Abstract
Transfer RNAs (tRNA) through their abundance and modification pattern significantly influence protein translation. Here, we present a systematic analysis of the tRNAome of Lactococcus lactis. Using the next-generation sequencing approach, we identified 40 tRNAs which carry 16 different posttranscriptional modifications as revealed by mass spectrometry analysis. While small modifications are located in the tRNA body, hypermodified nucleotides are mainly present in the anticodon loop, which through wobbling expand the decoding potential of the tRNAs. Using tRNA-based microarrays, we also determined the dynamics in tRNA abundance upon changes in the growth rate and heterologous protein overexpression stress. With a four-fold increase in the growth rate, the relative abundance of tRNAs cognate to low abundance codons decrease, while the tRNAs cognate to major codons remain mostly unchanged. Significant changes in the tRNA abundances are observed upon protein overexpression stress, which does not correlate with the codon usage of the overexpressed gene but rather reflects the altered expression of housekeeping genes.
Keywords: Lactococcus lactis, tRNA, tRNA modifications, Next generation sequencing, Mass spectrometry
INTRODUCTION
Transfer RNAs (tRNAs) act as a physical link between the nucleotide sequence on the messenger RNA (mRNA) and the amino acid sequence on the nascent polypeptide. tRNAs bear the most chemically diverse modifications among all types of nucleic acid entities: the presence of modified nucleotides in tRNA is ten-fold more than in the most abundant biological RNA, ribosomal RNAs (rRNAs), and in total 17 % of the tRNA nucleotides are modified (Giegé et al., 2012; Jackman and Alfonzo, 2012). These modifications play an important role and affect both structure and function of tRNAs. Modified nucleotides in the anticodon loop and some more distant to the anticodon loop affect the fidelity of decoding (Schmeing et al., 2011; Giegé et al., 2012). Modifications of nucleotide at position 34 alter the classical Watson-Crick pairing and allow one tRNA species to read more than one codon (Crick, 1966; Agris, 1991; Agris et al., 2007). Thus, much smaller set of tRNA isoacceptors (tRNAs that carry the same amino acid but read different codons) is needed to translate the 61 sense codons in each organism. Modifications in the acceptor stem influence tRNA aminoacylation and stability (Jackman and Alfonzo, 2012). The type and abundance of modifications can vary among the various isoacceptors. Alterations in modification patterns are increasingly recognized as molecular culprits of several diseases (Suzuki et al., 2011), although a thorough knowledge on the modification pattern in different organisms is lacking.
tRNAs deliver the amino acid to the translating ribosomes (Söll and RajBhandary, 1995). The rate of protein synthesis at each codon varies and is determined by the concentration of the cognate charged tRNA, the codon-anticodon interactions, the relative incorporation rates and the stereochemistry of the amino acid (Pavlov et al., 2009; Johansson et al., 2012). While tRNA accommodation in the ribosomal A-site, transpeptidation and translocation are much faster steps, the rate-limiting step is the diffusion of the aminoacyl-tRNA in a form of ternary complex with the elongation factor to the ribosome (Varenne et al., 1984). Thus, the rate of translation of each codon is directly proportional to the amount of the cognate tRNA present in the cellular pool, with translation rates maximized at codons pairing to high-abundance tRNAs and minimized at codons read by low-abundance tRNA (Zhang et al., 2009). For tRNAs of similar abundance, modifications in the anticodon loop can also change the rate of codon translation (Sørensen and Pedersen, 1991).
The abundance of tRNA isoacceptors vary more than 10-fold in the E. coli cell (Dong et al., 1996); in higher eukaryotes the tRNA abundance is subject to tighter regulation and differs among tissues and with the differentiation state of the cell (Dittmar et al., 2006). Although the mutation-selection-drift balance model proposes that rare codons are only an outcome of the mutation pressure and genetic drift (Hershberg and Petrov, 2008), it is evident that these codons are not randomly distributed within the coding sequence and rather tend to cluster over short stretches, thus causing ribosomes to transiently pause (Zhang and Ignatova, 2009). The stretches of slowly-translated codons are mostly located upstream of the domain boundaries of multi-domain proteins and coordinate their co-translational folding (Komar, 2009; Zhang et al., 2009). Of note, functionally analogous proteins have similar translation patterns, suggesting that the attenuation pattern is adjusted to the species-related variations in the tRNA concentration despite the lack of sequence identity (Zhang and Ignatova, 2009). Thus, failure in expressing many recombinant proteins in soluble, active form might in part due to significant differences between the translation pattern in the recombinant host and the parental strain. A stress-related alteration in the amount of aminoacyl-tRNA influences also the expressivity of each protein (Elf et al., 2003). Translation of the majority of the essential genes seems to be robust and insensitive to changes in the availability of aminoacyl-tRNAs as compared to genes expressed at much lower abundance and linked to regulation (Wohlgemuth et al., 2013). To accurately assess translation efficiency on a global cell-wide level in a parental strain and/or expression host, and to determine how alteration of tRNA availability influences the overall gene expressivity, knowledge of the tRNA abundance and modification pattern is required.
The abundance and modification patterns of a complete set of tRNAs are known only for a limited number of organisms; until now only for Escherichia coli (Dong et al., 1996) and Bacillus subtilis (Kanaya et al., 1999). Here, we present a systematic analysis of the tRNAome of Lactococcus lactis, a microorganism widely used in the biotechnology industry and important as a recombinant host for heterologous proteins, particularly membrane proteins (Kunji et al., 2003). We identified all tRNAs using a next-generation sequencing (NGS) approach and determined their modification patterns with enzymatic digestion coupled to LC-MS/MS analysis. In total, L. lactis expresses 40 different tRNAs that bear 16 different modifications. The majority of the hypermodified nucleotides are present at positions 34 and 37 in the anticodon loop which through wobbling increases the decoding spectrum of the tRNAs. Furthermore, using tRNA-specific microarrays we determined the dynamics in tRNA abundance as a function of growth rate and overexpression stress.
RESULTS
Genomic organization of L. lactis tRNA genes
64 tRNA genes decoding the codons for the 20 standard amino acids are predicted from the genome of L. lactis subsp. cremoris MG1363, hereafter named L. lactis (tRNA database (Chan and Lowe, 2008)). The amino acid carried by two of those tRNA genes is unknown. tRNAs carrying Ala, Pro, Val, Phe, Asn, Asp, Glu, His, Gln, Ile, Met, Tyr, Cys, and Trp are encoded as one isoacceptor for the different codons of each amino acid (Supplementary Materials, Table S1). For some other amino acids different isoacceptor tRNAs, i.e. tRNAs with different anticodons, are present in the genome. Gly and Lys are encoded by two isoacceptors each, Thr has three tRNA isoacceptors, Ser, Arg – four and Leu – five isoacceptors. In total, L. lactis has 34 tRNA isoacceptors with different anticodon sequences, which are not within one locus but rather scattered along the chromosome (Supplementary Materials, Table S1).
To purify tRNAs from the total RNA pool, we used two-dimensional polyacrylamide gel electrophoresis with varying acrylamide percentage and denaturant concentrations in the two dimensions. In the first dimension tRNAs were completely denatured in the 7M urea containing 10% polyacrylamide (PA) and were separated on the basis of their length. The length of tRNAs from L. lactis varies in a close range, from 61-90 nucleotides, thus they appear as a smear on the gel (Supplementary Materials, Fig. S1). In the second dimension, consisting of 20% PA and 4 M urea, the tRNAs were further resolved into 29 distinct spots. Under intermediate urea concentration, the tRNAs acquire partially secondary structure and thus are resolved based on their conformation (Supplementary Materials, Fig. S1). The number of the detected spots was lower than the total number of predicted different tRNAs (29 vs. 34) most likely due to the large degree of length and secondary structure similarity between the tRNAs and their co-migration in both direction. Although the 2D-PAGE provides a good resolution for small tRNA sets (Dong et al., 1996), the lack of information for the mobility and posttranscriptional modification of L. lactis tRNAs precluded their identification.
Next Generation Sequencing (NGS) approach to determine L. lactis tRNAome
To determine the relative abundance of the tRNAs, we analyzed the L. lactis tRNA pool using the Illumina platform for sequencing of small RNAs, which allows for detection of small RNA entities. We performed three independent tRNA isolations from exponentially growing L. lactis (S1, S2 and S3) and spiked them with tRNAs from either E. coli or yeast, which do not pair to any of the L. Lactis tRNAs. S1 and S2 were spiked with Phe-tRNAGAA from yeast and Lys-tRNAUUU from E. coli prior to cell lysis. S3 was spiked with Phe-tRNAGAA from yeast and Lys-tRNAUUU from E. coli before the library preparation to estimate possible tRNA loss during the extraction process. With the NGS approach we identified 40 different tRNA sequences (Table 1). Intriguingly, we detected different isodecoders (i.e. tRNAs with the same anticodon but different bodies) for two Leu-tRNAs and Lys-tRNAs and for the four MettRNAs (Table 1 and Supplementary Materials, Table S1).
