Summary
The Campylobacter jejuni flagellum exports both proteins that form the flagellar organelle for swimming motility and colonization and virulence factors that promote commensal colonization of the avian intestinal tract or invasion of human intestinal cells, respectively. We explored how the C. jejuni flagellum is a versatile secretory organelle by examining molecular determinants that allow colonization and virulence factors to exploit the flagellum for their own secretion. Flagellar biogenesis was observed to exert temporal control of secretion of these proteins, indicating that a bolus of secretion of colonization and virulence factors occurs during hook biogenesis with filament polymerization itself reducing secretion of these factors. Furthermore, we found that intramolecular and intermolecular requirements for flagellar-dependent secretion of these proteins were most reminiscent to those for flagellin secretion. Importantly, we discovered that secretion of one colonization and virluence factor, CiaI, was not required for invasion of human colonic cells, which counters previous hypotheses for how this protein functions during invasion. Instead, secretion of CiaI was essential for C. jejuni to facilitate commensal colonization of the natural avian host. Our work provides insight into the versatility of the bacterial flagellum as a secretory machine that can export proteins promoting diverse biological processes.
Keywords: Secretion, flagella, Campylobacter jejuni, Fed proteins
Introduction
The bacterial flagellum is a nanomachine constructed by dozens of different proteins to facilitate swimming motility. Whereas some structural proteins are present in just a few copies per a flagellum, other proteins, such as the flagellins that form the extracellular filament, number in the thousands. Flagellar biosynthesis occurs in discrete steps, with only certain proteins secreted during each step to ensure that formation of the rod occurs first, followed by the hook, and ending with the filament (see (Chevance and Hughes, 2008; Minamino et al., 2008; Buttner, 2012) for reviews). Flagellar biogenesis usually begins with formation of the flagellar type III secretion system (T3SS) that is surrounded by the MS ring in the inner membrane and the C ring in the cytoplasm (Kihara et al., 2001; Chen et al., 2011; Li and Sourjik, 2011; Morimoto et al., 2014). In addition, the FliI and FliH proteins form an ATPase ring complex that is associated with the T3SS and C ring in the cytoplasm (Claret et al., 2003; Chen et al., 2011). Most proteins forming components of the flagellum beyond the inner membrane are secreted via the T3SS. Specific flagellar chaperones and intramolecular domains within flagellar proteins direct flagellar subunits to the T3SS (Yokoseki et al., 1995; Fraser et al., 1999; Auvray et al., 2001; Bennett et al., 2001; Thomas et al., 2004; Evans et al., 2006; Vegh et al., 2006; Stafford et al., 2007; Dobo et al., 2010; Evans et al., 2013). The T3SS, ATPase ring complex, and chaperones function together to ensure that proteins are secreted in the proper order so that flagellar subunits can interact with each other correctly to construct a flagellum able to mediate motility.
Early studies with Yersinia enterocolitica revealed that the flagellar T3SS can promote secretion of non-effector and non-flagellar proteins (Young et al., 1999). These non-flagellar proteins, termed Fops, required a fully functional flagellar T3SS for secretion. The YplA phospholipase is one Fop that is required for full virulence of Y. enterocolitica and secretion of the protein is required for its phospholipase activity (Schmiel et al., 1998; Young et al., 1999). Further studies revealed that YplA is also a secretion substrate for two endogenous injectisome T3SSs in Y. enterocolitica and the first 20 amino acids are required for secretion via each T3SS (Young and Young, 2002; Warren and Young, 2005).
In addition to motility, the Campylobacter jejuni flagellum is required by the bacterium for commensal colonization of the intestinal tract of avian hosts and other animals and infection of human volunteers to promote diarrheal disease (Black et al., 1988; Nachamkin et al., 1993; Wassenaar et al., 1993; Hendrixson and DiRita, 2004; Wosten et al., 2004). The flagellum is necessary for C. jejuni to invade and penetrate human intestinal or colonic epithelium, which elicits host responses that contribute to the inflammatory enteritis characteristic of C. jejuni diarrheal disease (van Spreeuwel et al., 1985; Wassenaar et al., 1991; Grant et al., 1993; Yao et al., 1994). Upon invasion of eukaryotic cells, C. jejuni resides in a Campylobacter-containing vacuole (CCV) and alters its physiology for intracellular survival (Watson and Galan, 2008; Liu et al., 2012). The CCV does not fuse with lysosomes and allows C. jejuni a transient intracellular residence before eventual release from the infected cell.
Like other bacterial flagella, the C. jejuni flagellar T3SS secretes flagellar proteins for biosynthesis of the organelle. However, the C. jejuni flagellum has also been implicated as a secretory machine for proteins not required for motility, such as the Cia proteins. Flagellar-dependent secretion of Cia proteins has been reported to require a factor produced by human cells or a component of serum (Konkel et al., 1999; Rivera-Amill et al., 2001; Konkel et al., 2004). These proteins are required for WT levels of C. jejuni invasion of human intestinal epithelial cells or survival within these cells (Konkel et al., 1999; Konkel et al., 2004; Christensen et al., 2009; Buelow et al., 2011). At least eight Cia proteins have been proposed and four Cias – CiaB, CiaC, CiaD, and CiaI - have been characterized (Konkel et al., 1999; Christensen et al., 2009; Buelow et al., 2011; Neal-McKinney and Konkel, 2012; Samuelson et al., 2013; Samuelson and Konkel, 2013). CiaB is secreted by the flagellum and is also required for the flagellar-dependent secretion of other Cia proteins (Konkel et al., 1999). Although the exact role that CiaC performs in invasion is unknown, CiaD influences host cell signaling necessary for invasion and IL-8 production (Samuelson et al., 2013; Samuelson and Konkel, 2013). CiaI has been proposed to assist in the intracellular survival of C. jejuni by preventing the CCV from fusing with lysosomes (Buelow et al., 2011). Consistent with this observation, ectopically expressed GFP-CiaI in epithelial cells localized to late endosomal vesicles with a dileucine motif in CiaI mediating localization. However, a C. jejuni ciaI mutant with an altered dileucine motif was not impaired for invasion, which calls into question the mechanism by which CiaI assists in interactions with human epithelial cells (Barrero-Tobon and Hendrixson, 2012).
Production of CiaI and other Cia proteins was originally reported to be induced in the presence of the bile salt deoxycholate (Rivera-Amill et al., 2001; Malik-Kale et al., 2008; Buelow et al., 2011). However, we found that ciaI and four other genes (fedA-D) are members of the σ28 regulon (Barrero-Tobon and Hendrixson, 2012). In C. jejuni, the σ28 regulon largely includes flagellar genes encoding filament components such as the FlaA major flagellin (Carrillo et al., 2004). In addition, C. jejuni fspA1, which encodes a protein whose function is not fully understood, is also dependent on σ28 for expression (Poly et al., 2007). Although the Fed proteins, FspA1, and CiaI are co-expressed with FlaA and other flagellar filament proteins, these proteins are not required for motility (Poly et al., 2007; Barrero-Tobon and Hendrixson, 2012). Instead, we discovered that C. jejuni mutants lacking FedA, FedB, FedC, FedD or CiaI are attenuated 4- to 1000-fold for commensal colonization of the chick ceca (Barrero-Tobon and Hendrixson, 2012). In contrast, FspA1 is not required for colonization of chicks. Unlike previous observations where serum was required for secretion of CiaI and other Cia proteins via the flagellum, we and others observed that CiaI, FedB and FspA1 are secreted in a flagellar-dependent manner without any serum supplementation (Poly et al., 2007; Barrero-Tobon and Hendrixson, 2012). It is currently unclear if other Fed proteins are secreted.
The fact that the C. jejuni flagellum secretes proteins such as FedB, CiaI, and FspA1 not involved in motility presents many interesting questions regarding mechanisms by which these proteins are specifically and efficiently secreted by the organelle. It has been estimated that over 20,000 FlaA major flagellin monomers form a filament of one flagellum (Macnab, 2003; Minamino and Namba, 2004). In addition, FedB, CiaI, and FspA1 are produced simultaneously with FlaA, but transcribed from different promoters and produced at lower levels than FlaA (Barrero-Tobon and Hendrixson, 2012). Thus, these proteins must have a mechanism to effectively compete with copious amounts of flagellin for interactions with the flagellar T3SS that result in secretion. In previous studies examining secretion of proteins via the flagellum, intermolecular and intramolecular determinants were identified that impact flagellar-dependent secretion of substrates. For Cia proteins, an N-terminal region of up to 36 amino acids within the proteins can mediate secretion of a heterologous protein through the flagellum (Christensen et al., 2009). Additionally, a minimal flagellar structure that includes the hook and hook-filament junction is required for secretion of the Cias (Neal-McKinney and Konkel, 2012). For flagellins, a domain within the N-terminal 50 amino acids has been implicated as a secretion signal recognized by the flagellar T3SS, whereas other flagellar proteins require a different conserved motif within the N-terminus for T3SS recognition (Kuwajima et al., 1989; Vegh et al., 2006; Evans et al., 2013). Proteins that form minor components of the flagellum are often bound by a specific flagellar chaperone that enhances targeting to and recognition by the flagellar T3SS for secretion (Fraser et al., 1999; Bennett et al., 2001; Thomas et al., 2004). In addition, a specific chaperone interacts with flagellins to prevent premature polymerization of flagellin monomers into filaments in the cytoplasm prior to secretion (Auvray et al., 2001; Evdokimov et al., 2003). As such, we hypothesize that specific chaperones and intramolecular domains within FedB, CiaI, and FspA1 may assist these proteins to compete with flagellins for recognition and secretion by the T3SS. In addition, the process of flagellar biogenesis likely influences dynamics of secretion of these proteins. Furthermore, it is unknown whether secretion of these proteins, such as CiaI, is actually required for commensal colonization of chicks or invasion of human epithelial cells.
In this work, we investigated many of these questions regarding secretion of the C. jejuni FedB, CiaI, and FspA1 proteins. Among our findings is the discovery that flagellar biosynthesis in bacteria can temporally regulate secretion of colonization and virulence factors. We discovered that FedB, CiaI, and FspA1 are secreted during hook biogenesis, but the level of secretion of these proteins is reduced once filament polymerization begins. Thus, flagellar biogenesis and polymerization of the flagellar filament temporally control secretion of these proteins in C. jejuni. Among our other important findings are that both N- and C-terminal domains of FedB and CiaI influenced secretion via the flagellum, but no predicted C. jejuni flagellar chaperones were directly required for secretion of these proteins. Furthermore, we discovered that CiaI secretion was essential for commensal colonization of chicks, but not for invasion of human epithelial cells, which contradicts previous studies implying that CiaI secretion has a role vital for invasion. Our work provides new insights into the flagellum as a versatile secretion machine for virulence and colonization determinants, which may impact the biology of many bacteria.
Results
Intramolecular requirements for secretion of FedB and CiaI
Many secreted flagellar proteins contain intramolecular determinants necessary for secretion of the proteins. For example, different N-terminal domains of flagellar rod, hook, and flagellin proteins are recognized by the T3SS for translocation into the central channel within the nascent flagellum for transit to the growing tip of the organelle (Kuwajima et al., 1989; Vegh et al., 2006; Dobo et al., 2010; Neal-McKinney et al., 2010; Evans et al., 2013). In addition, a C-terminal domain of flagellins is bound by the FliS chaperone to prevent premature polymerization of flagellins into filaments in the cytoplasm prior to secretion (Auvray et al., 2001; Evdokimov et al., 2003; Muskotal et al., 2006). Proteins that are minor components of the flagellum, such as hook-filament junction proteins and the filament cap, rely on a cognate chaperone to bind to C-terminal regions of these proteins and target them to the T3SS, which has been proposed to increase their efficiency of secretion (Fraser et al., 1999; Bennett et al., 2001; Bange et al., 2010).
Based on this knowledge, we deleted N- and C-terminal domains of FedB and CiaI to determine whether these regions are required for flagellar-dependent secretion in C. jejuni. For these analyses, we arbitrarily divided the N- and C-terminal regions into 25-amino acid domains (e.g., domain 1 extends from residues 2–26; domain 2 extends from residues 27–51; domain 3 contains 26–50 residues from the C-terminus; and domain 4 contains the last 25 residues of the proteins; Fig. 1A and 1C). Domains 1 and 4 of FedB and CiaI were further divided into subdomains (e.g, subdomain 1A–D and 4E–H) composed of only six or seven amino acids. WT or mutant genes encoding FedB or CiaI with various domain deletions were expressed from plasmids in trans with native promoters and 5′ untranslated regions (UTRs) in C. jejuni 81-176 ΔfedB or ΔciaI and protein secretion was analyzed.
