Abstract
In response to acute myocardial infarction (MI), a complex series of cellular and molecular signaling events orchestrate the myocardial remodeling that ensues weeks to months after injury. Clinical, epidemiological, and pathological studies demonstrate that inadequate or impaired angiogenesis after myocardial injury is often associated with decreased left ventricular (LV) function and clinical outcomes. The microRNA family, miR-26, plays diverse roles in regulating key aspects of cellular growth, development, and activation. Recent evidence supports a central role for the miR-26 family in cardiovascular disease by controlling critical signaling pathways, such as BMP/SMAD1 signaling, and targets relevant to endothelial cell growth, angiogenesis, and LV function post-MI. Emerging studies of the miR-26 family in other cell types including vascular smooth muscle cells, cardiac fibroblasts, and cardiomyocytes suggest that miR-26 may bear important implications for a range of cardiovascular repair mechanisms. This review examines the current knowledge of the miR-26 family’s role in key cell types that critically control cardiovascular disease under pathological and physiological stimuli
Keywords: MicroRNA-26, Endothelial cells, Cardiac Myocytes, Fibroblasts, Vascular Smooth Muscle Cells
1. Introduction
Cardiovascular disease (CVD) and its complications including myocardial infarction (MI), stroke, and peripheral artery disease, are the leading cause of morbidity and mortality in Western Societies (WHO. Causes of Death 2008 Summary Tables. Geneva, Switzerland: World Health Organization; 2008). Patients experiencing a first MI are at a significantly higher risk of future cardiovascular events (Eapen et al., 2012; Goldstein et al., 2000; Milonas et al., 2010). Accumulating evidence indicates that impaired remodeling post-MI may also predispose to decreased or altered left ventricular (LV) function and consequently heart failure. Although substantial improvements have been made in the treatment of patients post-MI, including risk factor modification and pharmacologic therapies, significant residual cardiovascular risk remains and the mechanisms governing post-MI events are still poorly defined (Cohn et al., 2000; Frangogiannis, 2014; Pfeffer et al., 1985; Seropian et al., 2014).
MicroRNAs (miRNAs) are a class of small, evolutionarily conserved, 18–22 nucleotide long, non-coding single-stranded RNA molecules. They are important regulators of gene expression at the post-transcriptional level by inhibiting mRNA translation and/or promoting mRNA degradation. MiRNAs have been found to regulate various physiological and pathological processes involved in CVD (Quiat and Olson, 2013) such as, miR-143/145 in vascular injury and hypertension(Boettger et al., 2009; Xin et al., 2009), miR-21, miR-1, miR-133, miR-199, and miR-208a in cardiac hypertrophy(Callis et al., 2009; Montgomery et al., 2011; Thum et al., 2007; van Rooij et al., 2009; van Rooij et al., 2006; van Rooij et al., 2007), and miR-214, miR-499, and miR-92a in myocardial ischemia (Dorn et al., 2012; Icli et al., 2013; Roy et al., 2009; Shieh et al., 2011a). However, the role of miRNAs in post-MI repair mechanisms is not well-defined. In this review, we summarize the emerging roles of the miR-26 family members and their targets in a range of cell types important to post-MI repair mechanisms as well as other CVD states (Table 1).
Table 1.
Targets of the miR-26 family. Known cell type-specific targets of the miR-26 family associated with cardiovascular disease.
| MiR-26 family member | Cell type | Targets | Functions | References |
|---|---|---|---|---|
| miR-26a | EC | SMAD1 | Targets a BMP/SMAD1-Id1-p21WAF/CIP1/p27 signaling axis to promote an anti-angiogenic program in ECs. Regulates pathological and physiological angiogenesis in vivo. MiR-26a neutralization increases angiogenesis, decreases infarct size, and improves heart function post-MI. |
Icli, B. et al., 2013 |
| miR-26a/b | CM | GSK3β KCNJ2 |
Inhibits Ang-II induced expression of ANF and β-MHC in CMs in vitro. Overexpression of miR-26a suppresses KCNJ2 and KIR2.1 expression and reduces AF in response to an atrial tachypacing model in-vivo. |
Zhang, Z.H. et al., 2013 Luo, X. et al., 2013 |
| miR-26b | CM | PLCβ1 | In response to transverse aortic constriction (TAC), overexpression of cardiac-specific miR-26b increases LV wall thickness, but no effect on LV ejection fraction. Neutralization of miR-26a and miR-26b had no effect on TAC-induced LV hypertrophy or function. |
Han, M. et al., 2012 |
| miR-26a | SMC | SMAD1/SMAD4 | Inhibits VSMC cell differentiation and apoptosis in vitro, implicating miR-26a in VSMC phenotypic phenotypic switching. Expression is reduced in aortic aneurysms in mice at time point associated with VSMC de-differentiation. |
Dey, B.K. et al., 2012 |
| miR-26a | FB | CTGF Collagen I | Expression of miR-26a is reduced in response to Ang-II. Overexpression of miR-26a inhibits NF-κB activity. Decreased miR-26a expression in patients with AS. May regulate calcification related genes such as BMP2, SMAD1 and ALPL. Neutralization of miR-26a in acute MI resulted in improved LV function and reduced myocardial fibrosis (Figure 3). |
Wei, C. et al., 2013 Nigam, V. et al., 2010 |
SMAD1= mothers against decapentaplegic homolog 1; ID1= inhibitor of DNA binding 1, p21WAF1/CIP1= cyclin dependent kinase inhibitor 1A; p27=cyclin dependent kinase inhibitor 1B; GSK3β=glycogen synthase kinase 3 beta; Kir2.1=potassium inwardly-rectifying channel subfamily J; GATA4=GATA binding protein 4; PLCβ1= phospholipase C beta; SMAD4= mothers against decapentaplegic homolog 4; Ang-II, angiotensin II; CTGF=connective tissue growth factor; BMP2=bone morphogenetic protein 2; ALPL=alkaline phosphatase liver/bone/kidney; EC, endothelial cells; CM, cardiomyocytes; SMC, vascular smooth muscle cells; FB, fibroblasts.