Table 1.
Total number of reads aligned to each of the tRNA sequences in the three sequencing runs: S1, S2 and S3. To compensate for the errors due to modifications, a mismatch of 3 was allowed during the alignment. Yeast-Phe1, yeast-Phe2 and E. coli-Lys denote the spike-in tRNAs.
| tRNA | Anticodon | S1 | S2 | S3 | |||
|---|---|---|---|---|---|---|---|
| Total reads | % | Total reads | % | Total reads | % | ||
| Yeast-Phe1 | GAA | 415536 | 746408 | 86629 | |||
| Yeast-Phe2 | GAA | 20535 | 33741 | 7306 | |||
| E. coli-Lys | UUU | 33692 | 21375 | 16073 | |||
| Ala | UGC | 22444 | 0.256 | 12161 | 0.159 | 473 | 0.336 |
| Arg1 | ACG | 59683 | 0.680 | 25693 | 0.336 | 983 | 0.697 |
| Arg2 | CCG | 165 | 0.002 | 173 | 10023 | 11 | 0.008 |
| Arg3 | CCU | 312 | 0.004 | 175 | 0.002 | 18 | 0.013 |
| Arg4 | UCU | 1828 | 0.021 | 513 | 0.007 | 42 | 0.030 |
| Asn | GUU | 2601864 | 29.660 | 6564554 | 85.872 | 91573 | 64.967 |
| Asp | GUC | 756713 | 8.626 | 201852 | 2.640 | 5485 | 3.891 |
| Cys | GCA | 7514 | 0.086 | 4562 | 0.056 | 185 | 0.131 |
| Gln | UUG | 184109 | 2.099 | 42708 | 0.559 | 7656 | 5.432 |
| Glu | UUC | 21639 | 0.247 | 12489 | 0.163 | 822 | 0.583 |
| Gly1 | GCC | 113327 | 1.292 | 32436 | 0.424 | 3435 | 2.437 |
| Gly2 | UCC | 136225 | 1.553 | 57221 | 0.749 | 8634 | 6.125 |
| His | GUG | 120 | 0.001 | 55 | 0.0007 | 22 | 0.016 |
| Ile | GAU | 10788 | 0.123 | 3922 | 0.051 | 211 | 0.150 |
| Leu1 | AAG | 3606 | 0.041 | 2079 | 0.028 | 113 | 0.080 |
| Leu2 | AAG | 5830 | 0.066 | 2239 | 0.029 | 189 | 0.134 |
| Leu3 | CAA | 3080 | 0.035 | 1517 | 0.020 | 89 | 0.063 |
| Leu4 | CAG | 23301 | 0.266 | 6267 | 0.082 | 626 | 0.444 |
| Leu5 | UAA | 3432527 | 39.130 | 137062 | 1.793 | 1804 | 1.280 |
| Leu6 | UAG | 30 | 0.0003 | 3 | 0.0004 | 10 | 0.007 |
| Lys1 | CUU | 212643 | 2.424 | 49224 | 0.644 | 1742 | 1.236 |
| Lys2 | UUU | 22348 | 0.255 | 5645 | 0.074 | 476 | 0.338 |
| Lys3 | UUU | 33144 | 0.378 | 10635 | 0.139 | 211 | 0.150 |
| Met1 | CAU | 7269 | 0.083 | 3059 | 0.040 | 409 | 0.290 |
| Met2 | CAU | 10045 | 0.115 | 5502 | 0.072 | 126 | 0.089 |
| Met3 | CAU | 69730 | 0.795 | 21575 | 0.282 | 2375 | 1.685 |
| Met4 | CAU | 0 | 0 | 1 | 0.0001 | 0 | 0 |
| Phe | GAA | 9304 | 0.106 | 3855 | 0.050 | 235 | 0.167 |
| Pro | UGG | 277237 | 3.160 | 56638 | 0.741 | 1321 | 0.937 |
| Ser1 | CGA | 834 | 0.010 | 181 | 0.002 | 24 | 0.017 |
| Ser2 | GCU | 222 | 0.003 | 81 | 0.001 | 15 | 0.011 |
| Ser3 | GGA | 781 | 0.009 | 802 | 0.010 | 60 | 0.043 |
| Ser4 | UGA | 82156 | 0.937 | 32438 | 0.424 | 2366 | 1.679 |
| Ser5 | UGA | 1558 | 0.018 | 236 | 0.003 | 188 | 0.133 |
| Thr1 | CGU | 7739 | 0.088 | 7418 | 0.097 | 508 | 0.360 |
| Thr2 | GGU | 116 | 0.001 | 121 | 0.002 | 26 | 0.018 |
| Thr3 | UGU | 214805 | 2.449 | 207407 | 2.714 | 3315 | 2.352 |
| Trp | CCA | 389074 | 4.435 | 109560 | 1.433 | 2776 | 1.969 |
| Tyr | GUA | 38613 | 0.440 | 13782 | 0.180 | 1495 | 1.061 |
| Val | UAC | 9440 | 0.108 | 8757 | 0.115 | 906 | 0.643 |
The spiked-in Phe-tRNAGAA and Lys-tRNAUUU, both added at a ratio of 5:1 (w/v) in the S1 and S2 sequencing runs, yielded ratios of 13:1 and 37:1, respectively, indicating non-proportional losses between the sequencing runs. When added prior to the cDNA library preparation in S3, the spiked tRNAs yielded a ratio 5.8:1, which is in a reasonable agreement with the expected ratio of 5:1. Overall, these data indicate that a substantial loss of tRNAs occurred during the extraction process. The low reproducibility between the two sequencing sets might be in part due to posttranscriptional modifications which influence the fidelity of the reverse transcriptase used in the production of the cDNA library (Czech et al., 2010). In summary, the variations in the ratios of tRNAs (Table 1) suggest that NGS is a powerful approach for tRNA identification, including identification of various isodecoders, but cannot be used for quantification of tRNAs.
Identification of modifications of L. lactis tRNAs
More than 80 modifications on the nucleosides at various positions on tRNAs have been described for various species of eubacteria, archaea and eukaryotes (Söll and RajBhandary, 1995; Jackman and Alfonzo, 2012). However, information about the modifications of different species is scarce and completely lacking for L. lactis. To gain insight into the fully modified sequences of L. lactis tRNAs, the purified tRNAs were analysed by mass spectrometry (MS) on two different levels (Kowalak et al., 1993). First, to identify all modified bases a typical nucleoside analysis was performed. Briefly, the isolated pool of total tRNA was enzymatically digested down to a nucleoside level and then subjected to liquid chromatography-mass spectrometry (LC-MS) analysis. Modified bases were identified by their retention times in a LC-UV chromatogram and then confirmed by their unique mass-to-charge (m/z) ratios, including tandem mass spectrometry (MS/MS) base loss seen for most of the nucleosides except pseudouridine. Beyond the typical A, G, C, U we detected 16 modified nucleosides and note their typical location within the tRNA body (Table 2).
Table 2.
Summary of the detected nucleosides from the L. lactis tRNA pool based differences in their retention time in the HPLC analysis.
| tRNA modification | Designation | Position | Retention time (min) |
|---|---|---|---|
| Pseudouridine | ψ | Multiple | 3.6 |
| Dihydrouridine | D | D-Loop | 3.9 |
| Cytidine | C | Multiple | 5.0 |
| Uridine | U | Multiple | 6.5 |
| 5-carboxym ethylaminomethyl-2-thiouridine | cmnm5s2U | 34 | 11.6 |
| 2’-O-methylcytidine | Cm | 32 | 11.8 |
| 1-methyladenosine | m1A | 13.4 | |
| Inosine | I | 34 | 14.3 |
| 5-methyluridine | rT (m5U) | TψC-Loop | 15.0 |
| Guanosine | G | Multiple | 16.0 |
| 5-methoxyuridine | mo5U | 34 | 16.5 |
| 7-methylguanosine | m7G | V-Loop | 19.1 |
| 1-methylguanosine | m1G | 37 | 24.0 |
| N4-acetylcytidine | ac4C | 34 | 24.8 |
| Adenosine | A | Multiple | 26.1 |
| N6-threonylcarbamoyladenosine | t6A | 37 | 28.7 |
| 2-methyladenosine | m2A | 37 | 33.2 |
| N6-methyladenosine | m6A | 37 | 33.9 |
| N6-threoylcarbamoyl-2-methyladenosine | ms2t6A | 37 | 39.9 |
| N6-(3-methyl-2-butenyl)adenosine | i6A | 37 | 47.6 |
In an effort to place these modified nucleosides sequentially in each individual tRNA, a combination of de novo and targeted LC/MS/MS approaches were developed by modifying protocols as described earlier (Hossain and Limbach, 2006; Wetzel and Limbach, 2013). The approach used here involves digestion of tRNA with various ribonucleases (RNases), which cleave tRNAs at base- or structure-specific locations, yielding a multitude of digestion products many of which are unique to specific tRNAs. These RNase digests were analyzed using LC/MS/MS to obtain site- and sequence-specific data including information related to the modified nucleosides. By this approach, the locations of various modified nucleosides (Table 2) were first identified within individual RNase digestion products which were further compared against the 40 unique tRNA sequences to map the mass spectrometry results onto the relevant tRNA sequences (Supplementary Materials, Fig. S2).