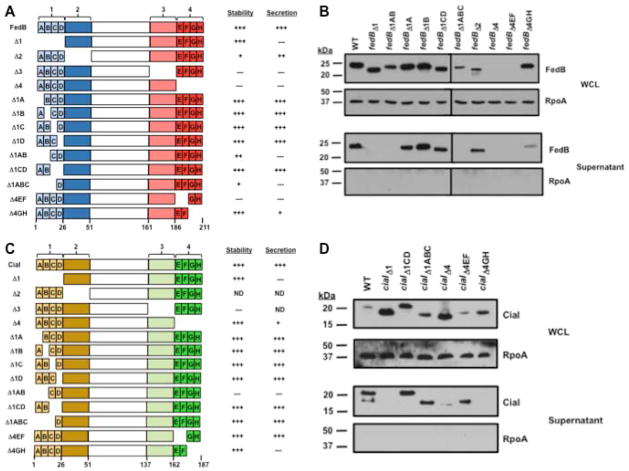
Figure 1. Identification of intramolecular domains of FedB and CiaI required for secretion.
(A) Domains and subdomains of C. jejuni FedB and a summary of their level of stability and secretion. The N- and C-terminal domains of FedB were divided into domains of 25 amino acids in length. FedB N-terminal domains are indicated in light blue (domain 1) or dark blue (domain 2). FedB C-terminal domains are indicated in pink (domain 3) or red (domain 4). Domains 1 and 4 were further divided into subdomains A–D or E–H, respectively, that were each six or seven amino acids in length. Each mutant protein that was constructed and analyzed is shown. A summary of the relative level of stability and secretion of mutant FedB proteins as assessed qualitatively by immunoblots are relative to WT FedB, which was set to “+++”. (B) Immunoblot analysis of WT and FedB mutant proteins in C. jejuni whole-cell lysates (WCL) and supernatants after growth in MH broth. WT and FedB mutant proteins were expressed from the native promoter in trans in C. jejuni 81-176 ΔfedB. All strains were grown in MH broth for 4 h at 37 °C in microaerobic conditions. WCL and supernatant proteins were recovered and analyzed by immunoblotting with antiserum specific for FedB or RpoA, which served as a control for a cytoplasmic protein. Molecular weight markers are indicated in kDa. (C) Domains and subdomains of C. jejuni CiaI and a summary of their level of stability and secretion. The N- and C-terminal domains of CiaI were divided into domains of 25 amino acids in length. CiaI N-terminal domains are indicated in light yellow (domain 1) or dark yellow (domain 2). CiaI C-terminal domains are indicated in light green (domain 3) or dark green (domain 4). Domains 1 and 4 were further divided into subdomains A–D or E–H, respectively, that were each six to seven amino acids in length. Each mutant protein that was constructed and analyzed is shown, except where indicated by “ND” (not determined). A summary of the relative level of stability and secretion of mutant CiaI proteins as assessed qualitatively by immunoblots are relative to WT CiaI, which was set to “+++”. (D) Immunoblot analysis of WT and CiaI mutant proteins in C. jejuni WCL and supernatants after growth in MH broth. WCL and supernatant proteins were recovered as described in (B) and analyzed by immunoblotting with antiserum specific for CiaI or RpoA, which served as a control for a cytoplasmic protein. Molecular weight markers are indicated in kDa
For FedB, we discovered that deletion of domain 1 (FedBΔ1) resulted in a stable protein that was not secreted by C. jejuni (Fig. 1A and 1B). In contrast, FedBΔ2 was produced less abundantly than WT FedB but still secreted, indicating that domain 2 was not required for secretion. When we analyzed subdomain deletions within domain 1 of FedB, each FedB mutant (e.g., FedBΔ1A - FedBΔ1D) was secreted at levels similar to those of WT C. jejuni (Fig. 1A and 1B; data not shown). However, deletion of both subdomains 1A and 1B (FedBΔ1AB) or domains 1A through 1C (FedBΔ1ABC) eliminated secretion, but deletion of subdomains 1C and 1D (FedBΔ1CD) did not impair secretion (Fig. 1A and 1B). These results implicate regions within the initial 26 residues – particularly residues 2–14 – of FedB in forming an intramolecular determinant that influences secretion. In these assays, we did not detect secretion of the cytoplasmic RNA polymerase subunit A (RpoA) protein in the supernatant (Fig. 1B), which verified that our procedures were adequate for analyzing C. jejuni secreted proteins rather than proteins from lysed cells.
Many FedB mutant proteins lacking various regions of the C-terminus were unstable (e.g., FedBΔ3, FedBΔ4, FedBΔ4EF; Figure 1A and 1B; data not shown), which hindered our analysis of whether these domains were required for secretion. We were able to create a stable FedB mutant lacking the last 12 residues (FedBΔ4GH). Secretion of this protein was reduced compared to WT FedB (Fig. 1A and 1B), suggesting that this C-terminal region of FedB influences secretion of the protein.
Intramolecular determinants required for secretion of CiaI were located in similar regions of the protein as in FedB. Deletion of domain 1 from CiaI (CiaIΔ1) abolished secretion with the protein accumulating in C. jejuni compared to WT CiaI (Fig. 1C and 1D). Like with FedB, deletion of each subdomain 1A–D individually did not hinder secretion of CiaI (Fig. 1C; data not shown). We were unable to create a stable protein by deleting both subdomains 1A and 1B, but deletion of subdomains 1C and 1D or subdomain 1A–C did not affect secretion of CiaI (Fig. 1C and 1D; data not shown). Curiously, CiaIΔ1CD migrated at slightly higher mobility than WT CiaI, which counters what would be predicted for a protein lacking 12 residues. This observation suggests a possible gross disruption of protein structure by deletion of these residues, which resulted in aberrant migration. In summary, our data showed that a region within residues 2–26 of CiaI was required for secretion, but deleting overlapping regions such as subdomains 1A–C or 1C–1D did not abolish secretion.
When we examined the C-terminus of CiaI, we were able to create a stable protein by deleting domain 4 (CiaIΔ4), which removes the C-terminal 25 residues (Fig. 1C and data not shown). CiaIΔ4 secretion was much lower than WT CiaI (Fig. 1D). Deletion of smaller subdomains within domain 4 revealed that CiaIΔ4EF was secreted, but CiaIΔ4GH was not (Fig. 1D), indicating that the C-terminal 12 residues of CiaI are required for secretion. As expected, RpoA was not secreted in any of the C. jejuni strains expressing ciaI mutants (Fig. 1C and 1D). Combined, our data suggest that regions of the N- and C-termini of FedB and CiaI influence secretion of these proteins.
N-terminal regions of FedB and CiaI are sufficient to promote secretion of C. jejuni flagellin
We next determined whether the N-terminus of FedB or CiaI was sufficient to mediate secretion of a heterologous protein via the flagellum. We first attempted to create chimeric proteins by fusing the N-terminal 26 residues of FedB (FedBN′26) or CiaI (CiaIN′26) to Cjj81176_0996. Like the FlaA major flagellin, CiaI, and the Fed proteins, Cjj81176_0996 requires σ28 for expression in C. jejuni (Goon et al., 2006; Barrero-Tobon and Hendrixson, 2012). This protein is required for motility in liquid medium, but is not secreted and remains associated with C. jejuni (Goon et al., 2006; Novik et al., 2010; Barrero-Tobon and Hendrixson, 2012). However, chimeric FedBN′26-0996 or CiaIN′26-0996 proteins were not secreted and remained associated with C. jejuni (data not shown). We suspect that the native folding of Cjj81176_0996 likely hindered secretion of the chimeras through the flagellum since flagellar proteins that are secreted by the flagellar T3SS remain in a semi-unfolded state during transit through the flagellar structure (Evans et al., 2013).
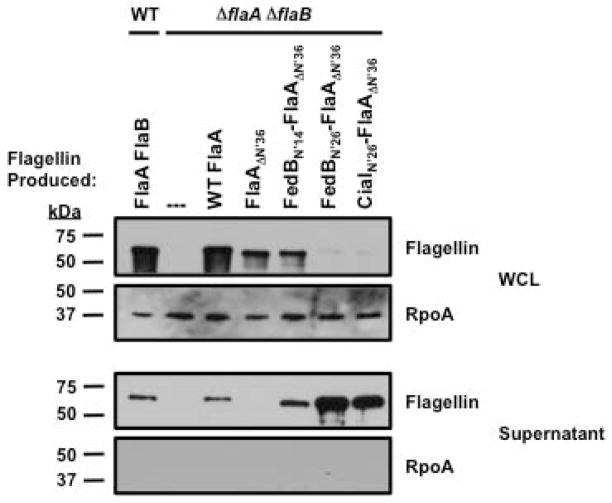
As an alternative, we constructed chimeric proteins by fusing the N-terminal regions of FedB and CiaI to a truncated FlaA flagellin monomer that cannot normally be secreted. Furthermore, flagellins are produced in an unfolded state prior to secretion, which would likely assist in our analysis of whether the N-terminal regions of FedB and CiaI are sufficient for secretion a heterologous protein. A previous study of C. jejuni FlaA identified an intramolecular secretion determinant within the initial 36 residues (Neal-McKinney et al., 2010). FlaA lacking these N-terminal residues (FlaAΔN′36) was not secreted. C. jejuni produces both FlaA and the FlaB minor flagellin, which are 95% identical and cross-react with flagellin antiserum (Nuijten et al., 1990; Wassenaar et al., 1994; Wosten et al., 2004). To analyze secretion of chimeric FlaA proteins, we first constructed a C. jejuni ΔflaA ΔflaB mutant and used the resulting strain to produce chimeric flagellin proteins and monitor secretion. We expressed WT FlaA and FlaAΔN′36 from the native flaA promoter and 5′ UTRs in cis by inserting these constructs within rdxA (Cjj81176_1083; encoding the RdxA nitroreductase; (Ribardo et al., 2010)) in the chromosome of a C. jejuni 81-176 ΔflaA ΔflaB. As expected, WT FlaA expressed in cis in the ΔflaA ΔflaB mutant was secreted at similar levels as FlaA in WT C. jejuni, but FlaAΔN′36 was not secreted (Fig. 2). When we examined strains producing chimeric proteins fusing the initial 26 amino acids of FedB or CiaI (i.e., domain 1 of these proteins; Fig. 1A and 1C), we noticed that the levels of FedBN′26-FlaAΔN′36 or CiaIN′26-FlaAΔN′36 within C. jejuni were much lower than WT FlaA (Fig. 2). However, these chimeric proteins were clearly secreted from C. jejuni, demonstrating that the N-terminal regions of FedB and CiaI were sufficient to restore secretion to FlaAΔN′36 (Fig. 2). Furthermore, fusion of only the initial 14 residues of FedB (subdomains 1A and 1B; Fig. 1A) to FlaAΔN′36, (FedBN′14-FlaAΔN′36) also facilitated secretion of the truncated flagellin. These results verify that the N-terminal domains of FedB and CiaI contain a determinant that is sufficient to mediate secretion of proteins via the C. jejuni flagellum.
Figure 2. Secretion of FedB-FlaA and CiaI-FlaA chimeric proteins from C. jejuni.
Immunoblot analysis of WT and chimeric proteins in whole-cell lysates (WCL) of C. jejuni strains or culture supernatants. WT C. jejuni and C. jejuni ΔflaA ΔflaB expressing WT FlaA, FlaAΔN′36, FedBN′14- FlaAΔN′36, FedBN′26- FlaAΔN′36 and CiaIN′26-FlaAΔN′36 from the flaA promoter in cis at the rdxA locus were grown in MH broth for 4 h at 37 °C in microaerobic conditions. The parental WT C. jejuni 81-176 SmR strain produces both the FlaA and FlaB flagellins. WCL and supernatant proteins were recovered and analyzed by immunoblotting with antisera specific for flagellin or RpoA, which served as a control for a cytoplasmic protein. Molecular weight markers are indicated in kDa.
Analysis of putative C. jejuni flagellar chaperones in secretion of FedB, CiaI, and FspA1
As mentioned above, some flagellar proteins are bound at their C-termini by specific chaperones that enhance recognition by the flagellar T3SS or prevent premature polymerization of the proteins into flagellar structures in the cytoplasm before secretion. Because we observed some involvement of the FedB and CiaI C-termini in secretion of these proteins, we assessed if any predicted C. jejuni flagellar chaperones are directly required for secretion of FedB, CiaI, or FspA1 (another secreted σ28-dependent protein; (Poly et al., 2007; Barrero-Tobon and Hendrixson, 2012)).