2. Genomic location of MiR-26 family members
Currently, there are more than 2,000 mature miRNAs in the human genome (http://www.mirbase.org). About 40% of the miRNAs are found between independent transcription units (intergenic), or in the intronic sequences of protein-coding genes and intronic/exonic regions of noncoding RNAs (intronic). (Rodriguez et al., 2004; Saini et al., 2007) Intergenic miRNAs genes have their own promoters and terminators, while the majority of intronic miRNAs share the same transcription elements as their host genes.
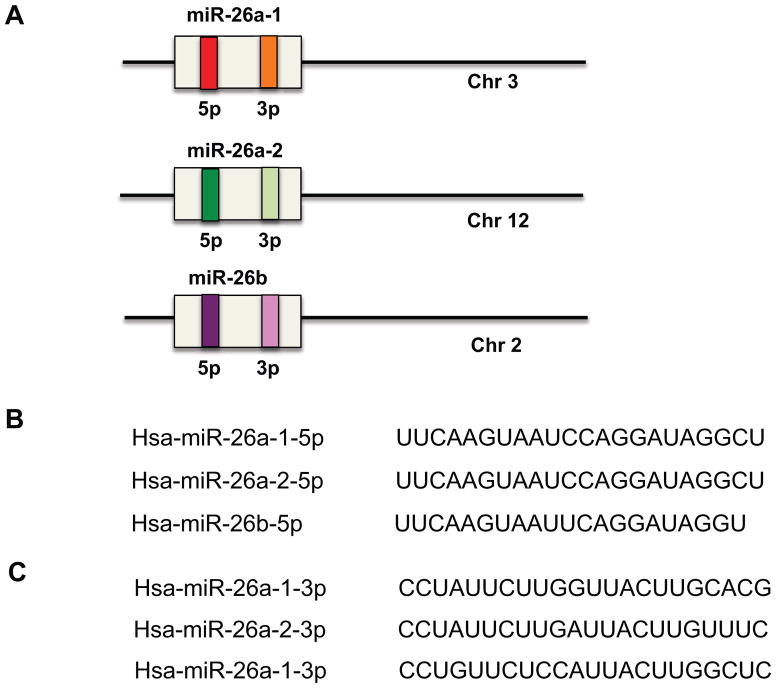
The human and mouse miR-26 family constitutes miR-26a-1, miR-26a-2, and miR-26b. MiR-26a-1 is localized at chromosome 3, miR-26a-2 is localized at chromosome 12, and miR-26b is localized at chromosome 2. The mature miRNA for miR-26a-1 and miR-26a-2 have the same sequence, which only differs from the mature miR-26b sequence by two nucleotides (Figure 1).
Figure 1. MiR-26 family members and their genomic locations.
(A) MiR-26a-1 is localized on chromosome 3, miR-26a-2 is localized on chromosome 12, and miR-26b is localized on chromosome 2. The mature miRNA for miR-26a-1 and miR-26a-2 have the same sequence which differ by 2 nucleotides from the mature miR-26b sequence. (B) Mature sequences of miR-26 family members that arise from the 5′ arm of the precursors. (C) Mature sequences of miR-26 family members that arise from the 3′ arm of the precursors.
MiR-26 family members are embedded within the introns of genes encoding for the proteins of carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase (CTDSP) family, which includes CTDSPL, CTDSP2, and CTDSP1. They can negatively regulate RNA polymerase II (RNAPII) by dephosphorylating its CTD on Ser-5 in vitro and function as transcriptional co-repressors that inhibit the transcription of neuronal genes in non-neuronal cells.(Yeo et al., 2005) CTDSP family can also act as phosphatases for SMAD1 and SMAD2/3 and snail (Sapkota et al., 2006; Wu et al., 2009). Under physiological conditions, miR-26 family and their host genes are expressed concurrently. In addition, miR-26 family and their host genes act synergistically to block G1/S phase transition in cancer cell lines derived from liver, lung, breast, and cervix and activate the Rb protein. Furthermore, c-Myc, the cell cycle progression gene, decreases both miR-26 and CTDSP families in tumor cell lines. (Zhu et al., 2012)
3. MiR-26 in Endothelial Cell Biology
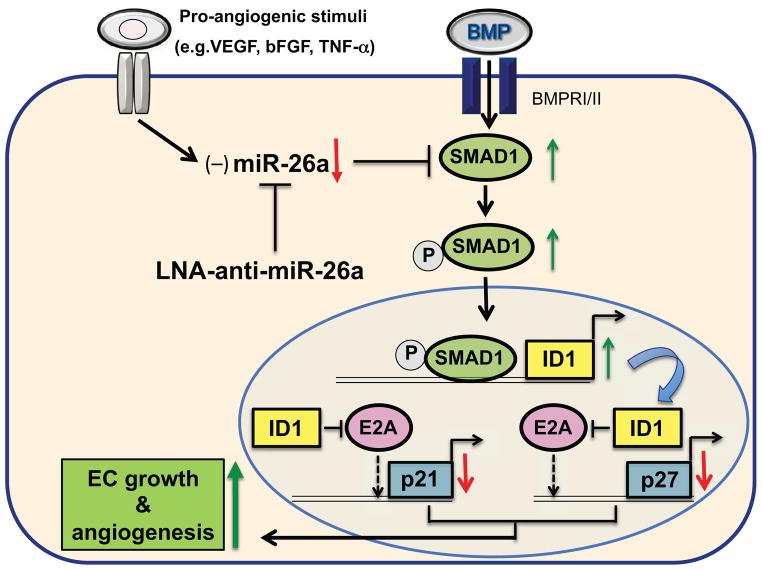
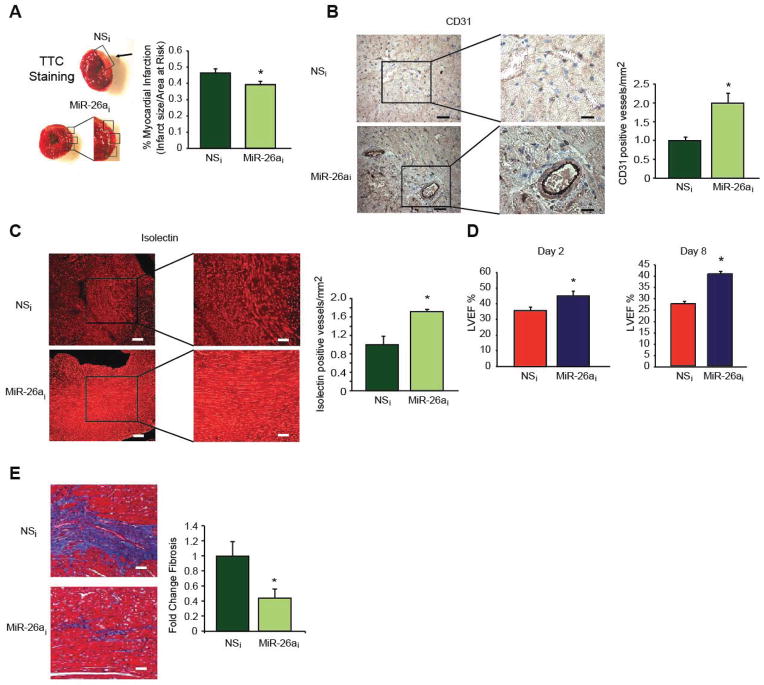
The role of miR-26a in endothelial cell biology has recently been studied in the context of angiogenesis.(Icli et al., 2013) Endothelial cells (ECs) play an important role in angiogenesis, where there is fine balance between pro- and anti-angiogenic factors. In response to pro-angiogenic stimuli, vascular ECs need to be rapidly activated to migrate to distant sites and proliferate to form new primary capillaries from existing ones.(Potente et al., 2011) Impaired EC angiogenic responses have been linked to the exacerbation of a wide range of disease states including poor cardiovascular function and outcomes.