In addition, different modifications (Table 2) can be placed onto specific tRNA gene sequences, i.e., prior to in silico predictions of anticipated RNase digestion products. These predicted digestion products were obtained using the Mongo Oligo Mass Calculator (http://rnamdb.cas.albany.edu/RNAmods/masspec/mongo.html), and by an in silico approach, different combinations of 16 anticipated modifications were mapped onto a variety of the 40 unique tRNA gene sequences (Table 3). Pseudouridine was not included in these determinations as it is an isomer of uridine that cannot be detected by standard mass spectrometry approaches. These predicted RNase digestion product results were then compared with the experimentally determined RNase digestion products to identify matches in the LC/MS/MS; the sites of posttranscriptional modification are annotated when the modification was confirmed in more than one RNase digest (Table 3). While it is difficult to uniquely characterize all of the expected isoacceptors by the method used in this work, we found a mixture of fully and partially modified sequences for certain tRNAs. For example, RNase T1 digestion yields both the fully modified digestion product unique to Val-tRNAUAC, (31)CCU[mo5U]AC[mA]AGp (39), as well its partially modified counterpart lacking mo5U, CCUUAC[mA]AGp. Another example is illustrated by the data arising from the anticodon region of Ala-tRNAUGC. When using RNase T1 for digestion, this tRNA will be cleaved in the middle of the anticodon at G35. In contrast, RNase V1 cleaves primarily at single stranded-loops thereby often releasing anticodon loops as digestion products. For Ala-tRNAUGC we detected both the fully modified product (32) pUU[mo5U]GC[mA] (37) as well as the same sequence region lacking a methylated adenosine at position 37, pUU[mo5U]GCA.
Table 3.
Post-transcriptional modifications (bold) identified in the L. lactis tRNAs. The anticodon is underlined in the sequence. Copy number denotes the number of genomic tRNAs. Nucleosides in brackets represent modified sequence locations. Dual nucleosides in brackets (e.g., [m6A/A]) represent sites of hypomodification where RNase digestion products were detected both with and without the identified modification. Methylated nucleosides are placed onto sequence locations, using known tRNA modification homology algorithms (Machnicka et al., 2013) as the LC-MS/MS approach cannot directly reveal the ring location of methylation except for 7-methylguanosine [m7G].
| tRNA | Anticodon | Copy number |
Sequence (5’ – 3’) |
|---|---|---|---|
| Ala | UGC | 6 | GGGGCCU[s4U]AGCUCAGC[D]GGGAGAGCGCCUGCUU[mo5U]GC[m6A/A]CGCAGGAGGUCAGCGG[m5U]UCGAUCCCGCUAGGCUCCACCA |
| Arg 1 | ACG | 2 | GCAGGCGUGGCUCAAC[D]GGA[D]AGAGUACCUGACU[I]CG[m2A/A]AUCAGGC[m7G]UUGUAGG[m5U]UCGAAUCCUACCGCUUGCACCA |
| Arg2 | CCG | 1 | GCGUCCG[s4U]AGUGUAA[D]GGA[D]AUCACGUAAGAUUCCGGUUCUUGAAUGGGGG[m5U]UCGAUUCCCUCCGGACGCACCA |
| Arg 3 | CCU | 1 | GGUCUCUUAGUUAAAUUGGA[D]AUAACUACUGCCUCCUAAGCAGUGAUUGCCAG[m5U]UCGAUUCUGGCAGGGGCCACCA |
| Arg 4 | UCU | 1 | GGUCCGAUAGCUCAGC[D]GGA[D]AGAGCAUUCGCCUUCUAAGCGAACG[m7G]UCGAGGG[m5U]UCGAAUCCCCCUCGGAUCACCA |
| Asn | GUU | 2 | UGCGGAU[s4U/U]AGCUCAGU[D]GG[D]AGUAGCGCAUGACUGUU[t6A]AUCAUGAU[m7G]UCGUCAG[m5U]UCGAGUCUGACAUCCGCAGCCA |
| Asp | GUC | 2 | GGUUCUGUAGUGUAGCGG[D][D]AUCACGUCGCCCUGUCACGGCGAAGAUCGCGGG[m5U]UCGAUUCCCGUCAGAACCGCCA |
| Cys | GCA | 1 | GGCGGCG[s4U]AGUGAAGUGG[D][m1A]ACACAUGGCUCUGCA[i6A]AAGCUUAAUCGUCGG[m5U]UCAAAUCCGACCGUCGCCUCCA |
| Gln | UUG | 2 | UGGGGUA[s4U]AGCCAAGCGG[D]AAGGCAAGGGACUUUGACUCCCUCAUGCGUUGG[m5U]UCGAAUCCAGCUACCCCAGCCA |
| Glu | UUC | 1 | GGUCCGUUGGUC[m1A/A]AGGGG[D][D]AAGACACCGCCUUUUCACGGCGGUAACACGGG[m5U]UCGAAUCCCGUACGGACUACCA |
| Gly 1 | GCC | 2 | GCGAACGUAGUUCAGGGG[D]AGAACACAACCUUGCCAUGGUUGGG[m7G]UCGCGAG[m5U]UCGAAUCUCGUCGUUCGCUCCA |
| Gly 2 | UCC | 2 | GCGGAUGUAGUUUAAUGGUAGAACCCCAGCCU[mo5U]CC[m6A/A]AGCUGGCUACGCGAG[m5U]UCGAUUCUCGUCAUCCGCUCCA |
| His | GUG | 1 | GCGGAUGUGGUGAAGUGG[D][D]AACACACCGGCUUGUGGCGCCGGCACUCGCGAG[m5U]UCGAUCCUCGUCAUCCGCCA |
| Ile | GAU | 2 | GGGAGUU[s4U/U]AGCUCAGU[D]GG[D][D]AGAGCACUGUGUUGAU[t6A]ACGCAGGG[m7G]UCCCAGG[m5U]UCGAAUCCUGGAAUUCCCACCA |
| Leu 1 | AAG | 2 | GCGGUGGUGGCGGAAU[D]GGCAGACGCGCAGGAUUAAG[m1G]AUCCUGUCGGGAAUUUUCUUGGUGCGGG[m5U]UCAAGUCCCGCCCACCGCACUACCA |
| Leu 2 | CAG | 1 | GGGGAAGUGGUGGAAUGGCAGACACGCAUGCUUCAGGUGCAUGUGCGAGAAAUCGCGUGAGGG[m5U]UCAAAUCCCUUCUUCCCCACCA |
| Leu 3 | UAG | 2 | GCGGAUGUGGCGGAAU[D]GGCAGACGCACUAGAUUUAG[m1G]AUCUAGCGCUUAACGGCGUGGGGG[m5U]UCAAGUCCCUUCAUCCGCACCA |
| Leu 4 | CAA | 1 | GCCGAAGUGGCGGAAUAGGCAGACGCGCCAGACU[Cm]AA[i6A]UCUGGUUCCUUCACGGGAGUGCCGG[m5U]UCGACCCCGGCCUUCGGCACCA |
| Leu 5 | UAA | 1 | CCCGGGGUGGCGGAAC[D/U]GGCAGACGCACAGGACU[cmnm5s2U]AA[i6A]AUCCUGCGAGGGGUAACCUUCGUACCGG[m5U]UCGAAACCGGUCCUCGGGACCA |
| Leu 6 | UAA2 | 1 | GCUCGAAUGGCGGAAU[D]GGCAGACGCUGCGGACUUAAAAUCCGUUGGUUAUUAAACCGUGAGGG[m5U]UCAAGUCCCUCUUUGAGCACCA |
| Lys 1 | CUU | 1 | GGCCCGGUAGCUCAGU[D]GG[D]AGAGCAGUAGACUCUU[t5A]AUCUAUGG[m7G]UCCAGGG[m5U]UCGAGCCCCUGCCGAGCCACCA |
| Lys 2 | UUU | 1 | GACUCGUUAGCUCAGU[D]GG[D]AGAGCAUUUGACU[cmnm5s2U]UU[t6A]AUCAAAGG[m7G]UCGCUGG[m5U]UCGAGCCCAGCACGGGUCACUACCA |
| Lys 3 | UUU2 | 2 | GACUCGUUAGCUCAGU[D]GG[D]AGAGCAUUUGACU[cmnm5s2U]UU[t6A]AUCAAAGG[m7G]UCGCUGG[m5U]UCGAGCCCAGCACGGGUCACCA |
| Met 1 | CAU | 1 | GGAUCUUUAGCUCAGU[D]GG[D][D]AGAGCUAUCGGCUCAU[m6A/A]ACCGAUCG[m7G]UCGCUGG[m5U]UCGAAUCCAGCAAGAUCCACCA |
| Met 2 | CAU | 1 | GGCGGUGUAGCUCAGCUGGCUAGAGCGUUCGGUU[ac4C]AU[m6A]CCCGAGAGGUCGGGGG[m5U]UCGAUCCCCUUCGCCGCUACCA |