The flagellar chaperones and substrates of well-characterized bacterial flagellar systems include FlgN (a chaperone for the hook-filament junction proteins FlgK and FlgL), FliT (a chaperone for the FliD filament cap), and FliS (a chaperone for flagellins) (Fraser et al., 1999; Auvray et al., 2001; Bennett et al., 2001; Evdokimov et al., 2003; Muskotal et al., 2006). FliJ is thought to be a chaperone for rod and hook proteins, but additional analyses revealed that this protein is also involved in the recycling of FlgN and FliT to assist these chaperones in binding free cognate substrates for subsequent secretion (Evans et al., 2006). We constructed C. jejuni 81-176 mutants lacking fliS, fliJ, and Cjj81176_1458 (Cjj1458; a putative flgN homologue). However, we were unable to identify an obvious fliT homologue encoded within the C. jejuni genome. We also analyzed a C. jejuni fliW (Cjj81176_1093) mutant. A FliW homologue in Treponema pallidum has been proposed to function as a chaperone to stabilize flagellins (Titz et al., 2006). However, FliW is also involved in the translational control of the Hag flagellin in Bacillus subtilis (Mukherjee et al., 2011; Mukherjee et al., 2013). The role of FliW in C. jejuni is unknown, but a transposon insertion in fliW reduced C. jejuni motility (Golden and Acheson, 2002). Because CiaB has been shown to be required for the flagellar-dependent secretion of some Cia proteins (Konkel et al., 1999), we analyzed a C. jejuni ciaB mutant to determine whether this protein was required for secretion of FedB, CiaI, or FspA1.
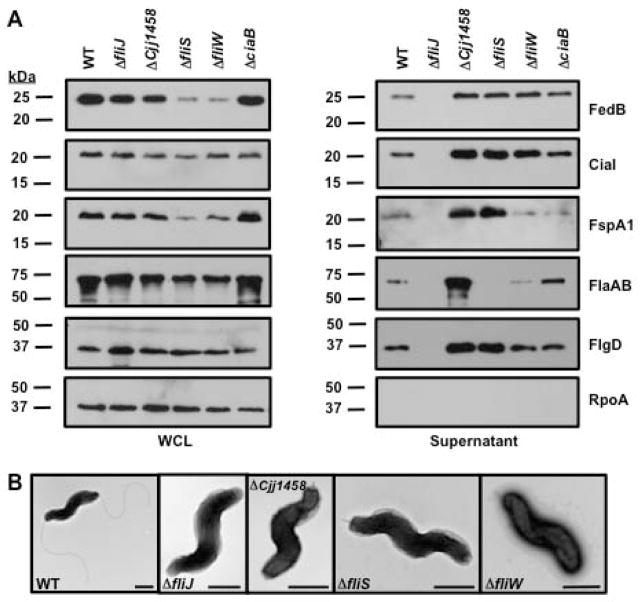
The presence of three flagellar proteins (the FlaA major flagellin, FlaB minor flagellin, and the FlgD hook cap), FedB, CiaI, and FspA1 were analyzed in culture supernatants of WT C. jejuni and isogenic chaperone mutants. Despite production of all these proteins in whole-lysates of C. jejuni ΔfliJ, these proteins were not present in the culture supernatants (Fig. 3A). This gross secretion defect of flagellar proteins is similar to what has been observed in a Salmonella fliJ mutant (Minamino et al., 2000). Examination of C. jejuni ΔfliJ by electron microscopy confirmed the lack of an extracellular flagellar hook at the poles of C. jejuni, which is consistent with the lack of secretion of the FlgD hook cap necessary for hook biosynthesis (Fig. 3A and 3B). These data suggest that FliJ is indirectly required for secretion of FedB, CiaI, and FspA1 by being required for hook biosynthesis and possibly an even earlier step such as rod formation.
Figure 3. Analysis of secretion of flagellar proteins, FedB, CiaI, and FspA1 in C. jejuni mutants lacking putative flagellar chaperones.
Immunoblot analysis of proteins in (A) whole-cell lysates of C. jejuni or in culture supernatants. WT and C. jejuni isogenic mutants lacking putative flagellar chaperones or CiaB, which has been previously reported to be required for secretion of Cia proteins, were grown in MH broth for 4 h at 37 °C in microaerobic conditions. Whole-cell lysates (WCL) and supernatant proteins were recovered and analyzed by immunoblotting with antisera specific for each protein or RpoA, which served as a control for a cytoplasmic protein. The secreted flagellar proteins include the FlgD flagellar hook cap, the FlaA major flagellin, and the FlaB minor flagellin. The antisera for flagellins recognizes both FlaA and FlaB, which are of similar size. Molecular weight markers are indicated in kDa. (B) Electron micrographs of WT and isogenic C. jejuni 81-176 mutants lacking genes encoding putative flagellar chaperones. The bar represents 0.5 μm.
Cjj1458 is a predicted FlgN homologue that chaperones the FlgK and FlgL hook-filament junction proteins, which link the hook to the filament. Consistent with this prediction, C. jejuni ΔCjj1458 only produced hook structures lacking filaments (Fig. 3B). In this mutant, FedB, CiaI, FspA1, and all flagellar proteins were present in the supernatant at higher levels than the WT strain (Fig. 3A). Flagellar hooks without filaments were also produced in ΔfliS and ΔfliW mutants (Fig. 3B). Very little to no flagellin was present in the supernatant of these mutants, which is consistent with the role of these proteins as possible flagellin chaperones (Fig. 3A). However, FliS and FliW were not required for secretion of FedB, CiaI, and FspA1 (Fig. 3A). In fact, we observed higher levels of FedB and CiaI in the supernatants of both C. jejuni ΔfliS and ΔfliW compared to WT C. jejuni. We also observed increased levels of FspA1 in the supernatant of the ΔfliS mutant, but similar amounts of secreted FspA1 in WT C. jejuni and the ΔfliW mutant (Fig. 3A). Curiously, we noticed lower levels of FedB and FspA1 in the whole-cell lysates of C. jejuni ΔfliS and ΔfliW, but these proteins were still efficiently secreted (Fig. 3A). Analysis of C. jejuni ΔciaB revealed no reductions in secretion of any flagellar protein, FedB, CiaI, or FspA1, which was surprising at least for CiaI considering that CiaB had been previously implicated in secretion of Cia proteins (Konkel et al., 1999). Combined, these results suggest that no predicted flagellar chaperones are directly required for secretion of FedB, CiaI, and FspA1.
Flagellar hook and filament biosynthesis oppositely regulate levels of secretion of FedB, CiaI, and FspA1
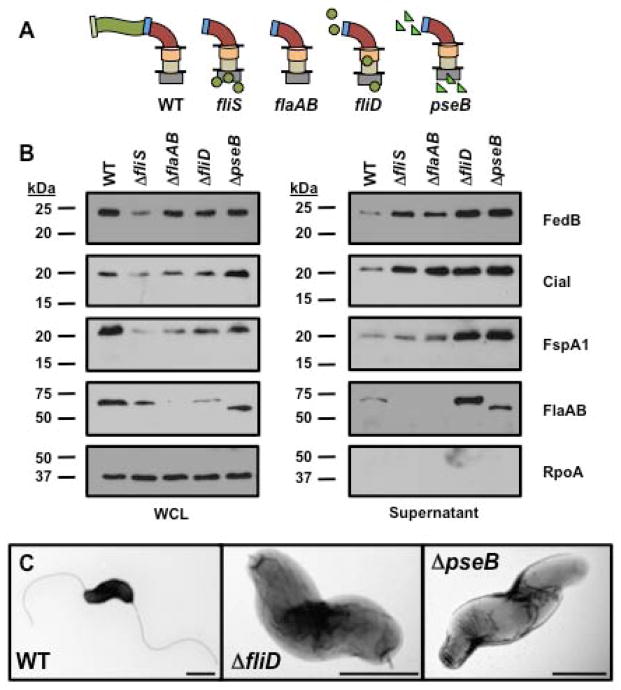
For some Cia proteins and FspA1, the FlgE hook protein, and the FlgK or FlgL hook-filament junction proteins were required for secretion (Poly et al., 2007; Neal-McKinney and Konkel, 2012). We hypothesized that the dynamics of secretion of FedB, CiaI, and FspA1 likely change during the various stages of flagellation as a nascent flagellum is constructed. Therefore, we generated a more comprehensive panel of C. jejuni mutants impaired in different steps or types of processes required for flagellation. Specifically, the mutants we generated were either blocked in distinct steps of flagellation (e.g., rod, hook, hook-filament mutants), defective in production of flagellins or unable to polymerize secreted flagellins into filaments.
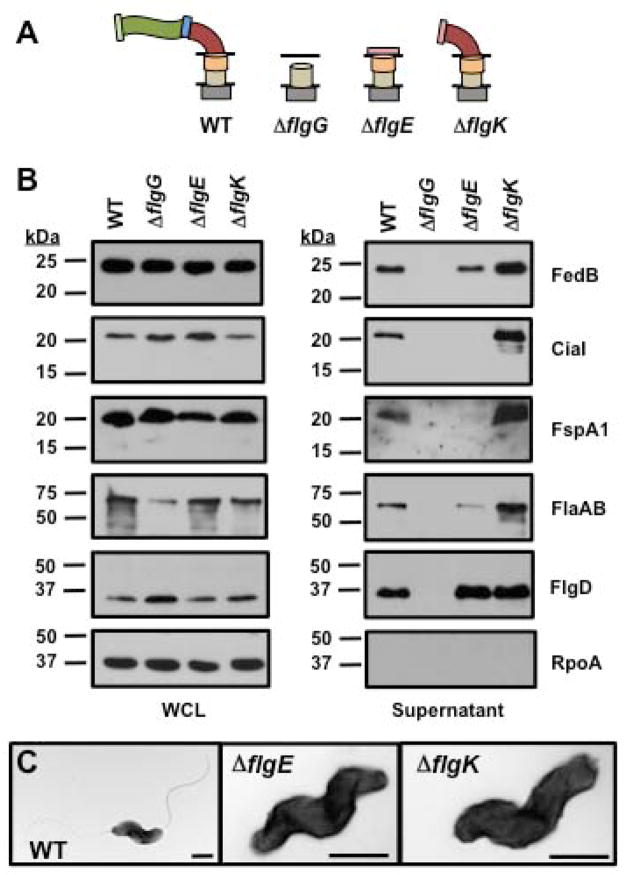
For analyzing C. jejuni mutants defective in formation of the flagellar rod, hook, or hook-filament junction, we constructed mutants that lacked FlgG (the distal rod protein), FlgE, and FlgK. We also attempted to create a flgD mutant that lacked the hook cap and should only produce a flagellar rod, but this mutation caused phase variation of the C. jejuni FlgSR two-component system that positively regulates flagellar rod and hook gene expression to switch to the phase ‘OFF’ state (Hendrixson, 2006, 2008). We observed that in C. jejuni ΔflgG, which would form an incomplete flagellar rod structure that consequently prevents hook biosynthesis, flagellar proteins, FedB, CiaI, or FspA1 were not secreted (Fig. 4A and 4B). In C. jejuni ΔflgE, CiaI and FspA1 were not secreted, but secretion of FedB and the flagellins did occur at slightly reduced levels compared to the WT strain. As expected, secretion of the FlgD hook cap, which occurs prior to FlgE secretion and hook biogenesis, was not affected in C. jejuni ΔflgE (Fig. 4A–C).
Figure 4. Secretion of flagellar proteins, FedB, CiaI, and FspA1 in C. jejuni rod and hook mutants.
(A) Diagram of representative flagellar structures produced by specific C. jejuni flagellar mutants. General substructures of the flagellum include flagellar T3SS, MS ring and C ring (grey box), proximal rod (tan cylinder), distal rod composed by FlgG (orange cylinder), hook (red curve), hook-filament junction (blue rectangle), filament (green wave), FliD filament cap (light green rectangle), and FlgD hook cap (pink rectangle). Lines indicate inner and outer membranes. (B) Immunoblot analysis of FedB, CiaI, FspA1, and flagellar proteins in whole-cell lysates (WCL) or culture supernatants. WT and C. jejuni isogenic mutants lacking flagellar rod and hook proteins were grown in MH broth for 4 h at 37 °C in microaerobic conditions. WCL and supernatant proteins were recovered and analyzed by immunoblotting with antisera specific for each protein or RpoA, which served as a control for a cytoplasmic protein. The secreted flagellar proteins include the FlgD flagellar hook cap, the FlaA major flagellin, and the FlaB minor flagellin. The antisera for flagellins recognized both the FlaA and FlaB, which are of similar size. Molecular weight markers are indicated in kDa. (C) Electron micrographs of WT and isogenic C. jejuni lacking flgE, encoding the flagellar hook, and flgK, encoding a hook-filament junction protein. The bar represents 0.5 μm.