(Potente et al., 2011; Wu et al., 1998) diabetic wound healing(Falanga, 2005), and neurodegenerative disorders.(Zlokovic, 2011) We identified that miR-26a targets a SMAD1-Id1-p21WAF/CIP1/p27 signaling axis and inhibits angiogenesis in ECs (Figure 2). Our study identified that miR-26a expression is increased in response to acute MI in mice and in human subjects with acute coronary syndromes. In contrast, pro-angiogenic stimuli such as VEGF or TNF-a reduced miR-26a expression in ECs. Furthermore, after one hour after myocardial ischemic injury, miR-26a expression is highest in the ischemic zone compared to the non-ischemic regions of the heart.(Icli et al., 2013) Inhibition of miR-26a rapidly induced angiogenesis and reduced acute MI size with improved heart function in a mouse model of acute myocardial infarction (Figure 3).(Icli et al., 2013) In addition, overexpression of miR-26a adversely affected physiological angiogenesis by impairing the formation of the caudal vein plexus (CVP), a BMP-responsive region in zebrafish. Furthermore, miR-26a overexpression blocked exercise-induced angiogenesis in skeletal muscle in mice.(Icli et al., 2013) Taken together, these findings provide cogent evidence that miR-26a serves an anti-angiogenic role in response to pathophysiological stimuli and that neutralization of miR-26a can markedly improve moyocardial angiogenesis and LV repair.
Figure 2. Mechanism leading to increased endothelial cell growth and angiogenesis.
MiR-26a expression is decreased by pro-angiogenic stimuli such as VEGF, bFGF and TNF-α in endothelial cells (ECs). MiR-26a inhibits SMAD1 by binding to its 3’-UTR, an effect that decreases ID1 and increases the expression of cell growth arrest proteins p21WAF1/CIP1 and p27 resulting in decreased endothelial cell growth and angiogenesis. Neutralization of miR-26a using LNA-antimiR-26a relieves the miR-26a-mediated repression of BMP/SMAD1 signaling and increases EC growth and angiogenesis.
Figure 3. Inhibition of miR-26a increases angiogenesis, decreases infarct size, and improves LV function and myocardial fibrosis in a mouse model of acute MI.
After a single tail vein injection in mice of LNA-anti-miR-26a (MiR-26ai) (24mg/kg) or scrambled non-specific control LNA-antimiRs (NSi) (n = 11–12 per group) on day 0, mice underwent acute myocardial infarction consisting of 45 minutes of ischemia and reperfusion of the left anterior descending artery (LAD) and infusion of fluorescent microbubbles on day 1. (A) TTC staining (top) demonstrates areas of infarct in the left ventricle. Myocardial infarction size was normalized to the area at risk. *P < 0.05 compared to NSi Angiogenesis was quantified by CD31 (B) or isolectin staining (C) in sections from the entire left ventricle on day 2. *P < 0.05 compared to NSi Scale bars, 500μm (left) and 250μm (right) in (B) and 100μm (left) and 50μm (right) in (C). (D) Left ventricular ejection fraction was measured by echocardiography on days 2 and 8. *P < 0.05 compared to NSi (E) Fibrosis was measured by Masson-Trichrome staining on day 8. *P < 0.05 compared to NSi. Scale bar 250μm.
