identify differentially expressed genes. Type I error was controlled using the Benjamini-Hochberg False Discovery Rate correction for multiple testing, with a cut-off p-value of 0.05. Cut-off for fold change was ±2. All microarray data used in this study are available through the National Center for Biotechnology Information’s Gene Expression Omnibus.
MicroRNA analysis
MiRNA expression profiling for melanoma cell lines and human primary melanocytes was performed using TaqMan human microRNA arrays v2.0A (Applied Biosystems) on Applied Biosystems 7900HT real-time PCR system per manufacturer’s protocol. Individual miRNA or pri-miRNA expression analysis was performed using Taqman MicroRNA or Pri-miRNA Assays in triplicate using the ΔΔCt method (Applied Biosystems). RNU44 and snoRNA202 (Applied Biosystems) were the internal standards for human and mouse, respectively.
mRNA expression analysis
Total RNA was prepared using TRIzol (Invitrogen) following manufacturer’s instructions. cDNA was synthesized using MMLV reverse transcriptase (Invitrogen) with random primers. Quantitative RT-PCR analysis was performed using SYBR Green PCR Master Mix in triplicate using the ΔΔCt method (Applied Biosystems). Eukaryotic 18S rRNA endogenous control (Applied Biosystems) was used as an internal standard. Primer sequences were as follows: Mouse Bnip3, F: TGAAGTGCAGTTCTACCCAGG; Mouse Bnip3, R: CCTGTCGCAGTTGGGTTC; Mouse Atf3, F: CAGACCCCTGGAGATGTCAGT; Mouse Atf3, R: TTCTTGTTTCGACACTTGGCA; human SERPINE1, F: AGCTCCTTGTACAGATGCCG; human SERPINE1, R: ACAACAGGAGGAGAAACCCA; human CA9, F: CAGCAACTGCTCATAGGCAC; human CA9, R: CTTTGCCAGAGTTGACGAGG.
Luciferase assays
4XHRE and 4XmHRE firefly luciferase reporters were constructed by inserting DNA oligonucleotides containing four tandem repeats of the wildtype or mutant human VEGF hypoxia response element into the pGL3-Promoter (Promega) between the NheI and XhoI sites. 100 ng 4XHRE or 4XmHRE along with 50 ng of renilla luciferase reporter (phRL-SV40; Promega) were transfected into cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Luciferase assays were performed 24 h after transfection using the Dual Luciferase Reporter Assay System (Promega). Each transfected well was assayed in triplicate. Firefly luciferase activity was normalized to renilla luciferase activity for each well to control for transfection efficiency.
Immunoblot analysis
The following antibodies were used: anti-HIF1A (610958, BD Biosciences), anti-HDAC2 (sc-7899, Santa Cruz), anti-BNIP3 (#3769, Cell Signaling) and anti-Tubulin (CP06, Calbiochem). To detect HIF1A, nuclear lysates were prepared using NE-PER Nuclear and Cytoplasmic Extraction Kit with Halt Protease Inhibitor Cocktail (Pierce) and used for immunoblot analysis.
MicroRNA sponge
The miRsponge vector was constructed by inserting a multiple cloning site (MCS) downstream of EGFP into the FerH-GFP lentiviral vector, which has been shown to provide stable cell labeling for in vivo melanoma study (Day et al., 2009). The Quad-miRsponge was constructed by inserting six tandem bulged binding sites for each of the four miRNAs (miR-210, miR-218, miR-224 and miR-452) at the MCS; this construct was then sequence verified. To produce lentiviral supernatant, HEK-293 cells were transfected with miRsponge and packaging vectors for 48 hours. Empty miRsponge vector was used as a control. To create melanoma cells with stable miRNA inhibition, cells were first transduced with high-titer lentiviral supernatant from Quad-miRsponge or empty miRsponge in the presence of polybrene for 8 hours. More than 24 hours after transduction, cells with high GFP expression were selected by fluorescence activated sorting for further analysis.
Cell cycle analysis
To confirm cell-cycle arrest, cells with or without nocodazole treatment were fixed in 70% ethanol overnight at −20°C. DNA content was then measured by propidium iodide (PI) staining followed by flow cytometry, as previously described (Hwang et al., 2009). Briefly, FACS data were collected on cells using a FACSCalibur (Becton Dickinson Biosciences, Franklin Lakes, NJ) equipped with Cell Quest software and cell cycle analysis was performed using FlowJo (TreeStar).
MicroRNA overexpression
Human miR-210, miR-218, miR-224 and miR-452 and their flanking genomic sequences were PCR amplified and then inserted between the BglII and HpaI sites of the pMSCV-PIG retroviral vector either individually (210-PIG, 218-PIG, 224-PIG, 452-PIG) or in combination (Quad-PIG) (Hemann et al., 2003). Sanger sequencing was used to verify all vectors. To produce viral supernatant, Phoenix cells were transfected with Quad-PIG or empty pMSCV-PIG for 48 hours. B16F10 cells were transduced with viral supernatant in the presence of polybrene for 8 hours and subsequently selected in growth media containing 1μg/mL puromycin for 1 week.