Relative to WT C. jejuni, higher levels of FedB, CiaI, FspA1 and flagellins were present in the supernatant of C. jejuni ΔflgK, which is locked into producing a hook without a filament (Fig. 4A–C). These results suggest that to a certain extent secretion of FedB can occur prior to hook biosynthesis, with secretion of CiaI and FspA1 occurring during hook formation. Furthermore, the hook-filament junction formed by FlgK and FlgL is not required for secretion of some Cia proteins and FspA1, which counters previous observations for secretion of these proteins (Poly et al., 2007; Neal-McKinney and Konkel, 2012). In summary, hook biogenesis positively influences the secretion of FedB, CiaI, and FspA1.
Considering the data described above, enhanced secretion of FedB, CiaI, and FspA1 was observed in mutants defective in formation of the hook-filament junction (i.e, ΔCjj1458 and ΔflgK; Fig. 3A and 4A), which also did not produce filaments. We next conducted secretion assays to discern how flagellin secretion and filament biosynthesis may specifically and separately influence the level of secretion of FedB, CiaI, and FspA1. For these experiments, we focused on two types of C. jejuni mutants. One mutant is a C. jejuni ΔflaA ΔflaB mutant that does not produce flagellins, which eliminates substrates that may compete with FedB, CiaI, and FspA1 for recognition and secretion by the flagellar T3SS. The other mutants produce flagellins normally, but are defective in filament synthesis. These mutants produce intact hooks and hook-filament junctions, but do not allow flagellins to polymerize into filaments. Instead, flagellins are abundantly secreted into the supernatant cultures of the filament synthesis mutants.
We first compared the level of FedB, CiaI, and FspA1 secretion from WT C. jejuni, a ΔfliS mutant, and a ΔflaA ΔflaB double mutant (which lacks all flagellins). As described above, flagellins are retained within C. jejuni ΔfliS, which lacks the chaperone to promote flagellin secretion and consequently filament biosynthesis (Fig 3A and 3B). C. jejuni lacking fliS or the flagellins secreted more FedB, CiaI, and FspA1 relative to WT C. jejuni (Fig. 5A and 5B). These data indicate that in the absence of flagellin secretion and filament biosynthesis, C. jejuni secretes a higher level of FedB, CiaI, and FspA1. We then analyzed a C. jejuni filament synthesis mutant that lacked FliD, which is the filament cap protein that traps flagellins at the growing end of the flagellar tip after hook-filament junction formation to promote polymerization of flagellins into the filament (Fig. 5A). As a consequence, C. jejuni ΔfliD produced flagellar hooks without filaments (Fig. 5C). Similar to fliD mutants in other motile bacteria, C. jejuni flagellins were present in increased levels in the supernatant of C. jejuni ΔfliD relative to WT C. jejuni (Fig. 5B) (Homma et al., 1984; Yokoseki et al., 1995). Even in the presence of persistent flagellin secretion and lack of filament polymerization, FedB, CiaI, and FspA1 were secreted at higher levels in C. jejuni ΔfliD relative to the WT strain (Fig. 5B). We also observed similarly high levels of FedB, CiaI, and FspA1 in the culture supernatant of another filament synthesis mutant, C. jejuni ΔpseB (Fig. 5B). PseB is one of many enzymes required for O-linked glycosylation specifically of C. jejuni flagellins with pseudaminic acid, which is required for C. jejuni flagellins to polymerize into the filament (Goon et al., 2003). In this mutant, higher levels of secretion of FedB, CiaI, and FspA1 occurred simultaneously with increased levels of unmodified flagellins (which are of smaller size than flagellin in WT C. jejuni) in the supernatant relative to WT C. jejuni (Fig. 5A–C). Thus, filament polymerization appears to reduce levels of secretion of FedB, CiaI, and FspA1 when comparing WT and isogenic filament synthesis mutants. Furthermore, our data suggest that FedB, CiaI, and FspA1 are not in competition with flagellins for secretion via the flagellar T3SS. Instead, these proteins are inherently secreted efficiently amid copious amounts of flagellins also present in the cytoplasm to be secreted. Taken together, our findings suggest that in relation to the various stages of flagellar biogenesis, the levels of secretion of FedB, CiaI, and FspA1 are temporally controlled, with hook synthesis promoting secretion and filament polymerization decreasing the level of secretion of these proteins.
Figure 5. Secretion of flagellins, FedB, CiaI, and FspA1 in C. jejuni mutants defective in filament biosynthesis.
(A) Diagram of representative flagellar structures produced by specific C. jejuni flagellar mutants. General substructures of the flagellum include flagellar T3SS, MS ring and C ring (grey box), proximal rod (tan cylinder), distal rod composed by FlgG (orange cylinder), hook (red curve), hook-filament junction (blue rectangle), filament (green wave), FliD filament cap (light green rectangle), FlgD hook cap (pink rectangle), glycosylated flagellins (green circles), and unglycosylated flagellins (green triangles). Lines indicate inner and outer membranes. (B) Immunoblot analysis of FedB, CiaI, FspA1 and flagellar proteins in whole-cell lysates (WCL) or culture supernatants. WT and C. jejuni isogenic mutants lacking proteins required for filament biosynthesis were grown in MH broth for 4 h at 37 °C in microaerobic conditions. WCL and supernatant proteins were recovered and analyzed by immunoblotting with antisera specific for each protein or RpoA, which served as a control for a cytoplasmic protein. The antisera for flagellins recognized both FlaA and FlaB, which are of similar size. Molecular weight markers are indicated in kDa. (C) Electron micrographs of WT and isogenic C. jejuni filament synthesis mutants lacking fliD, encoding the filament cap, and pseB, encoding an enzyme required for O-linked glycosylation of flagellin. The bar represents 0.5 μm.
Analysis of the requirement of CiaI secretion for host interactions
Although CiaI is secreted by the C. jejuni flagellum in vitro and is required for WT levels of invasion of human intestinal epithelial cells and commensal colonization of the avian host (Buelow et al., 2011; Barrero-Tobon and Hendrixson, 2012), it is unknown whether CiaI must be secreted from C. jejuni for either of these processes. To test the requirement for CiaI secretion for invasion of human colonic cells or commensal colonization of the chick ceca, we took advantage of two CiaI mutants, CiaIΔ1 and CiaIΔ4GH, which are stable proteins that are not secreted due to deletion of N- or C-terminal regions (Fig. 1C and 1D). Genes encoding WT CiaI or the CiaI mutant proteins were expressed with the native ciaI promoter in cis from the rdxA locus in C. jejuni ΔciaI. Similar to our findings in Fig. 1D, the CiaI mutant proteins were not secreted and instead remained associated with the bacterium (Fig. S1). As expected, C. jejuni ΔciaI complemented with WT ciaI in cis was secreted to the supernatant (Fig. S1).
When these strains were analyzed in invasion assays with human T84 colonic cells, approximately 14.5% of the WT C. jejuni inoculum was found intracellularly at the end of the 6-hour assay (Table 1). C. jejuni ΔciaI showed an approximately two-fold reduction in invasion, but WT levels of invasion were restored by in cis expression of WT ciaI. When we analyzed C. jejuni ΔciaI producing the non-secreted CiaIΔ1 or CiaIΔ4GH proteins, we found that CiaIΔ1 could not restore invasion. However, CiaIΔ4GH restored invasion to a level similar to C. jejuni ΔciaI complemented in cis with WT ciaI (Table 1). We interpret these data as suggesting that at least CiaIΔ4GH, which only lacks the last 12 residues, is a functional protein that does not need to be secreted to facilitate in vitro invasion of C. jejuni. We suspect that the more severe deletion of the N-terminal 25 amino acids of CiaI to create CiaIΔ1 may have structurally altered the protein to create a biologically inactive protein that cannot restore invasion to the ΔciaI mutant. Our data counter previous findings implying that CiaI secretion into eukaryotic cells is necessary for invasion or intracellular survival (Buelow et al., 2011). Instead, our data suggest that CiaI performs an intracellular function within C. jejuni for maximal invasion.
Table 1.
Invasion capacity of wild-type C. jejuni and isogenic ciaI mutants lacking domains for secretion.
| Strain | Invasion of T84 cells (% inoculum)a |
|---|---|
| Wild-type | 14.51 ± 1.43 |
| ΔciaI | 7.51 ± 0.74* |
| ΔciaI rdxA::ciaI | 12.89 ± 1.56 |
| ΔciaI rdxA::ciaI Δ1 | 7.12 ± 1.07* |
| ΔciaI rdxA::ciaI Δ4GH | 11.14 ± 1.01 |
Percent invasion was determined by comparing the number of intracellular bacteria surviving a 2 h gentamicin treatment of infected T84 cells compared to the number of bacteria in the infecting inoculum (approximately 3.0 × 106 cfu). Each assay was performed in triplicate, and at least three biological replicates were performed. The average percent invasion +/− standard error for each strain is presented. Statistically-significant differences in invasion between wild-type C. jejuni and mutant strains are indicated (* P-value < 0.05).
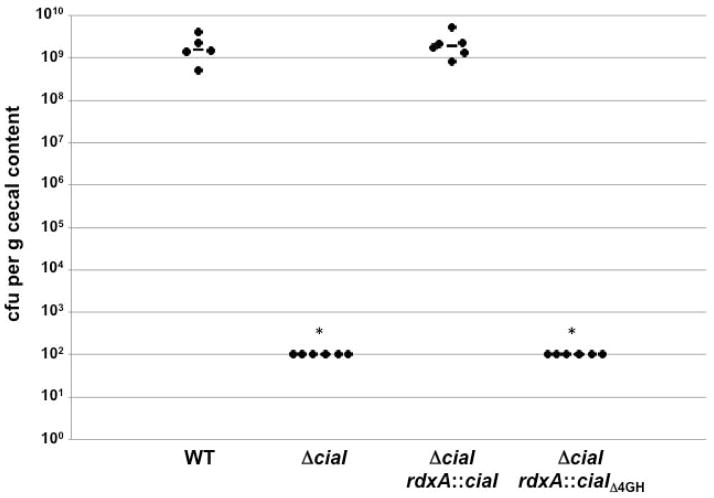
We then examined the ability of WT ciaI and ciaIΔ4GH expressed in cis to restore the commensal colonization capacity to C. jejuni ΔciaI for the chick ceca. One-day old chicks were orally gavaged with approximately 102 cfu of WT C. jejuni or ΔciaI mutant strains and the level of these bacteria in the chick ceca were determined 14-days post-infection. WT C. jejuni colonized chicks at levels between 3.4 × 108 – 2.9 × 109 cfu per gram of cecal content, but the level of colonization of the uncomplemented C. jejuni ΔciaI mutant was below the limit of detection of 102 cfu per gram of cecal content in all chicks (Fig. 6). WT CiaI expressed in cis fully restored the colonization capacity of the ΔciaI mutant to levels similar to WT C jejuni. However, CiaIΔ4GH did not restore colonization to C. jejuni ΔciaI (Fig. 6). Assuming that the small domain deletion in CiaIΔ4GH did not perturb the biological activity of the protein for commensal colonization, our data suggest that secretion of CiaI is essential for C. jejuni to promote commensal colonization of the chick intestinal tract. These data may indicate that CiaI performs an extracellular function in the cecal mucus layer or on the cecal epithelial surface for C. jejuni to promote colonization of the natural avian host.
Figure 6. Commensal colonization capacity of WT C. jejuni and C. jejuni ΔciaI mutants producing WT or secretion-deficient CiaI proteins.
One-day old chicks were orally inoculated with 102 cfu of C. jejuni strains. Each dot represents the amount of C. jejuni recovered from the ceca of chicks at day 14 post-infection. The geometric mean for each group is depicted by the horizontal bar. Statistical analysis was performed using the Mann-Whitney U test (*P < 0.01).