4. MiR-26 in Cardiomyocyte Biology
MiR-26 family is enriched in the heart but it is not cardiac specific.(Liang et al., 2007) The role of miR-26a in cardiac myocyte (CM) biology has been examined in the context of cardiac hypertrophy, oxidative stress, and atrial fibrillation. Cardiac hypertrophy is characterized by an increase in the size of individual cardiomyocytes leading to wall thickening of the LV.(Lorell and Carabello, 2000; Nishimura et al., 2003) This can occur in response to cardiac pressure overload or in response to primary myocyte pathology. MiR-26a expression is reduced in response to myocardial hypertrophy induced by transverse abdominal aortic constriction (TAAC) and in cardiomyocytes treated with angiotensin-II. (Wei et al., 2013; Zhang et al., 2013) Using a computational approach and transfection studies in HEK293T cells, Zhang et al demonstrated that miR-26a/b repressed the 3′-UTR of glycogen synthase kinase-3β (GSK3β) and reduced expression of ANF and β-MHC in CMs in vitro suggesting that GSK3β may be a relevant target for regulating cardiac hypertrophy. Future studies using relevant models will be required to assess whether miR-26 regulates LV hypertrophy in vivo via GSK3β.(Zhang et al., 2013)
Accumulating studies demonstrate that GATA4 mediates cardiomyocyte hypertrophy and gene expression.(Liang et al., 2001) Han et al.(Han et al., 2012) demonstrated in a pressure overload induced mouse model of cardiac hypertrophy that induction of GATA4 expression is post-transcriptionally regulated by reduced expression of miR-26b. Using 3′-UTR reporter studies, they demonstrated that GATA4 and phospholipase C beta (PLCβ1) are both targets of miR-26b. In addition, targeting of PLCβ1 by miR-26b in turn inhibits miR-26b expression in a negative feedback mechanism. Interestingly, in response to transverse aortic constriction (TAC), cardiac-specific transgenic mice overexpressing miR-26b showed a modest reduction in wall thickness, but no differences in LV ejection fraction.(Han et al., 2012) While the cardiac hypertrophic markers atrial natriuretic factor (ANF), PLCβ1, cardiac ankrin repeat protein (CARP), and alpha-skeletal actin (αSkAc) were not changed in TAC-treated transgenic miR-26b hearts, GATA4 and β-myosin heavy chain (MHC) expression was reduced suggesting potentially differential targets by miR-26b in this model system.(Han et al., 2012) However, neutralization of miR-26a and miR-26b using LNA-anti-miRs had no effect on inducing TAC-mediated cardiac hypertrophy in vivo. These results suggest a potential role for overexpression of miR-26b in regulating cardiomyocyte hypertrophy in which overexpression of miR-26b attenuates the development of cardiac hypertrophy; however, inhibition of miR-26a and miR-26b is not sufficient to induce hypertrophy. These findings are complementary to our findings that revealed increased angiogenesis and LV function with neutralization of miR-26a in the context of an acute MI. Thus, miR-26 family members may have more dominant functional roles in response to specific pathophysiological stimuli in the heart.
The regulation of the GSK3β by miR-26a has also been studied in the presence of reactive oxygen species (ROS). In disease states such as myocardial ischemia, generation of ROS triggers CM death via apoptosis or necrosis resulting in an irreversible injury to the heart.(Hori and Nishida, 2009) In response to H2O2 miR-26a expression increased in CMs. Overexpression of miR-26a in the presence of H2O2 induced CM apoptosis, whereas inhibition of miR-26a had the opposite effect. In addition, miR-26a overexpression in CM’s decreased GSK3β protein expression suggesting a role for GSK3β in miR-26a-mediated apoptosis in cardiac myocytes.(Suh et al., 2012) These data suggest that neutralization of miR-26a may increase CM cell survival.
Atrial fibrillation (AF) afflicts more than two million patients in the US and is a major contributor to cardioembolic stroke and exacerbation of heart failure.(Nattel, 2002; Nattel et al., 2007) AF reflects alterations in ion channels resulting in atrial electrical remodeling and arrhythmia with increased expression, for example, of KCNJ2 mRNA and its encoded protein KIR2.1. (Atienza et al., 2006; Bosch et al., 1999; Cha et al., 2004; Dobrev et al., 2002; Gaborit et al., 2005; Nattel et al., 2007; Workman et al., 2001; Zhang et al., 2005)
Human subjects and canine models with AF exhibited reduced expression of both miR-26a and miR-26b in CMs. In addition, miR-26 targeted KCNJ2 with 2 putative miR-26 binding sites in its 3′UTR.(Luo et al., 2013) Overexpression of miR-26a in H9C2 rat ventricular cells reduced Kir2.1 expression. In addition, knockdown of miR-26a promoted AF, whereas overexpression of miR-26a significantly reduced AF incidence in a mouse model of AF where AF was induced by intracardiac pacing. Mechanistically, nuclear factor of activated T cells (NFAT), which is activated in response to AF, was found to be a negative upstream regulator of miR-26a in cardiac myocytes. Consistent with this premise, inhibition of NFAT activity decreased Kir2.1 thereby allowing miR-26a to exert its protective effect over AF in a canine model of AF where AF was induced up to 6 weeks of atrial tachypacing.(Luo et al., 2013)
In summary, miR-26a may exhibit distinct roles in the heart. On the one hand, its expression is increased early post-MI in the ischemic zone and neutralization of miR-26a improved LV function by increasing angiogenesis and decreasing cardiomyocyte apoptosis, and increasing SMAD1 expression. In contrast, in a atrial pacing model of AF, miR-26a overexpression confers favorable effects on Kir2.1 expression and repressed AF. Future studies will be required to examine if there is propensity for AF or other arrhythmias in pathophysiological models of myocardial ischemia or hypertrophy.