Statistical analyses
All statistical analyses were performed using GraphPad Prism, with N = 3 for all samples.
Supplementary Material
Figure S1. Heatmap showing hierarchical cluster analysis of the expression of 377 miRNAs in the human melanoma cell lines 0380-MMU, UACC-3093, 0620-LNA, 0708-LND, and UACC-647. The miRNA expression separates the melanoma cell lines into the same two groups that were defined by mRNA expression (Figure 1A), indicating miRNA expression is distinctive between SOX10/MITF+ and TGFβ1+ groups.
Figure S2. Luciferase assays showing strong HIF1A transcriptional activity in the TGFβ1+ melanoma cell lines UACC-647 and 0620-LNA. RCC4, a renal carcinoma cell line that constitutively expresses HIF1A, and RCC4-VHL, a derivative of RCC4 that only expresses HIF1A in hypoxia, are positive and negative controls, respectively. Data are relative expression of luciferase reporter constructs: 4XHRE, HIF-responsive elements; 4XmHRE, mutated HIF-responsive elements. Error bars represent standard deviations from 2 independent experiments.
Table S1. TaqMan Low Density Array analysis of the miRNA expression patterns of primary human melanocytes and the human melanoma cell lines 0380-MMU, UACC-3093, 0620-LNA, 0708-LND, and UACC-647.
Significance.
To develop effective treatments for metastatic melanoma, a detailed understanding is needed of the changes in gene expression that occur as this devastating disease progresses. Previous work has shown that melanoma cells exhibit two different mRNA expression patterns that correlate with pathways regulated by either transforming growth factor, beta 1 (TGFβ1) or microphthalmia-associated transcription factor (MITF)/SRY-box containing gene 10 (SOX10). We discovered that melanoma cell lines exhibiting these expression programs also show distinctive expression of microRNAs (miRNAs). We also found that HIF1A expression in TGFβ1-pathway melanoma cells occurs independently of hypoxia, and this HIF1A expression may regulate the expression of specific miRNAs and their downstream gene expression programs.
Acknowledgments
The authors would like to thank Stacie Anderson for outstanding support in cell sorting and analysis by flow cytometry, Dr. Wendy Westbroek for help with primary human melanocyte culture and Dr. Chi-Ping Day for providing lentiviral vectors. Hun-Way Hwang is an HHMI Fellow of the Life Sciences Research Foundation. Funding of this research was provided by the National Human Genome Research Institute’s Intramural Research Program at the National Institutes of Health.
References
- Ameri K, Hammond EM, Culmsee C, Raida M, Katschinski DM, Wenger RH, et al. Induction of activating transcription factor 3 by anoxia is independent of p53 and the hypoxic HIF signalling pathway. Oncogene. 2007;26:284–289. doi: 10.1038/sj.onc.1209781. [DOI] [PubMed] [Google Scholar]
- Azad MB, Gibson SB. Role of BNIP3 in proliferation and hypoxia-induced autophagy: implications for personalized cancer therapies. Ann N Y Acad Sci. 2010;1210:8–16. doi: 10.1111/j.1749-6632.2010.05778.x. [DOI] [PubMed] [Google Scholar]
- Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, et al. Transcription Factor/microRNA Axis Blocks Melanoma Invasion Program by miR-211 Targeting NUAK1. Journal Invest Dermatol. 2014;134:441–51. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- Benita Y, Kikuchi H, Smith AD, Zhang MQ, Chung DC, Xavier RJ. An integrative genomics approach identifies Hypoxia Inducible Factor-1 (HIF-1)-target genes that form the core response to hypoxia. Nucleic Acids Res. 2009;37:4587–4602. doi: 10.1093/nar/gkp425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi VF, Stark MS, Hayward NK. MicroRNA regulation of melanoma progression. Melanoma Res. 2012;22:101–113. doi: 10.1097/CMR.0b013e32834f6fbb. [DOI] [PubMed] [Google Scholar]
- Chan SY, Loscalzo J. MicroRNA-210: a unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene. 2012;31:2461–2470. doi: 10.1038/onc.2011.425. [DOI] [PubMed] [Google Scholar]
- Day C-P, Carter J, Bonomi C, Esposito D, Crise B, Ortiz-Conde B, Hollingshead M, Merlino G. Lentivirus-mediated bifunctional cell labeling for in vivo melanoma study. Pigment Cell Melanoma Res. 2009;22:283–295. doi: 10.1111/j.1755-148X.2009.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. RNA. 2010;16:2043–2050. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon Y-J, Ngankeu A, et al. EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nature Med. 2012;18:74–82. doi: 10.1038/nm.2577. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerald JNF, Benjamin JM, Kron SJ. Robust G1 checkpoint arrest in budding yeast: dependence on DNA damage signaling and repair. J Cell Sci. 2002;115:1749–1757. doi: 10.1242/jcs.115.8.1749. [DOI] [PubMed] [Google Scholar]
- Hanna SC, Krishnan B, Bailey ST, Moschos SJ, Kuan P-F, Shimamura T, et al. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. Journal Clin Invest. 2013;123:2078–2093. doi: 10.1172/JCI66715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Fridman JS, Zilfou JT, Hernando E, Paddison PJ, Cordon-Cardo C, et al. An epi-allelic series of p53 hypomorphs created by stable RNAi produces distinct tumor phenotypes in vivo. Nat Genet. 2003;33:396–400. doi: 10.1038/ng1091. [DOI] [PubMed] [Google Scholar]