Discussion
The C. jejuni flagellum is a multipurpose organelle that functions in multiple biological processes for the bacterium. As with other motile bacteria, the C. jejuni flagellum confers swimming motility. We recently discovered that the C. jejuni flagellum also impacts cellular division by functioning in a mechanism to inhibit septation at polar regions of the bacterium so that division occurs at the cellular midpoint (Balaban and Hendrixson, 2011). The C. jejuni flagellum is a secretory machine for flagellar proteins, but has also been implicated in the secretion of the Cia proteins, FedB, and FspA1, many of which contribute to invasion of human epithelial cells and commensal colonization of chicks (Konkel et al., 1999; Konkel et al., 2004; Christensen et al., 2009; Buelow et al., 2011; Barrero-Tobon and Hendrixson, 2012). Although secretion of flagellar proteins has been studied in other motile bacteria, an understanding of mechanisms required to secrete proteins not involved in motility via the flagellum is fairly limited. Therefore, we investigated how the requirements and mechanisms for secretion of non-flagellar proteins compared to those for secretion of bona fide flagellar proteins. By performing these studies, we provided new knowledge regarding how the C. jejuni flagellum regulates secretion of virulence and colonization determinants and identified factors that allow the proteins to be efficiently secreted via the flagellum.
Many flagellar proteins possess an N-terminal region that is required for secretion. For rod and hook proteins, a conserved motif (FXXXϕ) located within the N-terminal 30–50 residues is recognized by the flagellar T3SS (Evans et al., 2013). However, flagellins do not contain this sequence and a structurally disordered region within the N-terminal 50 residues appears to be specifically recognized as a type of secretion signal recognized by the flagellar T3SS (Vegh et al., 2006; Dobo et al., 2010; Neal-McKinney et al., 2010). For Y. enterocolitica YplA, the first 20 residues of the protein as a whole are required for secretion through any T3SS produced by the bacterium, but this region of YplA also lacks the FXXXϕ motif (Warren and Young, 2005). Our analysis of FedB and CiaI suggest that a secretion signal at the N-terminus is most reminiscent of that of flagellins. Whereas we identified the N-terminal 14 or 26 residues as essential for secretion of FedB or CiaI and observed that these same regions promoted secretion of a mutant flagellin protein lacking its native secretion signal, removal of smaller subdomains within these N-terminal regions still allowed for secretion. If a specific amino acid sequence promoted secretion, we would have expected that deleting smaller domains within the N-terminal 14 residues of FedB or N-terminal 26 residues of CiaI would have abolished secretion, which was observed with Y. enterocolitica YplA. Thus, it is difficult to conclude that a conserved amino acid sequence, functionally similar to the FXXXϕ motif in flagellar rod and hook proteins, is required for the flagellar T3SS to recognize and secrete these proteins. Consistent with this hypothesis, we did not identify any conserved amino acid sequence within the N-terminal regions of FedB, CiaI, or FspA1 that could serve as an amino acid sequence-based secretion signal. Instead, we hypothesize that like flagellins, the N-terminus of FedB, CiaI, and FspA1 (mostly within the initial 14–26 residues) form a somewhat disordered structure of some minimal length for specific recognition by the flagellar T3SS. Considering that other proteins of C. jejuni likely have partially unfolded or disordered domains and are not secreted by the flagellar T3SS, we suspect that the flagellar T3SS can specifically detect this type of disordered motif at the N-terminus of FedB, CiaI, and FspA1 to secrete these proteins.
We also observed that the C-terminal regions of FedB and CiaI influence secretion. This requirement for a C-terminal domain for secretion is reminiscent of a chaperone-binding domain at the end of some flagellar proteins (Fraser et al., 1999; Auvray et al., 2001; Bennett et al., 2001). However, we were unable to demonstrate that any predicted C. jejuni flagellar chaperone was required for secretion of FedB, CiaI, or FspA1. Therefore, if a chaperone is required, it remains elusive. It is also possible that these proteins could be organized similarly as secreted type III effectors of injectisome systems, which are bound at their N-terminus by chaperones that assist in secretion via the respective T3SSs (Stebbins and Galan, 2001; Birtalan et al., 2002; Lilic et al., 2006; Costa et al., 2012). In this case, the C-terminal regions of FedB, CiaI, or FspA1 would be required for some other step in secretion. As such, additional analysis is necessary to identify other cellular factors that may also influence secretion of the FedB, CiaI, and FspA1 through the C. jejuni flagellum.
By analyzing elements of the flagellar structure required for secretion of FedB, CiaI, and FspA1, we provided evidence that flagellar biogenesis itself mechanistically imparts a type of temporal control of secretion of these non-flagellar proteins as the nascent flagellum polymerizes into the final organelle. We discovered that FedB can be secreted to a certain extent before hook biosynthesis initiates, but a bolus of FedB, CiaI, and FspA1 secretion appears to occur during or immediately after hook formation. Thus, hook biosynthesis positively influences secretion of FedB, CiaI, and FspA1, which is similar to the secretion of flagellins. After this stage, the C. jejuni flagellar T3SS is largely dedicated to the secretion of FlaA major flagellin monomers to complete flagellation. It has been estimated that 20,000 flagellin monomers must be secreted to form a single filament of a flagellum (Macnab, 2003; Minamino and Namba, 2004). As such, we thought that FedB, CiaI, and FspA1 may undergo significant competition with flagellins for secretion. Indeed, we observed enhanced secretion of these proteins in a flagellin mutant. However, in mutants that were unable to produce a filament due to mutation of fliD or the Pse flagellin glycosylation system but were competent for secretion of flagellin, we observed a significant increase in flagellins, FedB, CiaI, and FspA1 in the supernatant relative to WT C. jejuni. We surmise from these data that FedB, CiaI, and FspA1 compete quite well with flagellins for secretion via the flagellar T3SS, but filament polymerization itself gradually limits secretion of these proteins as the filament elongates. Thus, flagellar biogenesis gives temporal control of virulence and colonization determinants by promoting their secretion once the hook is synthesized and then reducing secretion of the proteins as the flagellins polymerize into the extracellular filament. We suspect that the FedB, CiaI, and FspA1 may be secreted at some low level after a filament is complete, which continues to support persistent colonization of the natural avian host. If a fully-formed filament completely inhibited secretion of these proteins in C. jejuni, then these proteins would only be secreted in new daughter cells that must form a flagellum at the new pole to complete the amphitrichous flagellation pattern after division. While the latter hypothesis is possible, it would appear to limit the ability to produce colonization factors that may benefit the C. jejuni population as a whole in the avian host to only C. jejuni cells that are constructing flagella.
Currently, we do not know the exact mechanisms used by the flagellum to decrease secretion of FedB, CiaI, and FspA1. It is unlikely that decreased secretion of these proteins is due to increasing steric hindrance during transit through the central channel of the flagellar filament as the filament grows. Flagellins, which also must transit through the same channel to polymerize on the growing flagellar tip, do not appear to experience steric hindrance. Alternatively, there may be a change in the efficiency of secretion of FedB, CiaI, and FspA1 once the flagellins polymerize into the filament. It is known that during different stages of flagellation, the substrate specificity of the flagellar T3SS changes from secreting rod and hook proteins to predominantly filament substrates (Fraser et al., 2003; Ferris et al., 2005; Minamino et al., 2009; Erhardt et al., 2010; Erhardt et al., 2011). It is conceivable that filament polymerization in C. jejuni triggers a change in the flagellar T3SS that alters its substrate specificity to reduce secretion of FedB, CiaI, and FspA1 and primarily secrete flagellins. Regardless of the mechanism, we propose a temporal regulation of secretion of virulence and colonization factors that is controlled during two stages of flagellation – promotion of secretion once hook biogenesis initiates and reduction of secretion as the flagellar filament polymerizes. To our knowledge, our work presents a new mechanism whereby the actual process of flagellar biosynthesis controls the levels of secretion of virulence and colonization factors of a bacterium.
We previously reported that mutation of C. jejuni flaA resulted in the absence of FspA1 in lysates and the lack of FedB and CiaI in culture supernatants (Barrero-Tobon and Hendrixson, 2012). We initially concluded from this work that the FlaA major flagellin was required for production, stability, and secretion of some of these proteins. For complete analysis in this work, we constructed a new C. jejuni mutant that lacked both FlaA and the FlaB minor flagellin. Our data herein clearly show that production and secretion of FedB, FspA1, or CiaI was not reduced in C. jejuni lacking both flagellins. To resolve this discrepancy, we reanalyzed our previous flaA mutant and found that it contained a phase-variable mutation in flgR, encoding the response regulator required for σ54-dependent expression of many flagellar rod and hook genes and consequently affects expression of σ28-dependent genes, including fed genes (data not shown; (Hendrixson and DiRita, 2003; Boll and Hendrixson, 2011; Barrero-Tobon and Hendrixson, 2012)). Because of this mutation, the previous flaA mutant failed to produce rod and hook proteins, which are required for secretion of many proteins via the flagellum, and reduced levels of the Fed proteins in lysates. Therefore, we can confidently conclude from the present work that flagellins are not required for stability or secretion of the Fed colonization and virulence determinants. Also, secretion of CiaI (along with FedB and FspA1) occurred without CiaB, addition of serum or host cell contact despite previous reports that Cia proteins need these factors for secretion (Konkel et al., 1999; Rivera-Amill et al., 2001; Konkel et al., 2004). Therefore, these factors originally implicated as requirements for secretion of Cia proteins via the flagellum do not appear to be broadly required for secretion of non-flagellar proteins through the organelle.
Our work combined with others clearly shows that CiaI is secreted from C. jejuni in vitro and is required for WT levels of invasion of human intestinal or colonic cells and commensal colonization of the chick cecum (Buelow et al., 2011; Barrero-Tobon and Hendrixson, 2012). In terms of invasion of human cells, other studies combining the use of a CiaI chimeric protein and CiaI expressed in eukaryotic cells suggested that CiaI secreted from C. jejuni may eventually reside within eukaryotic cells and localize with vesicles to prevent fusion of lysosomes with the CCV (Buelow et al., 2011; Neal-McKinney and Konkel, 2012). However, this hypothesis was weakened when a dileucine motif in CiaI previously indicated in localizing the protein to vesicles was found not to be required for invasion (Buelow et al., 2011; Barrero-Tobon and Hendrixson, 2012). Instead, mutation of a putative nucleotide-binding domain in CiaI caused a decrease in invasion, but it is currently unclear whether CiaI binds ATP or GTP and what role this activity may have in invasion (Barrero-Tobon and Hendrixson, 2012). Furthermore, it has never been tested whether native CiaI from C. jejuni actually must be secreted to mediate a function important in in vitro invasion or colonization of a natural host. We took advantage of two mutant CiaI proteins, CiaIΔ1 and CiaIΔ4GH, which are not secreted from C. jejuni to determine whether CiaI secretion from C. jejuni is essential for invasion of human colonic cells or in vivo growth in commensal colonization of chicks. We found that C. jejuni producing CiaIΔ4GH (which lacked only the last 12 residues) promoted WT levels of invasion of human colonic cells, indicating that this form of CiaI was able to support invasion when remaining associated with C. jejuni. Expression of CiaIΔ1 did not restore invasion to C. jejuni ΔciaI, but this protein may be biologically inactive when lacking the first 25 residues. However, CiaIΔ4GH did not restore the commensal colonization capacity of C. jejuni ΔciaI. Assuming that deletion of the last 12 residues of CiaI only disrupted its secretion and not an activity of the protein necessary for an in vivo function, these data suggest that secretion of CiaI is required for commensal colonization of chicks but not for invasion of human colonic cells. Therefore, our work supplies support that CiaI likely functions within C. jejuni for invasion, which counters a previously reported hypothesized role of secreted CiaI during invasion (Buelow et al., 2011). In relation to commensal colonization of chicks, C. jejuni resides in the mucus layer lining the ceca and intestinal tract with no invasion of the epithelium occurring. Currently, the fate of secreted CiaI during commensal colonization is unknown. The protein may exert a function at the mucus layer or the surface of an intestinal or cecal epithelial cell in the avian gastrointestinal tract. It is conceivable that secreted CiaI could eventually reside within avian epithelial cells and mediate a function important for C. jejuni colonization. This mechanism would depend on an unknown mechanism for uptake of CiaI by the target cecal cell or delivery into the cell. Regardless, our findings provide new information regarding whether a previously identified virulence and colonization factor must be secreted from C. jejuni for host interactions.
From a historical perspective, the Y. enterocolitica flagellum was the first flagellar organelle observed to secrete a non-flagellar protein (Young et al., 1999). Subsequently, the C. jejuni flagellum was found to be a versatile secretion machine by secreting flagellar proteins, Cia proteins, and some Fed virulence and colonization determinants. However, how the flagellum secretes non-flagellar proteins was not fully understood. In this work, we have revealed new knowledge regarding the versatility of the flagellum as a secretion machine. We discovered that flagellar biogenesis both positively and negatively regulates the level of secretion of C. jejuni virulence and colonization determinants. Furthermore, these proteins appear to have adopted traits common to proteins such as the flagellins, which are natural substrates for the flagellar secretion machinery, including possibly similar N-terminal structural regions and specific unidentified chaperones to assist in secretion. Importantly, we also uncovered new biological information about the requirement of secretion of one protein, CiaI, for interaction with different hosts. An interesting question to be addressed in future endeavors would be whether other motile bacteria commonly employ flagella to secrete non-flagellar proteins for various activities.