5. Role of MiR-26a in the Regulation of Smooth Muscle Biology
Vascular smooth muscle cells (VSMCs) play a crucial role in the pathogenesis of a variety of vascular injury disease states such as mechanical balloon injury, vein graft failure, abdominal aortic aneurysm (AAA), or atherosclerosis.(Curci, 2009; Gomez and Owens, 2012; Nguyen et al., 2013; Owens et al., 2004; Wan et al., 2012) VSMCs differ from the norm in their lack of terminal differentiation and their ability to assume multiple phenotypic profiles. This flexibility and diversity maintains homeostatic function and allows for rapid phenotypic switching in response to injury.(Rensen et al., 2007)
A recent study highlights a participatory role for miR-26a in VSMC phenotypic switching (Leeper et al., 2011). MiR-26a expression was significantly induced in VSMC differentiation in response to serum starvation. However, knockdown of miR-26a actually accelerated VSMC differentiation.(Leeper and Cooke, 2011) Conversely, overexpression of miR-26a reduced VSMC differentiation. MiR-26a was hypothesized to serve as an inhibitor of VSMC differentiation through a compensatory negative feedback mechanism.(Leeper et al., 2011) Indeed, VSMCs deficient in miR-26a were able to migrate less effectively towards a growth factor/serum gradient as compared to miR control VSMCs. In addition, VSMCs deficient in miR-26a displayed larger rates of apoptosis.(Leeper et al., 2011)
Given the role of TGF-β1 in inducing VSCMC phenotype cell switching, it was hypothesized that members of this signaling pathway may be targets of miR-26a. Consistent with this notion, VSMCs deficient in miR-26a expressed higher levels of SMAD1 and SMAD4 than control VSMCs.(Leeper et al., 2011) Conversely, VSMCs with miR-26a overexpression exhibited reduced expression for SMAD1 and SMAD4 (Leeper et al., 2011) Finally, in aortic aneurysm models in mice, expression of miR-26a was reduced at a time point when VSMC de-differentiation is pronounced. Interestingly, in the context of post-MI ischemic injury and neutralization of miR-26a, our studies found no differences in the expression of SM-α-actin expression in hearts compared to controls.(Icli et al., 2013) Future studies will need to define whether miR-26a directly targets these SMADs in VSMCs and whether miR-26a is necessary and sufficient to regulate the pathogenesis of AAA disease in vivo.
Taken together, miR-26a may be an important homeostatic regulator of VSMC phenotype by suppressing VSMC differentiation. Manipulation of miR-26a expression may thus have therapeutic application for AAA disease or mechanical vascular injury.
6. Role of MiR-26a in Cardiac Fibroblast Biology
Cardiac fibroblasts are the primary source of the synthesis of collagens, elastin, matrix metalloproteinases (MMP), fibronectin, and connective tissue growth factor (CTGF) expression in the heart and regulate the synthesis of the cardiac extracellular matrix (ECM).(Shieh et al., 2011b) Normal contractile function of the heart and cardiac relaxation rely on a balance between the contractile elements and the composition and structure of the ECM. An imbalance in the heart favoring the components of the ECM over the contractile elements results in cardiac fibrosis, an effect that may lead to cardiac stiffness, diastolic dysfunction, and arrhythmia. Therefore, strategies to prevent fibrosis are important for the management of cardiac dysfunction. MiRNAs have been shown to play a role in each of these cardiac pathologies and serve as potential therapeutic targets.(Divakaran and Mann, 2008)
Cardiac fibrosis may be induced by profibrotic factors such as TGF β,(Leask and Abraham, 2004) Angiotensin II (AngII),(Weber and Brilla, 1991) and NF-κB.(Kumar et al., 2011; Wei et al., 2013) The effects of miRNAs on cardiac fibrosis and relation to Ang-II have not been extensively studied until recently.(Adam et al., 2012; Castoldi et al., 2012; Thum and Lorenzen, 2012; Wang et al., 2014; Wei et al., 2013) One recent study by Wei et al. identified that miR-26a expression in mouse hearts is reduced in response to TAC at a time point that collagen I and CTGF is increased within cardiac fibroblasts.(Wei et al., 2013) In addition, expression of miR-26a was significantly reduced in Ang-II stimulated neonatal cardiac fibroblasts, an effect that was rescued using the NF-κB inhibitors, SN50 or a dominant-negative IκBα. Indeed, overexpression of miR-26a in neonatal cardiac fibroblasts suppressed CTGF and collagen I gene expression in the presence of Ang-II, and miR-26a suppressed the 3′-UTRs of these proteins in HEK293 cells.(Wei et al., 2013) The authors proposed a negative feedback regulation of NF-κB signaling by miR-26a: overexpression of miR-26a suppresses NF-κB activity via targeting of collagen I and CTGF genes. Ang-II leads to overexpression of NF-κB, which in turn, reduces miR-26a levels, in a potential feedback loop. Furthermore, Wei et al. explored the association of miR-26a and NF-κB in response to TAC. Using a TAC model in transgenic mice overexpressing a dominant-negative IκBα which is resistant to NF-κB activation they found that miR-26a expression was not repressed, suggesting the therapeutic potential for targeting miR-26a.(Wei et al., 2013) Interestingly, neutralization of miR-26a in an acute post-MI model was associated with increased Smad1 expression, angiogenesis, and decreased fibrosis (Icli et al., 2013) (Figure 3). Future studies will have to formally evaluate the in vivo role of miR-26a on cardiac fibrosis in pathophysiological models of LV remodeling and hypertrophy.
Finally, bicuspid aortic valve (BAV) is one of the major causes of aortic stenosis (AS) and aortic insufficiency (AI). AS is typically caused by calcific valve disease. Overexpression of miR-26a in cultured human aortic valve interstitial cells (AVICs), a fibroblast-like cell, resulted in reduced expression of calcification-related genes, including BMP2, SMAD1, and alkaline phosphatase (ALPL) expression. In patients with AS, miR-26a expression was also decreased compared to the patients with AI suggesting a role for miR-26a in the regulation of calcification-related genes.(Nigam et al., 2010) Thus, miR-26a may have differential functional effects on growth depending upon specific fibroblast subsets.