| Met 3 | CAU | 1 | CGCGGGA[s4U]GGAGCAGCUAGG[D]AGCUCGUCGGGCUCAUAACCCGAAG[m7G]UCAUAGG[m5U]UCAAAUCCUAUUCCCGCAACCA |
| Met 4 | CAU | 1 | AGUUCUUUAGCUUAGU[D]GG[D][D]AAAGUCCCCCGCUCAUAACGGGGUAAGCGCUGGUUUGAGUCCAGCAAGAACCA |
| Phe | GAA | 2 | GGCUCGG[s4U]AGCUCAGU[D]GG[D]AGAGCAAUGGAUUGAA[m1G]CUCCAUGU[m7G]UCGGCGG[m5U]UCGAUUCCGUCUCGCGCCACCA |
| Pro | UGG | 2 | CGGGAAGUAGCUCAGCUUGG[D]AGAGUACUUGGUU[mo5U]GG[m1G]ACCAAGGU[m7G]UCGCAGG[m5U]UCGAAUCCUGUCUUCCCGACCA |
| Ser 1 | GGA | 1 | GGAGAAGUGUCCGAG[D]GGCCGAAGGAGCACGCCUGGAAAGUGUGUAUACGUCACAAGCGUAUCGGGGG[m5U]UCGAAUCCCCUCUUCUCCUCCA |
| Ser 2 | CGA | 1 | GGAGCCAUGGCAGAGUGGUAAUGCAGCGGACUCGAAAUCCGUCGAACCGUGUAAAGCGGCGCAGGGG[m5U]UCAAAUCCCCUUGACUCCUCCA |
| Ser 3 | UGA | 1 | GGAGGAUUACCCAAGCCUGGCUGAAGGGAACGGUCU[mo5U]GA[i6A]AACCGUCAGGCAUGUAAAGGCGUGCGUGGG[m5U]UCGAAUCCCACAUCCUCCUCCA |
| Ser 4 | UGA2 | 1 | GGAGGAUUACCCAAGUCCGGCUGAAGGGAACGGUCU[mo5U]GA[i6A]AACCGUCAGGCGUGUAAAAGCGUGCGUGGG[m5U]UCGAAUCCCACAUCCUCCUCCA |
| Ser 5 | GCU | 1 | GGGGGGUUACUCAAGAGGCUGAAGAGGACGGUUUGCUAAAUCGUUAGGUCGGGAAACCGGCGCGAGGG[m5U]UCGAAUCCCUUACCCCCCUCCA |
| Thr 1 | GGU | 1 | GCCGUUGUAGCUCAGUCGGUAGAGCAGCACCAUGGUAAGGUGAAG[m7G]UCGACAG[m5U]UCGAUUCUGUUCAAUGGCACCA |
| Thr 2 | CGU | 1 | GCCGAGUUAGCUCAGUCGGUAGAGCACUUCACUCGUAACGAAGGG[m7G]UCACAGG[m5U]UCGAUUCCUGCACUCGGCACCA |
| Thr 3 | UGU | 2 | GCCGACU[s4U/U]AGCUCAGU[D]GG[D]AGAGCAUCUGAUU[mo5U]GU[t6A]AUCAGAGG[m7G]UCGCGUG[m5U]UCGAAUCAUGUAGUCGGCACCA |
| Trp | CCA | 1 | ACGGGCAUCGUAUAAAGGUAGUACAAAGGUCUCCA[i6A]AACCUUUAGUGUGGG[m5U]UCAAUUCCUGCUGCCCGUGCCA |
| Tyr | GUA | 1 | GGAAGGGUAGCGAAGAGGC[D]AA[m1A]CGCGGCGGACUGUA[i6A]AUCCGCUCCUUCGGG[m5U]UCGGGGGUUCGAAUCCCUCCCCUUCCACCA |
| Val | UAC | 3 | GGGAGUU[s4U/U]AGCUCAGC[D]GGGAGAGCAUCUGCCU[mo5U]AC[m1A]AGCAGAGG[m7G]UCAGCGG[m5U]UCGAUCCCGUUAACUCCCACCA |
Beyond the anticodon region, analysis with RNase U2, which cleaves at purines with A preferred over G, revealed the presence of 4-thiouridine modifications to U8 in a number of tRNAs (Table 3). In all cases, both the modified RNase U2 digestion product was detected along with a corresponding digestion product lacking s4U at position 8. Although we detected 16 modifications from the nucleoside digest of L. lactis total tRNA and placed most of these modifications onto tRNA isoacceptor sequences (except pseudouridine), we cannot rule out the possibility that L. lactis tRNAs contain additional modifications. It is possible that some modifications occur at a level below our detection limits or exists in mixed sequences such as [ms2t6A/t6A]. Furthermore, we are limited by availability of suitable digestion enzymes with defined nucleotide specificity to obtain complete nucleotide coverage.
tRNA levels are affected by growth rate and overexpression stress
The amount of aminoacyl-tRNAs is not a static parameter and depends on the growth conditions and nutrient availability (Elf et al., 2003; Dittmar et al., 2005).To assess the impact of the growth rate on the tRNA abundance in L. lactis, we used tRNA microarrays (Dittmar et al., 2005). Probes (Supplementary Materials, Table S2) were designed taking into account the different isoacceptors identified by the NGS approach (Table 1) and variations in their modification patterns (Table 3). Thus, for Lys-tRNAUUU two isoacceptors were detected which show similar modifications but differ in their sequences. Similarly, Met-tRNACAU has four probes as the Met-tRNACAU set differ in sequence and modification pattern.
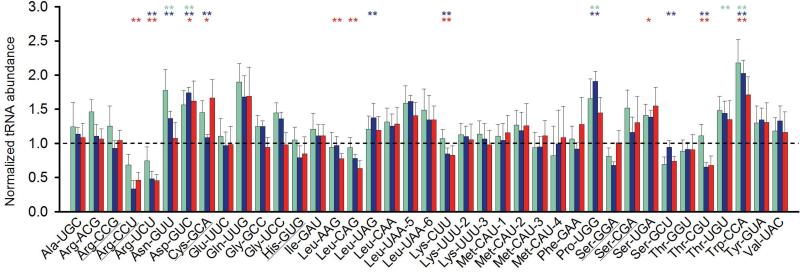
Total tRNA was isolated from cells grown in a glucose-limited chemostat cultures at a growth rate (μ) of 0.15 h−1, 0.3 h−1, 0.5 h−1 and 0.6h−1. Microarray technology allows for assessing the relative tRNA abundance, i.e., changes in the concentration of each isoacceptor compared to a control condition set (Dittmar et al., 2005), here the slowest growth rate of 0.15 h−1. At growth rates of 0.3 h−1, 0.5 h−1 and 0.6 h−1 Asp-tRNAGUC and TrptRNACCA were upregulated relative to μ = 0.15 h−1 (Fig. 1). Arg-tRNACCU, Arg-tRNAUCU, AsntRNAGUU, Asp-tRNAGUC, Cys-tRNAGCA, Leu-tRNAAAG, Leu-tRNACAG, Leu-tRNAUAG, LystRNACUU, Pro-tRNAUGG, Ser-tRNAUGA, Ser-tRNAGCU, Thr-tRNACGU and Thr-tRNAUGU displayed a change in at least one growth rate compared μ = 0.15 h−1 (Fig. 1). The levels of the majority of the tRNAs (i.e., 24 tRNA isoacceptors) remained unchanged (Fig. 1).
Figure 1.
Variations in the tRNA abundance of L. lactis grown at different growth rates. For comparison the slowest growth rate of 0.15 h−1 is set to 1, which is represented by the dotted horizontal line. The tRNA abundance at a growth rate of 0.3 h−1(green), 0.5 h−1 (blue) and 0.6 h−1 (red) are compared to the tRNA abundance at 0.15 h−1. * denotes p < 0.05 and ** denotes p < 0.01. tRNAs reading rare codons are underlined (for more details see Materials and Methods section).
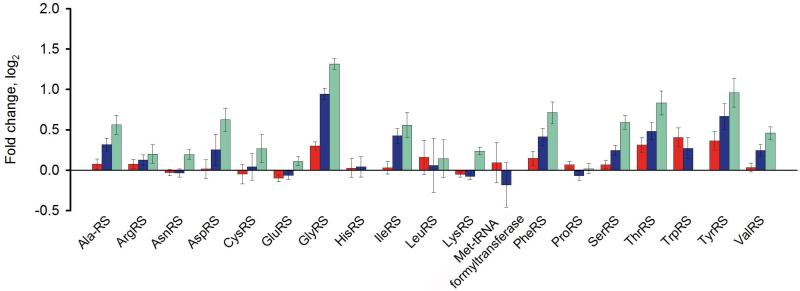
The same cells of L. lactis were also subjected to relative quantitative proteomics, using the iTRAQ approach. Again the protein levels at 0.3 h−1, 0.5 h−1 and 0.6 h−1 were benchmarked against those μ = 0.15 h−1. The levels of some aminoacyl-tRNA-synthetases (aaRS) were found to increase with increasing growth rate to maximally 50% of the amount at μ = 0.15 h−1 (Fig. 2). Upregulation of SerAS, AspRS, CysRS, TrpRS and ThrRS correlates with the increased levels of the cognate isoacceptors charged by these aaRSs, while the enhanced expression of AlaRS,GlyRS, PheRS, TyrRS, IleRS, SerRS and ValRS does not correlate (Fig. 1 and 2).