Experimental Procedures
Bacterial strains and plasmids
C. jejuni 81-176 strains used in this study are described in Tables S1 and S2 in Supporting Information. C. jejuni strain 81-176 was obtained from a patient with gastroenteritis (Korlath et al., 1985). Subsequent studies verified the capacity of this strain to infect human volunteers and promote commensal colonization of chicks (Black et al., 1988; Hendrixson and DiRita, 2004). C. jejuni was typically grown in microaerobic conditions (85% N2, 10% CO2, 5% O2) on Mueller-Hinton (MH) agar or in MH broth at 37 °C. As required, antibiotics were added to MH media at the following concentrations: 10 μg ml−1 trimethoprim (TMP), 20 μg ml−1 chloramphenicol, 100 μg ml−1 kanamycin, 30 μg ml−1 cefoperazone or 0.5, 1, 2, or 5 mg ml−1 streptomycin. All C. jejuni strains were stored at −80 °C in a 85% MH broth and 15% glycerol solution. Typical growth to perform most experiments required C. jejuni strains to be grown from frozen stocks for 48 h in microaerobic conditions at 37 °C, then streaked on MH agar and grown for additional 16 h in identical conditions. Escherichia coli DH5α and BL21were grown on Luria-Bertani (LB) agar or in LB broth containing 100 μg ml−1 ampicillin, 100 μg ml−1 kanamycin or 15 μg ml−1 chloramphenicol as appropriate. All E. coli strains were stored at −80 °C in a 80% LB broth and 20% glycerol solution.
Generation of antisera
Specific antiserum against C. jejuni FlgD was generated from purified recombinant protein with a glutathione-S-transferase (GST)-fusion. Primers containing 5′ in-frame BamHI restriction sites were used to amplify a flgD fragment from codons 58 to 225 from the C. jejuni 81-176 genome. The BamHI-digested PCR product was then ligated into BamHI-digested pGEX-4T-2 (GE Healthcare), generating pDRH2933. The resulting plasmid was then transformed into E. coli BL21(DE3) for protein induction and purification from the soluble fraction with glutathione Sepharose 4B, according to manufacturer’s instructions (GE Healthcare). Purified recombinant protein was then used to immunize five mice for production of polyclonal antisera (Cocalico Biologicals, Inc).
Construction of mutants
C. jejuni mutants were constructed by electroporation following previously described methods (Hendrixson et al., 2001). Genes to be deleted from the C. jejuni 81-176 SmR chromosome were first amplified by PCR using primers containing 5′ BamHI or EcoRI restriction sites. Each fragment contained the gene of interest with 750 bases of flanking sequence. Cloning of the fragments into the BamHI or EcoRI site of pUC19 resulted in the creation of the following plasmids: pDRH259 (pUC19::flgK), pDRH2429 (pUC19::fliDS), pDRH3235 (pUC19::pseBC), pLKB648 (pUC19::rdxA), pABT824 (pUC19::ciaB), pABT945 (pUC19:: fliW) and pABT1044 (pUC19::fliJ). For some genes, it was necessary to create restriction sites within the coding sequence by PCR-mediated mutagenesis or by SOEing mutagenesis (Higuchi, 1990; Makarova et al., 2000). The products of these PCRs created a SwaI site in fliJ (pABT1211), a StuI site in Cjj81176_1458 (pABT953) and fliW (pABT954), and an EcoRV site in pseB (pDRH3237), rdxA (pLKB653), fliD (pABT822), fliS (pABT823) and ciaB (pABT828). To create a flaAB double mutant, plasmid pDRH519 (pUC19::flaAB) was digested with EcoRV, which cuts within both flaA and flaB to remove a large 3′ region of flaA and the 5′ region of flaB. A SmaI-digested cat-rpsL cassette was obtained from pDRH265 and ligated into the appropriate restriction sites of the respective gene in each plasmid as listed in Table S2.
Each plasmid generated above was electroporated into C. jejuni 81-176 SmR (DRH212) to interrupt each respective gene on the chromosome with the cat-rpsL cassette. Transformants were recovered on MH agar containing chloramphenicol. Mutations were verified by colony PCR and the following isogenic mutants of 81-176 SmR were obtained: DRH1249 (flgK::cat-rpsL), DRH3240 (pseB::cat-rpsL), ABT857 (fliD::cat-rpsL), ABT861 (fliS::cat-rpsL), ABT919 (ciaB::cat-rpsL), ABT1019 (fliW::cat-rpsL), ABT1021 (Cjj81176_1458::cat-rpsL), ABT1246 (fliJ::cat-rpsL), and ABT1176 (flaAB::cat-rpsL).
In-frame deletions of genes originally cloned into pUC19 were created with specific primers using PCR-mediated mutagenesis (Makarova et al., 2000). Alternatively, some deletions were created by SOEing mutagenesis to result in fragments that were then cloned into the BamHI site of pUC19 (Higuchi, 1990). After sequencing to verify correct construction of in-frame deletions, the following plasmids were obtained: pABT931 (pUC19::ΔfliS), pABT1031 (pUC19::ΔCjj81176_1458) and pABT1057 (pUC19::ΔfliJ). To create C. jejuni strains containing deletions of specific genes, these plasmids were then electroporated into strains containing cat-rpsL interruptions of the respective genes on the chromosome. Transformants were recovered on MH agar with 0.5, 1, 2 or 5 mg/ml of streptomycin and then screened for chloramphenicol sensitivity. Deletion of each gene was verified by colony PCR, which resulted in creation of the following 81-176 SmR mutant strains: ABT952 (ΔfliS), ABT1055 (ΔCjj81176_1458), and ABT1266 (ΔfliJ).
Plasmids carrying wild-type ciaI or fedB were constructed by amplifying DNA fragments containing 258 bases upstream of the start codon and 23 bases downstream of the stop codon of ciaI or 252 bases upstream of the start codon and 25 bases downstream of the stop codon of fedB by PCR using primers containing 5′ BamHI restriction sites. These fragments were then cloned into the BamHI site of the E. coli- C. jejuni shuttle vector, pRY108, to result in the creation of pABT720 (pRY108::ciaI) and pABT951 (pRY108::fedB). Primers were designed to generate truncated mutants lacking 6 to 25 codons at the 5′ or the 3′ end of the coding sequence of ciaI or fedB by PCR-mediated or SOEing mutagenesis (Higuchi, 1990; Makarova et al., 2000). The plasmids were confirmed by sequencing. As a result the following plasmids were created pABT956 (pRY108::ciaIΔ1), pABT1075 (pRY108::ciaIΔ1A), pABT1069 (pRY108::ciaIΔ1B), pABT1047 (pRY108::ciaIΔ1C), pABT1122 (pRY108::ciaIΔ1D), pABT1127 (pRY108::ciaIΔ1AB), pABT1128 (pRY108::ciaIΔ1CD), pABT1354 (pRY108::ciaIΔ1ABC), pABT957 (pRY108::ciaIΔ3), pABT1032 (pRY108::ciaIΔ4), pABT1151 (pRY108::ciaIΔ4EF), pABT1152 (pRY108::ciaIΔ4GH), pABT1024 (pRY108::fedBΔ1), pABT1070 (pRY108::fedBΔ1A), pABT1056 (pRY108::fedBΔ1B), pABT1076 (pRY108::fedBΔ1C), pABT1048 (pRY108::fedBΔ1D), pABT1129 (pRY108::fedBΔ1AB), pABT1130 (pRY108::fedBΔ1CD), pABT1356 (pRY108::fedBΔ1ABC), pABT961 (pRY108::fedBΔ2), pABT955 (pRY108::fedBΔ3), pABT1025 (pRY108::fedBΔ4), pABT1135 (pRY108::fedBΔ4EF) and pABT1370 (pRY108::fedBΔ4GH). These plasmids were then transformed into the E. coli conjugation strain DH5α/RK212.1. The donor strains were used to conjugate the plasmids into the appropriate C. jejuni 81-176 SmR ΔciaI (ABT279) or ΔfedB (ABT473).
To create C. jejuni 81-176 SmR ΔciaI strains expressing wild-type CiaI or CiaI mutants in cis, pLKB653 (pUC19::rdxA) was first manipulated to create a PmeI restriction site using PCR-mediated mutagenesis to result in plasmid pABT1265. A SmaI-digested aphA-3 cassette obtained from pILL600 was ligated into the PmeI site of rdxA to create pABT1307 (pUC19::rdxA-aphA-3). The DNA encoding wild-type ciaI or mutant ciaI proteins from pRY108-based plasmids (Table S2) was released by BamHI-digestion and ligated into pABT1307 that had been digested with BglII. All inserts were confirmed by DNA sequencing. The resulting plasmids included pABT1342 (pUC19::rdxA-ciaI-aphA-3), pABT1322 (pUC19::rdxA-ciaIΔ1-aphA-3), and ABT1403 (pUC19::rdxA-ciaIΔ4GH-aphA-3). These plasmids were then electroporated into C. jejuni 81-176 SmR ΔciaI (ABT279). Transformants were recovered on MH agar containing kanamycin and confirmed by colony PCR. This resulted in the isolation of C. jejuni 81-176 SmR derivatives ABT1347 (ΔciaI rdxA::ciaI-aphA-3), ABT1331 (ΔciaI rdxA::ciaIΔ1-aphA-3), and ABT1413 (ΔciaI rdxA::ciaIΔ4GH-aphA-3).
SOEing mutagenesis was employed to construct chimeric proteins in which the N-terminus of FedB or CiaI was fused to FlaAΔN′36. First, a DNA fragment containing 203 bases upstream of the start codon through the stop codon of flaA was amplified by PCR using primers containing 5′ BamHI restriction sites. Additionally, primers were designed to amplify products to use in SOEing mutagenesis to result in an in-frame deletion of codons 2 to 36 of flaA. BamHI-digested flaA and flaAΔN′36 DNA fragments were ligated into BglII-digested pABT1307, resulting in plasmids ABT1465 (pUC19::rdxA-flaA-aphA-3) and ABT1412 (pUC19::rdxA-flaAΔN′36-aphA-3). To construct constructs encoding FedB-FlaA or CiaI-FlaA chimeric proteins, SOEing PCR was performed to create a DNA fragment that fused the promoter region of flaA, fedB or ciaI encoding the first 14 or 26 residues to flaA beginning at codon 37. The final PCR product contained 5′ BamHI restriction sites. The final fragment consisted of the promoter of flaA followed by chimeric genes encoding CiaIN′26-FlaAΔN′36, FedBN′14-FlaAΔN′36 or FedBN′26-FlaAΔN′36. The BamHI-digested PCR fragments were ligated into BglII-digested pABT1307. The resulting plasmids were sequenced for confirmation and included pABT1466 (pUC19::rdxA-ciaIN′26-flaAΔN′36-aphA-3), pABT1501 (pUC19::rdxA-fedBN′14-flaAΔN′36-aphA-3) and pABT1468 (pUC19::rdxA-fedBN′26-flaAΔN′36-aphA-3). Finally, pABT1412, pABT1465, pABT1466, pABT1468 and pABT1501 were electroporated into C. jejuni 81-176 SmR flaAB::cat-rpsL (ABT1176). Transformants were recovered on MH agar containing kanamycin and chloramphenicol and insertion was confirmed by colony PCR. This resulted in the isolation of C. jejuni 81-176 SmR derivatives ABT1470 (ΔflaAB rdxA::flaA-aphA-3), ABT1420 (ΔflaAB rdxA::flaAΔN′36-aphA-3), ABT1473 (ΔflaAB rdxA::ciaIN′26-flaAΔN′36-aphA-3), ABT1503 (ΔflaAB rdxA::fedBN′14-flaAΔN′36-aphA-3) and ABT1477 (ΔflaAB rdxA::fedBN′26-flaAΔN′36-aphA-3). Analysis of production and secretion of C. jejuni proteins. Proteins produced by or secreted from C. jejuni strains were collected as previously described (Barrero-Tobon and Hendrixson, 2012). Briefly, C. jejuni strains were suspended from MH agar plates into MH broth to an OD600 of 0.6. For each strain, 20 ml of diluted culture were incubated at 37 °C in microaerobic conditions without shaking for 4 h. In the case of C. jejuni strains that contained a plasmid, kanamycin was added to each culture for the duration of the 4 h. At the end of the incubation period, final OD600 measurements were obtained. For preparation of proteins from whole-cell lysates (WCL), 1 ml of culture for each strain was pelleted in a microcentrifuge at full speed for 3 min, washed once with PBS and resuspended in 25 μl of PBS and 25 μl of 2X SDS-PAGE loading buffer for a total volume of 50 μl. For recovery of supernatant proteins, the remaining 18 ml of culture were centrifuged for 30 min at 13,000 rpm. The supernatants were recovered and the centrifugation step was repeated twice to ensure removal of all bacteria. Proteins present in the supernatant were precipitated by combining 18 ml of supernatant with 2 ml of trichloroacetic acid (TCA; 10% final concentration) followed by a 30 min incubation on ice. Precipitated proteins were recovered by centrifugation for 15 min at 10,000 rpm. The protein pellets were rinsed with 1 ml of cold acetone and dried. Precipitated proteins were resuspended in 40 μl of 1M Tris, pH 8.0 and 40 μl of 2X SDS-PAGE loading buffer.