7. Conclusions and perspectives
The studies discussed above implicate critical roles for miR-26 family members in CVD. The emerging role for miR-26 family members especially in the regulation of EC-driven angiogenesis and VSMC differentiation indicate that miR-26 will likely have a central role in disease states such as post-acute and chronic MI and diabetic wound healing in which both angiogenesis and fibrosis contribute prominently to repair mechanisms, and vascular mechanical injury where SMC phenotype switching governs the response to injury. These observations also underscore several important and unresolved issues. For example, given the anti-angiogenic function of miR-26a in ECs (Icli et al., 2013) and its role in VSMC differentiation (Leeper et al., 2011), future studies will be required to identify upstream mechanisms governing the expression of miR-26a in ECs and in VSMCs in response to pathophysiological stimuli and pharmacological agents. By studying the putative ‘promoter’ regions of miR-26a and other family members, we may gain insight into how miR-26a expression itself is regulated at the transcriptional level. In addition, closer examination of genome wide association studies for identification of probable genetic variants in the promoter region of the miR-26 family may provide insight leading to its altered expression in CVD states.
The basis for cell-type specific expression of individual miR-26 family members and their ability to target specific genes also requires further investigation. For example, Kota et al. has reported in hepatoma cells that ectopic expression of miR-26a induces cell cycle arrest through directly targeting cyclins D2 and E2(Kota et al., 2009). In contrast, our study demonstrated that miR-26a overexpression in ECs targeted SMAD1 signaling and led to cell cycle arrest and decreased cell growth and angiogenesis in ECs. However this effect was independent of cyclins D2 and E2 and dependent on the induction of p21WAF1/CIP1 and p27. We should be mindful that ectopic expression of the same microRNA family member may result in different functional effects in transformed cells versus primary cells through the utilization of differentially expressed target genes. Indeed, an emerging paradigm has been appreciated for miRNA cell-specific function.(Sun et al., 2012; Sun et al., 2014)
Delivery of miRs or anti-miRs, including anti-miR-26a, to specific tissue or cell-specific niches remains an active area of investigation. As such, therapeutic targeting of miRNAs should consider: 1) the means of delivery; 2) the relevant miRNA-specific targets; and 3) the specific cell type to be targeted. In this context, while studies to date have used systemic approaches to achieve miR-26a inhibition in both cardiac and non-cardiac cell types, cell-type specific targeting may be considered depending on the relevant pathophysiologic process. For example, delivery of anti-miRs via the intravenous route may allow for enrichment of the anti-miR to the vasculature, thereby reducing off-target cell types as we reported using LNA-anti-miR-26a to enhance post-MI neovascularization.(Icli et al., 2013) Alternatively, “designer” miRNA therapeutics could be generated to target the vasculature using endothelial-specific peptides conjugated (e.g. using either cationic lipoparticles or nanoparticles) to miRNA mimics or anti-miRs. Indeed, liposomes incorporating an αvβ3-targeting cyclic RGD peptide was used to enrich vessel delivery of anti-miR-132 to regulate tumor angiogenesis.(Anand et al., 2010) Similar peptide-cell specific delivery strategies may be feasible for other relevant cell types in the cardiovascular system. Finally, the use of cell penetrating peptides (e.g. nanoparticles coated with penetratin) may be another promising method to enhance efficiency of miR or anti-miR cell delivery, albeit cell-specificity has not been verified using this technique.(Babar et al., 2012)
An insufficient blood supply of the heart muscle and hypoxia-related loss of the viable heart tissue is associated with considerable morbidity and mortality worldwide. By virtue of miR-26’s emerging roles in relevant cells types including ECs, VSMCs, CMs, and cardiac fibroblasts, it is likely that this miRNA will figure prominently in the pathogenesis and treatment for a range of CVD states.
Acknowledgments
Funding
This work was supported by funding from the National Institutes of Health (HL091076, HL115141, and HL117994) to M.W.F. an Institutional Ruth L. Kirschstein N.R.S.A. T32HL07604 to B.I.), and a Jonathan A. Levy Research Award (to M.W.F.).
Footnotes
Conflict of interest: none declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adam O, Lohfelm B, Thum T, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- Anand S, Majeti BK, Acevedo LM, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nature medicine. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atienza F, Almendral J, Moreno J, et al. Activation of inward rectifier potassium channels accelerates atrial fibrillation in humans: evidence for a reentrant mechanism. Circulation. 2006;114:2434–2442. doi: 10.1161/CIRCULATIONAHA.106.633735. [DOI] [PubMed] [Google Scholar]
- Babar IA, Cheng CJ, Booth CJ, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E1695–1704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. The Journal of clinical investigation. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch RF, Zeng X, Grammer JB, Popovic K, Mewis C, Kuhlkamp V. Ionic mechanisms of electrical remodeling in human atrial fibrillation. Cardiovascular research. 1999;44:121–131. doi: 10.1016/s0008-6363(99)00178-9. [DOI] [PubMed] [Google Scholar]
- Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. The Journal of clinical investigation. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi G, Di Gioia CR, Bombardi C, et al. MiR-133a regulates collagen 1A1: potential role of miR-133a in myocardial fibrosis in angiotensin II-dependent hypertension. J Cell Physiol. 2012;227:850–856. doi: 10.1002/jcp.22939. [DOI] [PubMed] [Google Scholar]
- Cha TJ, Ehrlich JR, Zhang L, Nattel S. Atrial ionic remodeling induced by atrial tachycardia in the presence of congestive heart failure. Circulation. 2004;110:1520–1526. doi: 10.1161/01.CIR.0000142052.03565.87. [DOI] [PubMed] [Google Scholar]