Figure 2.
Differences in the relative abundance of aminoacyl-tRNA-synthetases at 0.3 h−1(red), 0.5 h−1 (blue) and 0.6 h−1 (green). The differences at the given growth rate are compared to μ = 0.15 h−1; the values are presented as a fold change (log2)
Overexpression of proteins may alter the balance of aminoacyl-tRNAs that are available to translate housekeeping genes (Wohlgemuth et al., 2013). As L. lactis is increasingly used as a host for overexpression of heterologous proteins, we next sought to evaluate alterations in the tRNA levels when overexpressing the membrane protein OpuA, an ATP-binding cassette (ABC) transporter that couples ATP hydrolysis to import of compatible solutes. The L. lactis cells were grown in a pH-controlled bioreactor (2 L) and the expression of OpuA was induced at OD600 of 1.5 with 0.005 % (w/v) nisin A. Cells were harvested for tRNA isolation at after zero (OD600 = 0.5) and one hour of induction (OD600 = 1.5) with nisin A. The tRNA isolated from cells harvested at t=0 and t=1 h of induction were compared with that of non-induced cells (Fig. 3). In response to the overexpression, all tRNA isoacceptors for Met and Thr and Ala-tRNAUGC, Arg-tRNAACG, Arg-tRNACCU, His-tRNAGUG, Ile-tRNAGAU and ThrtRNACGU were increased compared to the non-induced sample. In contrast, other tRNAs bearing the amino acids Cys, Pro, Ser and Trp showed a decrease in abundance upon overexpression stress (Fig. 3). The opuA genes are expressed from a strong promoter, resulting in overproduction of the transporter. We hypothesized that an increased translational demand may be accompanied by a change in the relative abundance of tRNAs. Notably, we did not observe any correlation between the specifically elevated tRNAs and the codons overrepresented in the opuA gene. OpuA overexpression causes up- or downregulation of specific cellular genes (Marreddy et al., 2011). We thus determined whether those endogenous genes are enriched in codons that are read by the tRNAs for which we observed changes upon OpuA overexpression. We did not find such a correlation; we note that the codon usage of the endogenous (OpuA expression-affected) genes does not significantly deviate from those of a randomized gene sample of the same size (Supplementary Materials, Fig. S3A and B). However, when analyzing the genes with generic housekeeping functions of the cell, we observed a bias in usage of specific codons (Arg-tRNAAGG, Cys-tRNATGC, Cys-tRNATGT, Ser-tRNATCC, Pro-tRNACCC, Gln-tRNACAG, IletRNAATA Supplementary Materials, Fig. S3C). The downregulation of tRNAs for Cys, Ser and Pro upon OpuA overexpression (Fig. 3) correlates with the underrepresented usage of Cys-tRNATGT, Pro-tRNACCC and Ser-tRNATCC in housekeeping genes (Fig. 3). From the overrepresented codons only Ile-ATA correlates with upregulation of its cognate tRNA (Fig. 3). Together, these results suggest that upon overexpression of OpuA, the cell (partly) adjusts the tRNA concentrations to the codon usage of the housekeeping genes.
Figure 3.
Overexpression stress selectively alters the abundances of some tRNA species. The abundance of tRNA species isolated from cells growing in the mid-exponential phase (OD600 = 0.5), at the time of induction (green, OD600 = 1.5) and after 1 h of induction of the opuA genes (blue); the tRNA levels were normalized to the concentration of the corresponding tRNA isoacceptors of cells grown to the mid-exponential phase (OD600 = 0.5; dotted horizontal line); * denotes p < 0.05. tRNAs reading rare codons are underlined (for more details see the Materials and Methods section).
DISCUSSION
The tRNA concentration of only handful microorganisms is known at present. In addition, the entire (or nearly entire) modification pattern of a complete set of tRNAs has been determined for only a few organisms, including the bacteria E. coli (41 sequenced of 46 total tRNA species) and Mycoplasma capricolum (29 sequenced of 29 total tRNA species), the eukaryote Saccharomyces cerevisiae (37 sequenced of 42 total tRNA species), and the archaeon Haloferaxvolcanii (40 sequenced of 46 total tRNA species) (Yacoubi et al., 2012). We have now determined the tRNAome of L. lactis, an industrially important microorganism, that is frequently employed as an expression host for the production of therapeutic (Steidler et al., 2009) and membrane proteins (Kunji et al., 2003; Frelet-Barrand et al., 2010; Linares et al., 2010). The tRNA abundance is a critical parameter for determining the translation rate at a particular codon on the transcript (Zhang et al., 2009) or to assess the effect of overexpression burden on the housekeeping proteome (Wohlgemuth et al., 2013).
Here, we used next generation sequencing to identify various tRNAs transcribed in exponentially growing L. lactis cells. The L. lactis genome is predicted to contain 62 defined tRNA genes. 34 of these genes code for tRNA with a distinct anticodon. We found 40 different tRNA sequences in the total tRNA pool, indicating that various tRNA isoacceptors and isodecoders are present in L. lactis. The approach is, however, not quantitative as nucleoside modifications may influence the quantitative reverse transcription and thus alter the number of the reads for each tRNA species. tRNAs are the most extensively modified nucleic acids in the cell and all modifications are post-transcriptionally added. Thus, attaining the information regarding the position of modified nucleosides is important as it allows for introducing mismatch at that position to strengthen the alignments of sequenced tRNA, obtained by NGS and benchmarked against the genomic sequences available from the tRNA database.
All of the modified nucleosides detected in L. lactis encompass common modified nucleosides (e.g., Ψ, D, m5U, rT) and other methylations) as well as modified nucleosides known to be associated with the anticodon stem loop of tRNAs (e.g., cmnm5s2U, ac4C, mo5U). Modifications to L. lactis tRNAs are similar to those identified in other bacteria whose complete (or nearly complete) complement of tRNAs has been characterized at the posttranscriptional level (Table 4), with the body of the tRNA modified with methyl and dihydrouridine groups and the anticodon containing the more complex modifications. No previously undiscovered modifications were found in L. lactis. On the basis of the Modomics database (Machnika et al., 2013), B. subtilis has six other modified nucleosides not found in L. lactis: cmnm5U (which could arise from lack of thiolation found in cmnm5s2U), Gm, Q, k2C, m2G, and ms2i6A. E. coli has 12 other modified nucleosides not found in L. lactis: acp3U, mnm5U, s2C, m5C, cmnm5Um, Um, Gm, Q, oQ, k2C, m2G, and ms2i6A.
Table 4.