All protein samples were boiled for 5 min prior to loading on 12.5% SDS-PAGE gels. For the wild-type C. jejuni WCL, 10 μl (for analysis of CiaI and RpoA), 15 μl (for analysis of FspA1 and FlgD), and 2 μl (for analysis of FedB) were analyzed. For detection of FlaA and FlaB in WCL, 2 μl of a 1:5 dilution of WCL were used. The volumes of WCL of mutant strains analyzed were normalized based on the final OD600 readings of WT and mutant strains to ensure that equal amounts of proteins between strains were analyzed. For supernatant samples, 4 μl (for analysis of FlaAB and FedB), 10 μl (for analysis of CiaI and RpoA) or 15 μl (for analysis of FspA1 and FlgD) of precipitated proteins were separated by 12.5% SDS-PAGE. For immunoblot analysis, primary murine antisera was used at the following concentrations to detect proteins: α-FlgD M92, 1:750; α-FspA1 M140, 1:2000 (Barrero-Tobon and Hendrixson, 2012); α-CiaI M157, 1:2000 (Barrero-Tobon and Hendrixson, 2012); and α-RpoA M60, 1:2500 (Sommerlad and Hendrixson, 2007). For detection of FedB or the FlaA and FlaB flagellins, polyclonal rabbit antiserum was used at a dilution of 1:10,000 or 1:22,000 respectively (Lee et al., 1999; Barrero-Tobon and Hendrixson, 2012). A 1:10,000 dilution of a horseradish peroxidase (HRP)-conjugated goat anti-mouse or anti-rabbit antiserum (Bio-Rad) was used as the secondary antibody. For FlaA and FlaB, a 1:20,000 dilution of anti-rabbit secondary antibody was used. Immunoblots were developed by using the Western Lightning Plus ECL kit (Perkin-Elmer). Differences in levels of proteins in lysates or supernatants were compared visually.
Chick colonization assays
All use of animals in experimentation has been approved by IACUC at the University of Texas Southwestern Medical Center. The ability of WT or mutant C. jejuni 81-176 SmR strains to colonize the ceca of chicks after oral inoculation was determined as previously described (Hendrixson and DiRita, 2004). Briefly, fertilized chicken eggs (SPAFAS) were incubated for 21 d at 37.8 °C with appropriate humidity and rotation in a Sportsman II model 1502 incubator (Georgia Quail Farms Manufacturing Company). Approximately 12 to 24 h after hatching, chicks were orally infected with 100 μl of MH broth containing approximately 102 cfu of a single WT C. jejuni or mutant strain. To prepare strains for infection, C. jejuni strains were suspended from plates after growth at 37 °C in microaerobic conditions and diluted in MH broth to obtain the appropriate inoculum for oral gavage of chicks. Dilutions of the inocula were spread on MH agar to determine the number of bacteria in each inoculum. Fourteen days post-infection, chicks were sacrificed and the cecal contents were recovered and suspended in MH broth. Serial dilutions were spread on MH agar containing TMP and cefoperazone. Bacteria were grown for 72 h at 37 °C in microaerobic conditions and then counted to determine the cfu per gram of cecal contents.
In vitro invasion assays
Internalization of C. jejuni into T84 colonic epithelial cells was determined using a gentamicin-protection assay (Barrero-Tobon and Hendrixson, 2012). Semi-confluent monolayers of T84 cells (2.5 × 105 cells/ml) were seeded 24 h prior to infection using a 24-well tissue culture plates and DME/F12 (HyClone) with 5% FBS. Wild-type and mutant C. jejuni 81-176 SmR strains were suspended from plates in MH broth and diluted to an OD600 0.4. The culture was further diluted 1:10 in MH broth and this final dilution served as the inoculum. Prior to infection, media was removed from the T84 cells and 300 μl of tissue culture media were added back to the cells. Monolayers were then infected with 15 μl of each diluted bacterial culture (~3 × 106 cfu per monolayer). Serial dilutions of the inoculum were plated on MH agar to verify the actual number of bacteria used to infect each monolayer. Tissue culture plates were then centrifuged for 5 min at 960 rpm at room temperature to enhance contact between C. jejuni and colonic epithelial cells. The plates were then incubated for 4 h at 37 °C in a 5% CO2 incubator. T84 cells were washed three times with PBS and fresh tissue culture media containing 250 μg ml−1 of gentamicin was added to the monolayer. After a 2 h incubation at 37 °C in 5% CO2, cells were rinsed three times with PBS. Monolayers were released from the plates with 0.25% trypsin and cells were disrupted by repeated pipetting. Serial dilutions were then plated on MH agar. After incubation for 72 h at 37 °C in microaerobic conditions, the number of internalized bacteria were determined. Percent invasion was determined by dividing the number of internalized bacteria by the number of bacteria in the inoculum.
Electron microscopy analysis
C. jejuni strains were prepared as previously described for electron microscopy (Hendrixson, 2006). Briefly, bacteria were grown for 16 h then suspended from MH agar plates with PBS and diluted to an OD600 of 0.8. One milliliter of the diluted culture was then pelleted for 3 min at full speed in a microcentrifuge and the pellet was washed twice with PBS. The pellet was resuspended in a solution of 2% gluteraldehyde in 0.1 M cacodylate and incubated on ice for 1 h to allow the cells to be fixed. Cells were then stained with 2% uranyl acetate and visualized using a FEI Technai G2 Spirit BioTWIN transmission electron microscope.
Statistical analysis
Tests for statistical significance in invasion assays were conducted by using the Student’s t test (two-tailed distribution with two-sample, equal variance calculations). Statistically-significant differences between relevant strains possessed P-values < 0.05. For chick colonization assays, statistical analyses were performed by the Mann-Whitney U test, with statistically-significant differences between wild-type and mutant strains indicated with P-values < 0.01 or 0.05.
Supplementary Material
Acknowledgments
We thank Dr. Deborah Ribardo for assistance with chick colonization assays and Connor (CJ) Gulbronson for assistance with electron microscopy. This work was supported by NIH grants R01AI065539 and R21AI103643.
References
- Auvray F, Thomas J, Fraser GM, Hughes C. Flagellin polymerisation control by a cytosolic export chaperone. J Mol Biol. 2001;308:221–229. doi: 10.1006/jmbi.2001.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban M, Hendrixson DR. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog. 2011;7:e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, Sinning I. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc Natl Acad Sci U S A. 2010;107:11295–11300. doi: 10.1073/pnas.1001383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero-Tobon AM, Hendrixson DR. Identification and analysis of flagellar coexpressed determinants (Feds) of Campylobacter jejuni involved in colonization. Mol Microbiol. 2012;84:352–369. doi: 10.1111/j.1365-2958.2012.08027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JC, Thomas J, Fraser GM, Hughes C. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol Microbiol. 2001;39:781–791. doi: 10.1046/j.1365-2958.2001.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtalan SC, Phillips RM, Ghosh P. Three-dimensional secretion signals in chaperone-effector complexes of bacterial pathogens. Mol Cell. 2002;9:971–980. doi: 10.1016/s1097-2765(02)00529-4. [DOI] [PubMed] [Google Scholar]
- Black RE, Levine MM, Clements ML, Hughes TP, Blaser MJ. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- Boll JM, Hendrixson DR. A specificity determinant for phosphorylation in a response regulator prevents in vivo cross-talk and modification by acetyl phosphate. Proc Natl Acad Sci U S A. 2011;108:20160–20165. doi: 10.1073/pnas.1113013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow DR, Christensen JE, Neal-McKinney JM, Konkel ME. Campylobacter jejuni survival within human epithelial cells is enhanced by the secreted protein CiaI. Mol Microbiol. 2011;80:1296–1312. doi: 10.1111/j.1365-2958.2011.07645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner D. Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant- and animal-pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. doi: 10.1128/MMBR.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo CD, Taboada E, Nash JH, Lanthier P, Kelly J, Lau PC, Verhulp R, Mykytczuk O, Sy J, Findlay WA, Amoako K, Gomis S, Willson P, Austin JW, Potter A, Babiuk L, Allan B, Szymanski CM. Genome-wide expression analyses of Campylobacter jejuni NCTC11168 reveals coordinate regulation of motility and virulence by flhA. J Biol Chem. 2004;279:20327–20338. doi: 10.1074/jbc.M401134200. [DOI] [PubMed] [Google Scholar]
- Chen S, Beeby M, Murphy GE, Leadbetter JR, Hendrixson DR, Briegel A, Li Z, Shi J, Tocheva EI, Muller A, Dobro MJ, Jensen GJ. Structural diversity of bacterial flagellar motors. Embo J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6:455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JE, Pacheco SA, Konkel ME. Identification of a Campylobacter jejuni-secreted protein required for maximal invasion of host cells. Mol Microbiol. 2009;73:650–662. doi: 10.1111/j.1365-2958.2009.06797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claret L, Calder SR, Higgins M, Hughes C. Oligomerization and activation of the FliI ATPase central to bacterial flagellum assembly. Mol Microbiol. 2003;48:1349–1355. doi: 10.1046/j.1365-2958.2003.03506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa SC, Schmitz AM, Jahufar FF, Boyd JD, Cho MY, Glicksman MA, Lesser CF. A new means to identify type 3 secreted effectors: functionally interchangeable class IB chaperones recognize a conserved sequence. mBio. 2012;3:e00243–11. doi: 10.1128/mBio.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobo J, Varga J, Sajo R, Vegh BM, Gal P, Zavodszky P, Vonderviszt F. Application of a short, disordered N-terminal flagellin segment, a fully functional flagellar type III export signal, to expression of secreted proteins. Appl Environ Microbiol. 2010;76:891–899. doi: 10.1128/AEM.00858-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Hirano T, Su Y, Paul K, Wee DH, Mizuno S, Aizawa S, Hughes KT. The role of the FliK molecular ruler in hook-length control in Salmonella enterica. Mol Microbiol. 2010;75:1272–1284. doi: 10.1111/j.1365-2958.2010.07050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Singer HM, Wee DH, Keener JP, Hughes KT. An infrequent molecular ruler controls flagellar hook length in Salmonella enterica. EMBO J. 2011;30:2948–2961. doi: 10.1038/emboj.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LD, Poulter S, Terentjev EM, Hughes C, Fraser GM. A chain mechanism for flagellum growth. Nature. 2013;504:287–290. doi: 10.1038/nature12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans LD, Stafford GP, Ahmed S, Fraser GM, Hughes C. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc Natl Acad Sci U S A. 2006;103:17474–17479. doi: 10.1073/pnas.0605197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov AG, Phan J, Tropea JE, Routzahn KM, Peters HK, Pokross M, Waugh DS. Similar modes of polypeptide recognition by export chaperones in flagellar biosynthesis and type III secretion. Nat Struct Biol. 2003;10:789–793. doi: 10.1038/nsb982. [DOI] [PubMed] [Google Scholar]
- Ferris HU, Furukawa Y, Minamino T, Kroetz MB, Kihara M, Namba K, Macnab RM. FlhB regulates ordered export of flagellar components via autocleavage mechanism. J Biol Chem. 2005;280:41236–41242. doi: 10.1074/jbc.M509438200. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Bennett JC, Hughes C. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol Microbiol. 1999;32:569–580. doi: 10.1046/j.1365-2958.1999.01372.x. [DOI] [PubMed] [Google Scholar]
- Fraser GM, Hirano T, Ferris HU, Devgan LL, Kihara M, Macnab RM. Substrate specificity of type III flagellar protein export in Salmonella is controlled by subdomain interactions in FlhB. Mol Microbiol. 2003;48:1043–1057. doi: 10.1046/j.1365-2958.2003.03487.x. [DOI] [PubMed] [Google Scholar]