- Cohn JN, Ferrari R, Sharpe N. Cardiac remodeling--concepts and clinical implications: a consensus paper from an international forum on cardiac remodeling. Behalf of an International Forum on Cardiac Remodeling. J Am Coll Cardiol. 2000;35:569–582. doi: 10.1016/s0735-1097(99)00630-0. [DOI] [PubMed] [Google Scholar]
- Curci JA. Digging in the “soil” of the aorta to understand the growth of abdominal aortic aneurysms. Vascular. 2009;17(Suppl 1):S21–29. doi: 10.2310/6670.2008.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divakaran V, Mann DL. The emerging role of microRNAs in cardiac remodeling and heart failure. Circulation research. 2008;103:1072–1083. doi: 10.1161/CIRCRESAHA.108.183087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrev D, Wettwer E, Kortner A, Knaut M, Schuler S, Ravens U. Human inward rectifier potassium channels in chronic and postoperative atrial fibrillation. Cardiovascular research. 2002;54:397–404. doi: 10.1016/s0008-6363(01)00555-7. [DOI] [PubMed] [Google Scholar]
- Dorn GW, 2nd, Matkovich SJ, Eschenbacher WH, Zhang Y. A human 3′ miR-499 mutation alters cardiac mRNA targeting and function. Circulation research. 2012;110:958–967. doi: 10.1161/CIRCRESAHA.111.260752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen ZJ, Tang WH, Felker GM, et al. Defining heart failure end points in ST-segment elevation myocardial infarction trials: integrating past experiences to chart a path forward. Circ Cardiovasc Qual Outcomes. 2012;5:594–600. doi: 10.1161/CIRCOUTCOMES.112.966150. [DOI] [PubMed] [Google Scholar]
- Falanga V. Wound healing and its impairment in the diabetic foot. Lancet. 2005;366:1736–1743. doi: 10.1016/S0140-6736(05)67700-8. [DOI] [PubMed] [Google Scholar]
- Frangogiannis NG. The immune system and the remodeling infarcted heart: cell biological insights and therapeutic opportunities. J Cardiovasc Pharmacol. 2014;63:185–195. doi: 10.1097/FJC.0000000000000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborit N, Steenman M, Lamirault G, et al. Human atrial ion channel and transporter subunit gene-expression remodeling associated with valvular heart disease and atrial fibrillation. Circulation. 2005;112:471–481. doi: 10.1161/CIRCULATIONAHA.104.506857. [DOI] [PubMed] [Google Scholar]
- Goldstein JA, Demetriou D, Grines CL, Pica M, Shoukfeh M, O’Neill WW. Multiple complex coronary plaques in patients with acute myocardial infarction. N Engl J Med. 2000;343:915–922. doi: 10.1056/NEJM200009283431303. [DOI] [PubMed] [Google Scholar]
- Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc Res. 2012;95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M, Yang Z, Sayed D, et al. GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovascular research. 2012;93:645–654. doi: 10.1093/cvr/cvs001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori M, Nishida K. Oxidative stress and left ventricular remodelling after myocardial infarction. Cardiovascular research. 2009;81:457–464. doi: 10.1093/cvr/cvn335. [DOI] [PubMed] [Google Scholar]
- Icli B, Wara AK, Moslehi J, et al. MicroRNA-26a Regulates Pathological And Physiological Angiogenesis by Targeting BMP/SMAD1 Signaling. Circulation research. 2013 doi: 10.1161/CIRCRESAHA.113.301780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O’Donnell KA, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Seqqat R, Chigurupati S, et al. Inhibition of nuclear factor kappaB regresses cardiac hypertrophy by modulating the expression of extracellular matrix and adhesion molecules. Free radical biology & medicine. 2011;50:206–215. doi: 10.1016/j.freeradbiomed.2010.10.711. [DOI] [PubMed] [Google Scholar]
- Leask A, Abraham DJ. TGF-beta signaling and the fibrotic response. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2004;18:816–827. doi: 10.1096/fj.03-1273rev. [DOI] [PubMed] [Google Scholar]
- Leeper NJ, Cooke JP. MicroRNA and mechanisms of impaired angiogenesis in diabetes mellitus. Circulation. 2011;123:236–238. doi: 10.1161/CIRCULATIONAHA.110.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeper NJ, Raiesdana A, Kojima Y, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. Journal of cellular physiology. 2011;226:1035–1043. doi: 10.1002/jcp.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, De Windt LJ, Witt SA, Kimball TR, Markham BE, Molkentin JD. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem. 2001;276:30245–30253. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorell BH, Carabello BA. Left ventricular hypertrophy: pathogenesis, detection, and prognosis. Circulation. 2000;102:470–479. doi: 10.1161/01.cir.102.4.470. [DOI] [PubMed] [Google Scholar]
- Luo X, Pan Z, Shan H, et al. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. The Journal of clinical investigation. 2013;123:1939–1951. doi: 10.1172/JCI62185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milonas C, Jernberg T, Lindback J, Agewall S, Wallentin L, Stenestrand U. Effect of Angiotensin-converting enzyme inhibition on one-year mortality and frequency of repeat acute myocardial infarction in patients with acute myocardial infarction. Am J Cardiol. 2010;105:1229–1234. doi: 10.1016/j.amjcard.2009.12.032. [DOI] [PubMed] [Google Scholar]
- Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattel S. New ideas about atrial fibrillation 50 years on. Nature. 2002;415:219–226. doi: 10.1038/415219a. [DOI] [PubMed] [Google Scholar]
- Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiological reviews. 2007;87:425–456. doi: 10.1152/physrev.00014.2006. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Gomez D, Bell RD, et al. Smooth muscle cell plasticity: fact or fiction? Circ Res. 2013;112:17–22. doi: 10.1161/CIRCRESAHA.112.281048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigam V, Sievers HH, Jensen BC, et al. Altered microRNAs in bicuspid aortic valve: a comparison between stenotic and insufficient valves. The Journal of heart valve disease. 2010;19:459–465. [PMC free article] [PubMed] [Google Scholar]
- Nishimura RA, Ommen SR, Tajik AJ. Cardiology patient page. Hypertrophic cardiomyopathy: a patient perspective. Circulation. 2003;108:e133–135. doi: 10.1161/01.CIR.0000097621.97566.96. [DOI] [PubMed] [Google Scholar]
- Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- Pfeffer JM, Pfeffer MA, Braunwald E. Influence of chronic captopril therapy on the infarcted left ventricle of the rat. Circ Res. 1985;57:84–95. doi: 10.1161/01.res.57.1.84. [DOI] [PubMed] [Google Scholar]
- Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. The Journal of clinical investigation. 2013;123:11–18. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensen SS, Doevendans PA, van Eys GJ. Regulation and characteristics of vascular smooth muscle cell phenotypic diversity. Netherlands heart journal : monthly journal of the Netherlands Society of Cardiology and the Netherlands Heart Foundation. 2007;15:100–108. doi: 10.1007/BF03085963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome research. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Khanna S, Hussain SR, et al. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovascular research. 2009;82:21–29. doi: 10.1093/cvr/cvp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HK, Griffiths-Jones S, Enright AJ. Genomic analysis of human microRNA transcripts. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17719–17724. doi: 10.1073/pnas.0703890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massague J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. The Journal of biological chemistry. 2006;281:40412–40419. doi: 10.1074/jbc.M610172200. [DOI] [PubMed] [Google Scholar]
- Seropian IM, Toldo S, Van Tassell BW, Abbate A. Anti-inflammatory strategies for ST-elevation acute myocardial infarction ventricular remodeling. J Am Coll Cardiol. 2014 doi: 10.1016/j.jacc.2014.01.014. [DOI] [PubMed] [Google Scholar]
- Shieh JM, Cheng TH, Shi MD, et al. alpha-Tomatine suppresses invasion and migration of human non-small cell lung cancer NCI-H460 cells through inactivating FAK/PI3K/Akt signaling pathway and reducing binding activity of NF-kappaB. Cell biochemistry and biophysics. 2011a;60:297–310. doi: 10.1007/s12013-011-9152-1. [DOI] [PubMed] [Google Scholar]
- Shieh JT, Huang Y, Gilmore J, Srivastava D. Elevated miR-499 levels blunt the cardiac stress response. PloS one. 2011b;6:e19481. doi: 10.1371/journal.pone.0019481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh JH, Choi E, Cha MJ, et al. Up-regulation of miR-26a promotes apoptosis of hypoxic rat neonatal cardiomyocytes by repressing GSK-3beta protein expression. Biochemical and biophysical research communications. 2012;423:404–410. doi: 10.1016/j.bbrc.2012.05.138. [DOI] [PubMed] [Google Scholar]
- Sun X, Icli B, Wara AK, et al. MicroRNA-181b regulates NF-kappaB-mediated vascular inflammation. J Clin Invest. 2012;122:1973–1990. doi: 10.1172/JCI61495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Sit A, Feinberg MW. Role of miR-181 family in regulating vascular inflammation and immunity. Trends Cardiovasc Med. 2014;24:105–112. doi: 10.1016/j.tcm.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thum T, Galuppo P, Wolf C, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- Thum T, Lorenzen JM. Cardiac fibrosis revisited by microRNA therapeutics. Circulation. 2012;126:800–802. doi: 10.1161/CIRCULATIONAHA.112.125013. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Quiat D, Johnson BA, et al. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Developmental cell. 2009;17:662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wan S, George SJ, Berry C, Baker AH. Vein graft failure: current clinical practice and potential for gene therapeutics. Gene Ther. 2012;19:630–636. doi: 10.1038/gt.2012.29. [DOI] [PubMed] [Google Scholar]
- Wang BW, Wu GJ, Cheng WP, Shyu KG. MicroRNA-208a increases myocardial fibrosis via endoglin in volume overloading heart. PLoS One. 2014;9:e84188. doi: 10.1371/journal.pone.0084188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KT, Brilla CG. Pathological hypertrophy and cardiac interstitium. Fibrosis and renin-angiotensin-aldosterone system. Circulation. 1991;83:1849–1865. doi: 10.1161/01.cir.83.6.1849. [DOI] [PubMed] [Google Scholar]
- Wei C, Kim IK, Kumar S, et al. NF-kappaB mediated miR-26a regulation in cardiac fibrosis. Journal of cellular physiology. 2013;228:1433–1442. doi: 10.1002/jcp.24296. [DOI] [PubMed] [Google Scholar]
- Workman AJ, Kane KA, Rankin AC. The contribution of ionic currents to changes in refractoriness of human atrial myocytes associated with chronic atrial fibrillation. Cardiovascular research. 2001;52:226–235. doi: 10.1016/s0008-6363(01)00380-7. [DOI] [PubMed] [Google Scholar]
- Wu KC, Zerhouni EA, Judd RM, et al. Prognostic significance of microvascular obstruction by magnetic resonance imaging in patients with acute myocardial infarction. Circulation. 1998;97:765–772. doi: 10.1161/01.cir.97.8.765. [DOI] [PubMed] [Google Scholar]
- Wu Y, Evers BM, Zhou BP. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. The Journal of biological chemistry. 2009;284:640–648. doi: 10.1074/jbc.M806916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin M, Small EM, Sutherland LB, et al. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes & development. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo M, Lee SK, Lee B, Ruiz EC, Pfaff SL, Gill GN. Small CTD phosphatases function in silencing neuronal gene expression. Science. 2005;307:596–600. doi: 10.1126/science.1100801. [DOI] [PubMed] [Google Scholar]
- Zhang H, Garratt CJ, Zhu J, Holden AV. Role of up-regulation of IK1 in action potential shortening associated with atrial fibrillation in humans. Cardiovascular research. 2005;66:493–502. doi: 10.1016/j.cardiores.2005.01.020. [DOI] [PubMed] [Google Scholar]
- Zhang ZH, Li J, Liu BR, et al. MicroRNA-26 Was Decreased in Rat Cardiac Hypertrophy Model and May Be a Promising Therapeutic Target. Journal of cardiovascular pharmacology. 2013;62:312–319. doi: 10.1097/FJC.0b013e31829b82e6. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lu Y, Zhang Q, et al. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic acids research. 2012;40:4615–4625. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlokovic BV. Neurovascular pathways to neurodegeneration in Alzheimer’s disease and other disorders. Nature reviews Neuroscience. 2011;12:723–738. doi: 10.1038/nrn3114. [DOI] [PMC free article] [PubMed] [Google Scholar]