Comparison of modifications and their sequence locations between L. lactis, B. subtilis and E. coli
| Modification | L. lactis | B. subtilis | E. coli | Comments |
|---|---|---|---|---|
| D | ✓ | ✓ | ✓ | Universally conserved in bacteria – found at U20 |
| ψ | ✓ | ✓ | ✓ | Universally conserved in bacteria – found at U55 |
| xm5s2U | ✓ (x=cmn) | ✓ (x=cmn) | ✓ (x=mn) | Usually found at U34 in tRNAs that decode codons ending in G or A |
| s4U | ✓* | ✓ | ✓ | *Not found in total nucleoside analysis but identified in RNA modification mapping studies |
| Cm | ✓ | ✓ | ✓ | Found at C34 of Leu-tRNACAA |
| m5U | ✓ | ✓ | ✓ | Universally conserved in bacteria – found at U54 |
| I | ✓ | ✓ | ✓ | Found at A34 of Arg-tRNAACG |
| xo5U | ✓ (x=m) | ✓ (x=m) | ✓ (x=cm) | Usually found at U34 in 4 or 6-box tRNAs (Ala, Pro, Ser, Thr, Val); 5-methoxy derivatives found in gram positive bacteria; 5-carboxy derivatives found in gram negative bacteria |
| m7G | ✓ | ✓ | ✓ | Universally conserved in bacteria – found at G46 |
| m1G | ✓ | ✓ | ✓ | Usually found at G37 in tRNAs-Pro, Arg, Leu |
| ac4C | ✓ | ✓ | ✓ | Found at C34 of Met-tRNACAU |
| t6A | ✓ | ✓ | ✓ | Universally conserved in bacteria – found at A37 of tRNAs decoding codons beginning with A. |
| m2A | ✓ | ✓ | ✓ | |
| m6A | ✓ | ✓ | ✓ | Universally conserved in bacteria – found at A37 |
| i6A | ✓ | ✓ | ✓ | Highly conserved in bacteria – found at A37 of tRNAs decoding codons beginning with U. |
| m1A | ✓ | ✓ | ✓ | Highly conserved in bacteria although not seen in E. coli – found at A58 |
| ms2t6A | ✓ | ✓ | [m6t6A] | Commonly identified in bacteria – found at A37 |
A comparison of existing tRNA sequences and modification patterns reveals that some anticodons might be modified in L. lactis. For example, 5-methoxyuridine (mo5U) has previously been found at the wobble base (U34) of Ala, Pro, Thr, Val, and Ser-tRNAs (Machnicka et al., 2013). Similarly, U34 of Gln, Glu and Lys tend to possess modified uridines from the xm5(s2)U family (Machnicka et al., 2013). Modifications found adjacent to the anticodon at position 37 also have characteristics that facilitate placing these identifications onto specific tRNA sequences. For example, N6-(3-methyl-2-butenyl)adenosine (i6A) is invariably found in bacteria at A37 of almost every tRNA reading UNN codons (N being one of the four canonical nucleotides A, C, G, or U)(Marck and Grosjean, 2002). Similarly, N6-threonylcarbamoyladenosine (t6A) is found at A37 of almost every tRNA reading ANN codons (Marck and Grosjean, 2002). The majority of the hypermodified nucleosides are present only in the positions 34 and 37, covering the anticodon loop, while relatively simple modifications like methylation and thiolated derivatives exist outside the anticodon region (Söll and RajBhandary, 1995). The modifications present at the anticodon regions of these tRNAs, primarily positions 32, 34 and 37, affect translation (Phelps et al., 2004). For example, we detected mo5U (34) at all the expected family box tRNAs (Ala, Pro, Ser, Thr and Val). This modification is suggested to increase the base wobbling beyond the typical Watson-Crick base pairing, which could expand the codon recognition of these tRNAs.
tRNA composition and concentration in different organisms is a dynamic factor (Dong et al., 1996; Dittmar et al., 2006), which might serve as an additional tool for the cells to optimally adjust protein synthesis to their physiology needs. In rapidly growing E. coli, the levels of some major tRNAs, i.e., those pairing to highly abundant codons, increase compared to suboptimal growth conditions (Dong et al., 1996). This occurs most likely in response to the enhanced translation of abundant proteins that use those major tRNAs. At a four-fold increase in the growth rate of L. lactis, some tRNAs are upregulated 1.5 to 2-fold, but the majority of the tRNAs remain at a constant level. In E. coli the tRNA levels change maximally two-fold for some of the tRNAs over a six-fold difference in the growth rate (Dong et al., 1996): In E. coli tRNAs that correspond to abundant codons increase, while tRNAs cognate to low-abundant tRNAs remain unchanged. In contrast, we find that in L. lactis among the various synonymous codons, some tRNAs cognate to low-abundance codons decrease in abundance as a function of growth rate (the underlined tRNAs in Fig. 1 are those pairing to rare codons). In line with the observations made in E. coli, many abundant tRNAs in L. lactis show an increase ranging from 1.2- to 2-fold with increasing growth rate. The relative proteomic quantification of various aaRS also indicates an increase of maximally 1.5-fold for the majority of the enzymes with increasing growth rate. A study inSalmonella typhimurium and E. coli shows an increase in the specific activity of ArgRS, GlnRS, SerRS and ValRS with growth rate, whereas the activity of SerRS remains constant (McKeever and Neidhardt., 1976). An increase of aaRS rather than tRNA levels suggests that more efficient aminoacylation of the existing tRNA is sufficient to support faster growth.
The overexpression stress caused more variations in the tRNA levels than the variations in the growth rate did. As overexpressing a membrane protein in L. lactis leads to upregulation of chaperones, proteases and a general and cell envelope-specific stress response (Marreddy et al., 2011), changes in tRNA abundance may not be specific to overexpression of a membrane protein alone but rather serve the changed expression of housekeeping genes. And this is what we find. The most significant change in the tRNA concentration was registered for Met-tRNAs (elongator and initiator tRNAs). An increase of the initiator Met-tRNA is indicative of enhanced translation (Pavon-Eternod et al., 2013). The major fraction of the housekeeping genes of L. lactis participates in transcription and translation. Increasing the expression of the corresponding genes could boost the synthesis of proteins, here the membrane transporter OpuA. Another part of the housekeeping genes are involved in the folding and clearance of proteins from the cell. Thus, overexpression of a membrane protein may activate cellular stress response programs, including chaperones to facilitate protein folding and proteases to diminish the burden of any misfolded protein formed.
In summary, we present the first experimental discovery of all predicted tRNAs in L. lactis, including the positions of 16 different posttranscriptional modifications. Although L. lactis has a small tRNA set, in some tRNAs bulky hypermodified nucleosides are present (at positions U34 and A37) that most likely expand the codon specificity of those tRNAs. Membrane protein overexpression stress alters the tRNA abundance significantly, which is independent of the tRNAs used for the synthesis of the target protein. Rather the changes seem to reflect an enhanced synthesis of housekeeping proteins to counteract the stress.
MATERIAL AND METHODS
Strain and growth medium
L. lactis ssp. cremoris MG 1363 was grown on chemically defined medium for prolonged cultivation (CDMPC) containing 25 mM glucose as the limiting nutrient and the medium composition as detailed before (Goel et al., 2012). Glucose-limited chemostat cultures were grown in 2 L bioreactors with a working volume of 1.2 L at 30°C and pH of 6.5 under continuous stirring. L. lactis harbouring plasmid pNZ8048 (de Ruyter et al., 1996), carrying the opuA gene under the control of the nisA promoter, was grown in M17 media in pH-controlled 2 L batch bioreactors. The opuA expression was induced 0.005% (w/v) nisin A for 1 h after the cells reached an OD600 of 1.5. The cells from the chemostats and batch cultures were harvested for tRNA isolation by instantaneous quenching with methanol (40 % v/v) pre-chilled at −40 °C.
Purification of tRNA
Cells were pelleted and washed with suspension buffer (1 mM Tris-HCl pH 7.2 containing 10 mM MgCl2). Next, the cells were mixed with 2 volumes of phenol (pre-saturated with suspension buffer) and lysed by vortexing with glass beads (100 μm diameter) in an ice-cold tissue lyser (Qiagen, Venlo, The Netherlands). The phases were separated by centrifugation at 21,000xg for 15 min at 4°C. The aqueous phase was precipitated with 1 volume of isopropanol plus 0.1 volume of potassium acetate (pH 5) upon incubation at −80°C for 30 min. The RNA pellet was extracted upon centrifugation at 21,000 × g for 30 min at 4°C. The pellet was dissolved in 1 volume of 1 M NaCl and precipitated again using 1 volume of isopropanol upon incubation at −80°C for 30 min. The total tRNA pellet was then deacylated by dissolving it in 0.5 M Tris-HCl (pH 8.8) and incubating the mixture at 37°C for 1h. The total tRNA was precipitated again by adding 1 volume of isopropanol plus 0.1 volume of potassium acetate (pH 5.0). The obtained pellet was washed with 70% ethanol and precipitated again by centrifugation at 21,000 × g for 15 min at 4°C. The RNA was re-suspended in RNAse/DNAse free water.
Analysis of the nucleoside composition of tRNAs
A Nucleobond (Machery Nagel) anion exchange column was used to purify and desalt the tRNA prior to enzyme digestion. The tRNA was loaded onto the column in a solution of 200 mM KCl, 100 mMTris-acetate (pH 6.3) and 15% ethanol then washed with the same solution. For a second wash 400 mM KCl, 100 mM Tris-acetate (pH 6.3) plus 15% ethanol was used and then tRNA was eluted with 750 mM KCl, 100 mM Tris-acetate (pH6.3) containing 15% ethanol. Isopropanol was used to precipitate the tRNA. The purified tRNA was washed with 70 % ethanol and resuspended in nuclease-free water.
Prior to enzymatic digestion, the tRNA was denatured at 100°C for 3 min then chilled in an ice water bath. To lower the pH, 1/10 volume of 0.1 M ammonium acetate (pH 5.3) was added. For each 0.5 absorbance unit (AU) of tRNA, 2 units Nuclease P1 were added and the mixture was incubated at 45°C for 2 h. The pH was readjusted by adding 1/10 volume of 1.0 M ammonium bicarbonate, then 0.002 units of snake venom phosphodiesterase was added and incubated at 37°C for 2 h. Finally, 0.5 units of antartic phosphatase was added and incubated at 37 °C for 1 h. The nucleoside digests were stored at -80°C for further analysis (Crain, 1990).
The nucleoside digests were analysed using a Hitachi D-7000 HPLC system with a Hitachi D-7400 UV detector set at 260 nm. A LC-18-S (Supelco) 2.1 x 250 mm, 5 μm particle column was used at a flow rate of 300 μL/min. Mobile phases used were as follows:5mM ammonium acetate, pH 5.3 and 40% acetonitrile in water. The column eluent was split immediately after the column, 1/3 to the electrospray ion source and 2/3 to the UV detector. The gradient used follows that previously described (Mandal et al., 2010).