- Golden NJ, Acheson DW. Identification of motility and autoagglutination Campylobacter jejuni mutants by random transposon mutagenesis. Infect Immun. 2002;70:1761–1771. doi: 10.1128/IAI.70.4.1761-1771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Ewing CP, Lorenzo M, Pattarini D, Majam G, Guerry P. A σ28-regulated nonflagella gene contributes to virulence of Campylobacter jejuni 81-176. Infect Immun. 2006;74:769–772. doi: 10.1128/IAI.74.1.769-772.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol Microbiol. 2003;50:659–671. doi: 10.1046/j.1365-2958.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- Grant CCR, Konkel ME, Cieplak W, Jr, Tompkins LS. Role of flagella in adherence, internalization, and translocation of Campylobacter jejuni in nonpolarized and polarized epithelial cell cultures. Infect Immun. 1993;61:1764–1771. doi: 10.1128/iai.61.5.1764-1771.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR. A phase-variable mechanism controlling the Campylobacter jejuni FlgR response regulator influences commensalism. Mol Microbiol. 2006;61:1646–1659. doi: 10.1111/j.1365-2958.2006.05336.x. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR. Restoration of flagellar biosynthesis by varied mutational events in Campylobacter jejuni. Mol Microbiol. 2008;70:519–536. doi: 10.1111/j.1365-2958.2008.06428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrixson DR, Akerley BJ, DiRita VJ. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol. 2001;40:214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, V, DiRita J. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol. 2003;50:687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- Hendrixson DR, V, DiRita J. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol. 2004;52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- Higuchi R. Recombinant PCR. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: a Guide to Methods and Applications. London: Academic Press; 1990. pp. 77–183. [Google Scholar]
- Homma M, Fujita H, Yamaguchi S, Iino T. Excretion of unassembled flagellin by Salmonella typhimurium mutants deficient in hook-associated proteins. J Bacteriol. 1984;159:1056–1059. doi: 10.1128/jb.159.3.1056-1059.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara M, Minamino T, Yamaguchi S, Macnab RM. Intergenic suppression between the flagellar MS ring protein FliF of Salmonella and FlhA, a membrane component of its export apparatus. J Bacteriol. 2001;183:1655–1662. doi: 10.1128/JB.183.5.1655-1662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel ME, Kim BJ, Rivera-Amill V, Garvis SG. Bacterial secreted proteins are required for the internaliztion of Campylobacter jejuni into cultured mammalian cells. Mol Microbiol. 1999;32:691–701. doi: 10.1046/j.1365-2958.1999.01376.x. [DOI] [PubMed] [Google Scholar]
- Konkel ME, Klena JD, Rivera-Amill V, Monteville MR, Biswas D, Raphael B, Mickelson J. Secretion of virulence proteins from Campylobacter jejuni is dependent on a functional flagellar export apparatus. J Bacteriol. 2004;186:3296–3303. doi: 10.1128/JB.186.11.3296-3303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152:592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- Kuwajima G, Kawagishi I, Homma M, Asaka J, Kondo E, Macnab RM. Export of an N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci U S A. 1989;86:4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LH, Burg E, 3rd, Baqar S, Bourgeois AL, Burr DH, Ewing CP, Trust TJ, Guerry P. Evaluation of a truncated recombinant flagellin subunit vaccine against Campylobacter jejuni. Infect Immun. 1999;67:5799–5805. doi: 10.1128/iai.67.11.5799-5805.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Sourjik V. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol. 2011;80:886–899. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- Lilic M, Vujanac M, Stebbins CE. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol Cell. 2006;21:653–664. doi: 10.1016/j.molcel.2006.01.026. [DOI] [PubMed] [Google Scholar]
- Liu X, Gao B, Novik V, Galan JE. Quantitative proteomics of intracellular Campylobacter jejuni reveals metabolic reprogramming. PLoS Pathog. 2012;8:e1002562. doi: 10.1371/journal.ppat.1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macnab RM. How bacteria assemble flagella. Annu Rev Microbiol. 2003;57:77–100. doi: 10.1146/annurev.micro.57.030502.090832. [DOI] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- Malik-Kale P, Parker CT, Konkel ME. Culture of Campylobacter jejuni with sodium deoxycholate induces virulence gene expression. J Bacteriol. 2008;190:2286–2297. doi: 10.1128/JB.01736-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Chu R, Yamaguchi S, Macnab RM. Role of FliJ in flagellar protein export in Salmonella. J Bacteriol. 2000;182:4207–4215. doi: 10.1128/jb.182.15.4207-4215.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Imada K, Namba K. Mechanisms of type III protein export for bacterial flagellar assembly. Mol bioSystems. 2008;4:1105–1115. doi: 10.1039/b808065h. [DOI] [PubMed] [Google Scholar]
- Minamino T, Moriya N, Hirano T, Hughes KT, Namba K. Interaction of FliK with the bacterial flagellar hook is required for efficient export specificity switching. Mol Microbiol. 2009;74:239–251. doi: 10.1111/j.1365-2958.2009.06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T, Namba K. Self-assembly and type III protein export of the bacterial flagellum. J Mol Microbiol Biotechnol. 2004;7:5–17. doi: 10.1159/000077865. [DOI] [PubMed] [Google Scholar]
- Morimoto YV, Ito M, Hiraoka KD, Che YS, Bai F, Kami-Ike N, Namba K, Minamino T. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol. 2014;91:1214–1226. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Babitzke P, Kearns DB. FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J Bacteriol. 2013;195:297–306. doi: 10.1128/JB.01654-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol Microbiol. 2011;82:447–461. doi: 10.1111/j.1365-2958.2011.07822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskotal A, Kiraly R, Sebestyen A, Gugolya Z, Vegh BM, Vonderviszt F. Interaction of FliS flagellar chaperone with flagellin. FEBS Lett. 2006;580:3916–3920. doi: 10.1016/j.febslet.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Nachamkin I, Yang XH, Stern NJ. Role of Campylobacter jejuni flagella as colonization factors for three-day-old chicks: analysis with flagellar mutants. Appl Environ Microbiol. 1993;59:1269–1273. doi: 10.1128/aem.59.5.1269-1273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal-McKinney JM, Christensen JE, Konkel ME. Amino-terminal residues dictate the export efficiency of the Campylobacter jejuni filament proteins via the flagellum. Mol Microbiol. 2010;76:918–931. doi: 10.1111/j.1365-2958.2010.07144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal-McKinney JM, Konkel ME. The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front Cell Infect Microbiol. 2012;2:31. doi: 10.3389/fcimb.2012.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novik V, Hofreuter D, Galan JE. Identification of Campylobacter jejuni genes involved in its interaction with epithelial cells. Infect Immun. 2010;78:3540–3553. doi: 10.1128/IAI.00109-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuijten PJ, van Asten FJ, Gaastra W, van der Zeijst BA. Structural and functional analysis of two Campylobacter jejuni flagellin genes. J Biol Chem. 1990;265:17798–17804. [PubMed] [Google Scholar]
- Poly F, Ewing C, Goon S, Hickey TE, Rockabrand D, Majam G, Lee L, Phan J, Savarino NJ, Guerry P. Heterogeneity of a Campylobacter jejuni protein that is secreted through the flagellar filament. Infect Immun. 2007;75:3859–3867. doi: 10.1128/IAI.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribardo DA, Bingham-Ramos LK, Hendrixson DR. Functional analysis of the RdxA and RdxB nitroreductases of Campylobacter jejuni reveals that mutations in rdxA confer metronidazole resistance. J Bacteriol. 2010;192:1890–1901. doi: 10.1128/JB.01638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Amill V, Kim BJ, Seshu J, Konkel ME. Secretion of the virulence-associated Campylobacter invasion antigens from Campylobacter jejuni requires a stimulatory signal. J Infect Dis. 2001;183:1607–1616. doi: 10.1086/320704. [DOI] [PubMed] [Google Scholar]
- Samuelson DR, Eucker TP, Bell JA, Dybas L, Mansfield LS, Konkel ME. The Campylobacter jejuni CiaD effector protein activates MAP kinase signaling pathways and is required for the development of disease. Cell Commun Signal: CCS. 2013;11:79. doi: 10.1186/1478-811X-11-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson DR, Konkel ME. Serine phosphorylation of cortactin is required for maximal host cell invasion by Campylobacter jejuni. Cell Commun Signal: CCS. 2013;11:82. doi: 10.1186/1478-811X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiel DH, Wagar E, Karamanou L, Weeks D, Miller VL. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommerlad SM, Hendrixson DR. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol. 2007;189:179–186. doi: 10.1128/JB.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford GP, Evans LD, Krumscheid R, Dhillon P, Fraser GM, Hughes C. Sorting of early and late flagellar subunits after docking at the membrane ATPase of the type III export pathway. J Mol Biol. 2007;374:877–882. doi: 10.1016/j.jmb.2007.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. Maintenance of an unfolded polypeptide by a cognate chaperone in bacterial type III secretion. Nature. 2001;414:77–81. doi: 10.1038/35102073. [DOI] [PubMed] [Google Scholar]
- Thomas J, Stafford GP, Hughes C. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc Natl Acad Sci U S A. 2004;101:3945–3950. doi: 10.1073/pnas.0307223101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titz B, Rajagopala SV, Ester C, Hauser R, Uetz P. Novel conserved assembly factor of the bacterial flagellum. J Bacteriol. 2006;188:7700–7706. doi: 10.1128/JB.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spreeuwel JP, Duursma GC, Meijer CJ, Bax R, Rosekrans PC, Lindeman J. Campylobacter colitis: histological immunohistochemical and ultrastructural findings. Gut. 1985;26:945–951. doi: 10.1136/gut.26.9.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegh BM, Gal P, Dobo J, Zavodszky P, Vonderviszt F. Localization of the flagellum-specific secretion signal in Salmonella flagellin. Biochem Biophys Res Commun. 2006;345:93–98. doi: 10.1016/j.bbrc.2006.04.055. [DOI] [PubMed] [Google Scholar]
- Warren SM, Young GM. An amino-terminal secretion signal is required for YplA export by the Ysa, Ysc, and flagellar type III secretion systems of Yersinia enterocolitica biovar 1B. J Bacteriol. 2005;187:6075–6083. doi: 10.1128/JB.187.17.6075-6083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, Bleumink-Pluym NMC, Newell DG, Nuijten PJ, van der Zeijst BAM. Differential flagellin expression in a flaA flaB+ mutant of Campylobacter jejuni. Infect Immun. 1994;62:3901–3906. doi: 10.1128/iai.62.9.3901-3906.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, Bleumink-Pluym NMC, van der Zeijst BAM. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. Embo J. 1991;10:2055–2061. doi: 10.1002/j.1460-2075.1991.tb07736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TM, van der Zeijst BAM, Ayling R, Newell DG. Colonization of chicks by motility mutants of Campylobacter jejuni demonstrates the importance of flagellin A expression. J Gen Microbiol. 1993;139:1171–1175. doi: 10.1099/00221287-139-6-1171. [DOI] [PubMed] [Google Scholar]
- Watson RO, Galan JE. Campylobacter jejuni survives within epithelial cells by avoiding delivery to lysosomes. PLoS Pathog. 2008;4:e14. doi: 10.1371/journal.ppat.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wosten MMSM, Wagenaar JA, van Putten JPM. The FlgS/FlgR two-component signal transduction system regulates the fla regulon in Campylobacter jejuni. J Biol Chem. 2004;279:16214–16222. doi: 10.1074/jbc.M400357200. [DOI] [PubMed] [Google Scholar]
- Yao R, Burr DH, Doig P, Trust TJ, Niu H, Guerry P. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol Microbiol. 1994;14:883–893. doi: 10.1111/j.1365-2958.1994.tb01324.x. [DOI] [PubMed] [Google Scholar]
- Yokoseki T, Kutsukake K, Ohnishi K, Iino T. Functional analysis of the flagellar genes in the fliD operon of Salmonella typhimurium. Microbiology. 1995;141(Pt 7):1715–1722. doi: 10.1099/13500872-141-7-1715. [DOI] [PubMed] [Google Scholar]
- Young BM, Young GM. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J Bacteriol. 2002;184:1324–1334. doi: 10.1128/JB.184.5.1324-1334.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young GM, Schmiel DH, Miller VL. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci U S A. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.