- Hill VK, Gartner JJ, Samuels Y, Goldstein AM. The Genetics of Melanoma: Recent Advances. Annu Rev Genomics Hum Genet. 2013;14:257–79. doi: 10.1146/annurev-genom-091212-153429. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Goding CR. Cancer stem cells versus phenotype-switching in melanoma. Pigment Cell Melanoma Res. 2010;23:746–759. doi: 10.1111/j.1755-148X.2010.00757.x. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Schlegel NC, Brafford P, Sucker A, Ugurel S, Kumar R, et al. Metastatic potential of melanomas defined by specific gene expression profiles with no BRAF signature. Pigment Cell Res. 2006;19:290–302. doi: 10.1111/j.1600-0749.2006.00322.x. [DOI] [PubMed] [Google Scholar]
- Huang X, Le Q-T, Giaccia AJ. MiR-210--micromanager of the hypoxia pathway. Trends Mol Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H-W, Wentzel EA, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs AR, Glover AC, Slobbe LJ, Wang L, He S, Hazlett JA, et al. A gene expression signature of invasive potential in metastatic melanoma cells. PLoS ONE. 2009;4:e8461. doi: 10.1371/journal.pone.0008461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–864. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1 alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Bio. 2011;31:2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, et al. A microRNA signature of hypoxia. Mol Cell Bio. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Bio. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C, Khaled M, Robinson KC, Veguilla RA, Chen P-H, Yokoyama S, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W-X, Laurent LC, Agent S, Hodges J, Chen D-B. Human placental expression of SLIT/ROBO signaling cues: effects of preeclampsia and hypoxia. Biol Reprod. 2012;86:111. doi: 10.1095/biolreprod.110.088138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Lujambio A, Lowe SW. The microcosmos of cancer. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Molecular Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor HR, Harris AL. The role of the hypoxia-inducible BH3-only proteins BNIP3 and BNIP3L in cancer. Cancer Metastasis Rev. 2007;26:553–566. doi: 10.1007/s10555-007-9080-0. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaou V, Stratigos AJ. Emerging trends in the epidemiology of melanoma. The Br J Dermatol. 2014;170:11–19. doi: 10.1111/bjd.12492. [DOI] [PubMed] [Google Scholar]
- Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151:1068–1082. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Sheehy NT, Cordes KR, White MP, Ivey KN, Srivastava D. The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development. 2010;137:4307–4316. doi: 10.1242/dev.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- Sullivan R, Graham CH. Hypoxia-driven selection of the metastatic phenotype. Cancer Metastasis Rev. 2007;26:319–331. doi: 10.1007/s10555-007-9062-2. [DOI] [PubMed] [Google Scholar]
- Thompson MR, Xu D, Williams BRG. ATF3 transcription factor and its emerging roles in immunity and cancer. J Mol Med. 2009;87:1053–1060. doi: 10.1007/s00109-009-0520-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbroek W, Klar A, Cullinane AR, Ziegler SG, Hurvitz H, Ganem A, et al. Cellular and clinical report of new Griscelli syndrome type III cases. Pigment Cell Melanoma Res. 2012;25:47–56. doi: 10.1111/j.1755-148X.2011.00901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer DS, Hoek KS, Cheng PF, Eichhoff OM, Biedermann T, Raaijmakers MIG, Hemmi S, Dummer R, Levesque M. Hypoxia Contributes to Melanoma Heterogeneity by Triggering HIF1α-Dependent Phenotype Switching. J Invest Dermatol. 2013;133:2436–2443. doi: 10.1038/jid.2013.115. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Heatmap showing hierarchical cluster analysis of the expression of 377 miRNAs in the human melanoma cell lines 0380-MMU, UACC-3093, 0620-LNA, 0708-LND, and UACC-647. The miRNA expression separates the melanoma cell lines into the same two groups that were defined by mRNA expression (Figure 1A), indicating miRNA expression is distinctive between SOX10/MITF+ and TGFβ1+ groups.
Figure S2. Luciferase assays showing strong HIF1A transcriptional activity in the TGFβ1+ melanoma cell lines UACC-647 and 0620-LNA. RCC4, a renal carcinoma cell line that constitutively expresses HIF1A, and RCC4-VHL, a derivative of RCC4 that only expresses HIF1A in hypoxia, are positive and negative controls, respectively. Data are relative expression of luciferase reporter constructs: 4XHRE, HIF-responsive elements; 4XmHRE, mutated HIF-responsive elements. Error bars represent standard deviations from 2 independent experiments.
Table S1. TaqMan Low Density Array analysis of the miRNA expression patterns of primary human melanocytes and the human melanoma cell lines 0380-MMU, UACC-3093, 0620-LNA, 0708-LND, and UACC-647.