A Thermo LTQ-XL ion trap mass spectrometer equipped with an ion max electrospray ionization source was used for the low-resolution LC/MS and LC/MS/MS analyses. Mass spectra were recorded in the positive ion mode with a capillary temperature of 275°C, spray voltage, 3.7 to 4.0 kV and sheath gas, auxiliary gas and sweep gas of 45, 25 and 10 arbitrary units, respectively. Data dependent MS/MS of each of the two most abundant ions were recorded throughout the LC/MS run.
Mass spectrometry analysis of tRNA
tRNA was digested with three different enzymes. tRNA was incubated with 50 U of RNaseT1 (Sigma-Aldrich, St. Louis, MO) per mg of tRNA in 220 mM ammonium acetate for 2 h at 37°C. In parallel, tRNA was incubated with0.2 U of RNase V1 (Ambion, Grand Island, NY) per mg of tRNA in 150 mM ammonium bicarbonate containing 5 mM MgCl2 for 4 h at 37°C. RNase U2 (overexpressed and purified in house, (Martínez-Ruiz et al., 2000)) digestion of tRNA was performed by incubating approximately 0.1 mg of purified protein per mg of tRNA in 220 mM ammonium acetate for 15 min at 55°C. The reactions were then extracted with phenol, chloroform, and evaporated in a vacuum drier before analysis.
Digestion products were separated using an XBridge™ C18 column (150 mm × 1 mm; 3.5μm, 120Å) from Waters (Santa Clara, CA.). Before each run the column was equilibrated for 10 min at 95% Buffer A (400 mM 1,1,1,3,3,3 hexafluoroisopropanol (HFIP), 16.3 mM triethylamine (TEA), pH 7) and 5% Buffer B (400 mM HFIP, 16.3 mM TEA: methanol, 50:50 v:v, pH 7). The optimal mobile phase gradient started at 10 %B and increased at 8 %/min for 10 min. The mobile phase was then increased to 95% for 5 min before re-equilibrating prior to the next analysis. LC/MS/MS analyses were performed using a Hitachi La Chrome Ultra UPLC containing two L-216OU pumps, L-2455U diode array detector and an L-2300 column oven (35°C) connected in-line with a Thermo Scientific (Waltham, MA) LTQ™ linear ion trap mass spectrometer. The mass spectrometer operating parameters included a capillary temperature of 275°C, spray voltage of 4 kV, source current of 100 μA, and sheath, auxiliary and sweep gases set to 40, 10 and 10 arbitrary units, respectively. Each instrumental segment consisted of a full scan range restricted from m/z 600 to 2000 (scan 1), collected in negative polarity, followed by three product ion scans (scans 2-4). Product ion scans were obtained using collision-induced dissociation (CID) at a normalized collision energy 35% with an activation time of 15 ms. In data-dependent mode, scans 2-4 were triggered by the three most abundant ions from scan 1 and isolated by a mass width of 2 (±1 m/z). Each ion selected for CID was analyzed for up to 10 scans before it was added to a dynamic exclusion list for 15 s (typical chromatographic fwhm) for both modes of data acquisition.
Five micrograms of each RNase digestion reaction were analyzed three times using different MS scan ranges including m/z 600-1000, 1000-1500 and 1500-2000. The three scan ranges were used to maximize the number of digestion products which triggered MS/MS (data dependent) scans, thus increasing the chance detecting lower abundant products and yielding the maximum sequence coverage for each tRNA. RNase T1 data was analyzed using a targeted approach, where tRNA modification positions were predicted and the data was filtered searching for the m/z of all expected digestion products based on in silico digestion. If unexpected digestion products were encountered, manual de novo sequencing was performed, based on MS/MS fragmentation. For RNAse V1 and RNase U2 samples, only manual de novo sequencing methods were applicable due to unspecific digestion sites and missed cleavages. In all cases a digestion product was only considered identified if a minimum of 80% sequence coverage was obtained. Once a modified sequence was determined it was then mapped back to the specific tRNA(s) that could contain this modified sequence. A more extensive description of the various mass spectrometry approaches used to map modifications onto the primary tRNA sequences will be the subject of a following paper (Gaston et al., manuscript in preparation).
tRNA library preparation for next generation sequencing (NGS) and analysis
tRNA library for NGS was prepared using the Small RNA sample Prep Kit FC-102-1009 from Illumina. The barcoded adapters were ligated to both ends of the resulting fragments, according the standard Illumina protocol. Fragments with an insert size of 70bp on average were excised using the Invitrogen E-gel system and the extracted DNA was amplified with PCR. After amplification of the enriched products with PCR the quality of the products was verified on the Biorad Experion instrument and sequenced on the HiSeq2000 with 100 bp single reads. Image files were processed using standard Illumina base calling software and the generated reads were ready for downstream processing.
The sequencing reads so obtained were pre-filtered with the following criteria (i) The adapter sequence (TCGTATGCCGTC) was removed if detected with or without maximum of one mismatch, (ii) sequences were truncated if Phred quality of base <20 (i.e. the confidence of nucleotide <99 %), (iii) the length of the read should be greater than or equal to 50, and (iv) at the position of modified bases in the sequenced tRNA, modified nucleotide will match to any of the 4 nucleotide. Reads shorter than 50 nt were discarded.
Reads that aligned to rRNA sequences with less than 3 mismatches were considered as rRNA reads and were removed from the sequencing dataset before the alignment to the tRNA sequences. The detected rRNA percentage reaches a plateau when mismatch was set to >=3, showing that allowing many mismatches cannot detect more rRNA. The remaining reads were aligned to the tRNA reference sequences using modified Smith-Waterman algorithm that considers modified nucleotides, i.e, a modified nucleotide can be matched by all 4 nucleotides from the reference sequences. Zero to three errors (including mismatches, insertions and deletions) was allowed in the alignment. tRNA alignment using different settings for the rRNA removal or direct alignment without pre-filtering of the rRNA reads gave similar results, confirming that the rRNA removal did not discard any tRNA reads. The Spearman correlation coefficient between the two biological replicates spiked-in with the identical tRNAs, S1 and S2, were 0.87, 0.97, 0.98 and 0.98 for mismatch of 0, 1, 2 and 3 respectively, thus suggesting a good correlation for mismatch value varying between 1 and 3. At mismatch of 0, the reproducibility was quite poor, showing unstable sequencing quality. After the modification pattern of the tRNAs was resolved, we realigned the tRNA sequences from the NGS runs to the tRNA reference sequences considering the modified nucleotides, i.e, allowing a modified nucleotide to be matched by all 4 nucleotides from the reference sequences. NGS data has been deposited with the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE55392.
tRNA microarray and analysis
tRNA microarrays were performed with 39 tDNA probes (Supplementary Materials, Table S2) designed for L. lactis. As the two Ser-tRNAUGA differ by only one nucleotide in the body sequence, only one probe was designed to detect both of them. Each microarray consisted of 12 identical blocks with two replicates of each tDNA probe within each block, i.e. 24 samples for each tRNA. The total RNA was extracted using phenol extraction method and deacetylated for 45 min at 37°C with 100 mM Tris-HCl (pH 9.0). Fluorescent stem-loop RNA/DNA oligonucleotide (Dittmar et al., 2005), bearing a Cy3 or Atto 653 fluorescent dye, was ligated to all deacetylated tRNAs overnight at 16°C using T4 DNA ligase (NEB). The ligation efficiency was analyzed by resolving the samples on denaturing 10% PAGE and detected by fluorescence (Fujifilm LAS-4000) and SYBR gold (Invitrogen) staining. tRNAs, differentially labelled with Cy3 and Atto-653, were simultaneously hybridized on the same microarray for 16 h at 60°C, using the Hyb4 microarray hybridization system (Digilab). Subsequently, the microarrays were washed three times in 6x SSC (35°C), once in 2x SSC (30°C) and once in 0.2xSSC (30°C), and then recorded on a GenPIX 4200A scanner (Molecular Devices). For normalization, identical amounts of in vitro synthesized tRNA standards were added to each tRNA mixture prior to deacetylation. Quantification was performed by normalizing the median of the Atto-653 tRNA signal of each tRNA species (here Atto-653 reports the amount of tRNA of the control condition) to the corresponding Cy3-labled tRNA (which gives the amount of tRNA value of the analyzed condition). The significance of the change in tRNA at different growth rates or upon opuA overexpression was assessed with the one-sample t-test test. Each array contains 24 spots for each tRNA, which showed a normal distribution. For each condition two independent biological replicates were analyzed. For each tRNA in each biological replicate the SD was calculated from the signal of all 24 probes on each microarray. tRNAs were designated as significantly up- or downregulated compared to the corresponding control condition when the same trend is observed in both biological replicates with p<0.05.
Microarray data of tRNA expression levels have been deposited with the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE55336.
Sequence and codon usage analysis
From the codon usage table of L. lactis we selected the 10 codons with the lowest usage in the L. Lactis (Cys-tRNATGC, Arg-tRNAAGG, Arg-tRNACGG, Pro-tRNACCG, Pro-tRNACCC, SertRNATCC, Cys-tRNATGT, Ser-tRNATCG, Arg-tRNACGC, His-tRNACAC). Seven of them are read by single tRNA: Arg-tRNACCG for Arg CGG codon, Arg-tRNACCU for Arg AGG codon, CystRNAGCA for both TGC and TGT codons, His-tRNAGUG for His CAC, Ser-tRNAGGA for Ser TCC and Ser-tRNACGA for Ser TCG. Pro-tRNAUGG probe on the microarray pairs to all Pro tRNAs pairs to all four Pro codons, Arg-tRNAACG reads CGC and CGU codons thus Pro CCG, Pro CCC and Arg CGC codons cannot be judged from the microarrays.
Mass spectrometry analysis revealed that seven genes of L. lactis are upregulated and 92 downregulated by OpuA overexpression (Marreddy et al., 2011). The codon usage of these two gene sets was compared to the codon usage of a randomly sampled gene group of the same size. Similarly, we analysed the codon usage of 63 housekeeping genes (including ribosomal proteins, transcription and translation factors, chaperones and proteases). The datasets were correlated using the Spearman- Correlation method as their distribution deviated from a normal distribution. The Wilcoxon-Mann-Whitney test was applied to identify codons that are used differently in the analyzed set.
Supplementary Material
ACKNOWLEDGEMENT
We thank R. Lukoszek (Univ. Potsdam) for help with probes design for the microarrays, E. Ehrentreich-Förster for microarray printing and scanning, and Prof. B. Addepalli (Univ. Cincinnati) for the gift of RNase U2.
FUNDING
This work is supported by the Dutch Technology Foundation STW [08080], which is part of the Netherlands Organization for Scientific Research (NWO) and NWO-TOP GO subsidy [700.10.53 to BP]), the Deutsche Forschungsgemeinschaft [FOR1805 to ZI], the National Science Foundation [CHE0910751 and CHE1212625 to PAL] and the National Institutes of Health [GM58843 to PAL]. BP is additionally supported by the Netherlands Proteomics Centre.
REFERENCES
- Agris PF. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie. 1991;73:1345–1349. doi: 10.1016/0300-9084(91)90163-u. [DOI] [PubMed] [Google Scholar]
- Agris PF. Decoding the genome: a modified view. Nucleic Acids Research. 2003;32:223–238. doi: 10.1093/nar/gkh185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Vendeix FAP, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol. 2007;366:1–13. doi: 10.1016/j.jmb.2006.11.046. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Research. 2008;37:D93–D97. doi: 10.1093/nar/gkn787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crain PF. Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Meth Enzymol. 1990;193:782–790. doi: 10.1016/0076-6879(90)93450-y. [DOI] [PubMed] [Google Scholar]
- Crick FH. Codon--anticodon pairing: the wobble hypothesis. J Mol Biol. 1966;19:548–555. doi: 10.1016/s0022-2836(66)80022-0. [DOI] [PubMed] [Google Scholar]
- Czech A, Fedyunin I, Zhang G, Ignatova Z. Silent mutations in sight: co variations in tRNA abundance as a key to unravel consequences of silent mutations. Mol BioSyst. 2010;6:1767. doi: 10.1039/c004796c. [DOI] [PubMed] [Google Scholar]
- de Ruyter PG, Kuipers OP, de Vos WM. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J Mol Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–1722. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- Frelet-Barrand A, Boutigny S, Kunji E. Membrane protein expression in Lactococcus lactis. Methids Mol Biol. 2010;601:67–85. doi: 10.1007/978-1-60761-344-2_5. [DOI] [PubMed] [Google Scholar]
- Giegé R, Jühling F, Pütz J, Stadler P, Sauter C, Florentz C. Structure of transfer RNAs: similarity and variability. WIREs RNA. 2012;3:37–61. doi: 10.1002/wrna.103. [DOI] [PubMed] [Google Scholar]
- Goel A, Santos F, Vos W.M. de, Teusink B, Molenaar D. Standardized assay medium to measure Lactococcus lactis enzyme activities while mimicking intracellular conditions. Appl Environ Microbiol. 2012;78:134–143. doi: 10.1128/AEM.05276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberg R, Petrov DA. Selection on Codon Bias. Genetics. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- Hossain M, Limbach PA. Mass spectrometry-based detection of transfer RNAs by their signature endonuclease digestion products. RNA. 2006;13:295–303. doi: 10.1261/rna.272507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. WIREs RNA. 2012;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M, Zhang J, Ehrenberg M. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci USA. 2012;109:131–136. doi: 10.1073/pnas.1116480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kudo Y, Ikemura T. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene. 1999;238:143–155. doi: 10.1016/s0378-1119(99)00225-5. [DOI] [PubMed] [Google Scholar]
- Komar A. A pause for thought along the co-translational folding pathway. Trends in Biochemical Sciences. 2009;34:16–24. doi: 10.1016/j.tibs.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Research. 1993;21:4577–4585. doi: 10.1093/nar/21.19.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunji ER, Slotboom DJ, Poolman B. Lactococcus lactis as host for overproduction of functional membrane proteins. ACTA-BIOENERG. 2003;1610:97–108. doi: 10.1016/s0005-2736(02)00712-5. [DOI] [PubMed] [Google Scholar]
- Linares DM, Geertsma ER, Poolman B. Evolved Lactococcus lactis Strains for Enhanced Expression of Recombinant Membrane Proteins. J Mol Biol. 2010;401:11–11. doi: 10.1016/j.jmb.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, et al. MODOMICS: a database of RNA modification pathways--2013 update. Nucleic Acids Research. 2013;41:D262–7. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal D, Köhrer C, Su D, Russell SP, Krivos K, Castleberry CM, et al. Agmatidine, a modified cytidine in the anticodon of archaeal tRNA(Ile), base pairs with adenosine but not with guanosine. Proc Natl Acad Sci USA. 2010;107:2872–2877. doi: 10.1073/pnas.0914869107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marreddy RK, Pinto JPC, Wolters JC, Geertsma ER, Fusetti F, Permentier HP, Kuipers OP, Kok J, Poolman B. The response of Lactococcus lactis to membrane protein production. PLoS ONE. 2011;6:e24060. doi: 10.1371/journal.pone.0024060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Ruiz A, García-Ortega L, Kao R, Oñaderra M, Mancheño JM, Davies J, et al. Ribonuclease U2: cloning, production in Pichia pastoris and affinity chromatography purification of the active recombinant protein. FEMS Microbiol Lett. 2000;189:165–169. doi: 10.1111/j.1574-6968.2000.tb09224.x. [DOI] [PubMed] [Google Scholar]
- Novoa EM, de Pouplana LR. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Pavlov MY, Watts RE, Tan Z, Cornish VW, Ehrenberg M, Forster AC. Slow peptide bond formation by proline and other N-alkylamino acids in translation. Proc Natl Acad Sci USA. 2009;106:50–54. doi: 10.1073/pnas.0809211106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Rosner MR, Pan T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA. 2013;19:461–466. doi: 10.1261/rna.037507.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps SS, Malkiewicz A, Agris PF, Joseph S. Modified nucleotides in tRNA(Lys) and tRNA(Val) are important for translocation. J Mol Biol. 2004;338:439–444. doi: 10.1016/j.jmb.2004.02.070. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol. 2011;18:432–436. doi: 10.1038/nsmb.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söll D, RajBhandary UL. TRNA. 1995 [Google Scholar]
- Steidler L, Rottiers P, Coulie B. Actobiotics as a novel method for cytokine delivery. Ann N Y Acad Sci. 2009;1182:135–145. doi: 10.1111/j.1749-6632.2009.05067.x. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet. 2011;45:299–329. doi: 10.1146/annurev-genet-110410-132531. [DOI] [PubMed] [Google Scholar]
- Sørensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J Mol Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- Takai K, Takaku H, Yokoyama S. Codon-reading specificity of an unmodified form of Escherichia coli tRNA1Ser in cell-free protein synthesis. Nucleic Acids Research. 1996;24:2894–2899. doi: 10.1093/nar/24.15.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Wetzel C, Limbach PA. The global identification of tRNA isoacceptors by targeted tandem mass spectrometry. Analyst. 2013;138:6063–6072. doi: 10.1039/c3an01224g. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth SE, Gorochowski TE, Roubos JA. Translational sensitivity of the Escherichia coli genome to fluctuating tRNA availability. Nucleic Acids Research. 2013;41:8021–8033. doi: 10.1093/nar/gkt602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacoubi El, B., Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- Yacoubi El, B., Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, et al. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Research. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z. Generic algorithm to predict the speed of translational elongation: implications for protein biogenesis. PLoS ONE. 2009;4:e5036. doi: 10.1371/journal.pone.0005036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.