Summary
A modern general theory of sex determination and sexual differentiation identifies the factors that cause sexual bias in gene networks, leading to sex differences in physiology and disease. The primary sex-biasing factors are those encoded on the sex chromosomes that are inherently different in the male and female zygote. These factors, and downstream factors such as gonadal hormones, act directly on tissues to produce sex differences, and to antagonize each other to reduce sex differences. Recent study of mouse models such as the Four Core Genotypes has begun to distinguish between direct effects of sex chromosome complement (XX vs. XY) and hormonal effects. Several lines of evidence implicate epigenetic processes in the control of sex differences, although a great deal of more information is needed about sex differences in the epigenome.
Keywords: sex chromosome, Y chromosome, X chromosome, Four Core Genotypes, sexome, gene networks, hormones
1. Introduction: Why study sex differences?
For much of the 20th century, the study of sex differences focused on large sexual dimorphisms that are functionally related to reproduction. Most investigators in this field wanted to discover the genetic and hormonal factors that cause sex differences in the gonads, external and internal genitalia, and brain. Although the study of sexual differentiation was seen as a subfield of the study of reproduction, these studies served to define basic ideas about the factors that cause sex differences in tissues. Those ideas, and more modern ideas that derive from them, represent a general theory of sexual differentiation, discussed in the next section. In the last 20 years or more, however, the realization has dawned that many tissues and diseases, not overtly related to reproduction, also differ in males and females (Voskuhl, 2011; Ober et al., 2008; Regitz-Zagrosek et al., 2006; Arnold, 2010). This means that the best course of treatment of disease might proceed differently in the two sexes. Moreover, one sex may be protected from a disease, or may experience a milder disease course. In some cases, the protection offered by the individual’s sex can be greater than that offered by drugs or other therapies. The awareness that sex-biasing factors can protect from disease has drawn attention to the need to identify these factors, with the aim of exploiting this knowledge to develop novel targets of therapy. Thus, in increasing numbers, investigators are interested in animal models and conceptual frameworks for designing investigations that will help identify factors that make the two sexes different.
At the same time, there is increasing attention paid to the fact that most experimental subjects in biomedical research have been males, both in clinical and pre-clinical studies (Beery and Zucker, 2011; Taylor et al., 2011). The choice of males is more than just a social bias of the experimenters, who are more often male than female. In animal studies, for example, females have been viewed as more variable than males, because of the changes caused by the estrous cycle. From one perspective, in which physiology is viewed as not likely to be much different between the sexes, it might make sense to study the most experimentally tractable (least variable) sex, with the expectation that the physiology of the male kidney (for example) tells us what we need to know about the physiology of the female kidney. This perspective has two important flaws, the first being that females are not necessarily more variable than males (e.g., see Mogil and Chanda, 2005); and, even if they are, then the female’s physiology is not the same as the male’s. Secondly, independent of the issue of variability, numerous aspects of physiology and disease differ in the two sexes. The non-equivalence of the sexes is a strong argument to shift the balance of studies so that females are studied more than in the past, lest the research in physiology and medicine be relevant only to the male half of the human population.
An important point, however, is that more study of females is not enough. There is also a need to compare the sexes directly. The comparison of the sexes can uncover important questions and answers that would not otherwise be investigated. For example, a comparison of the death rates in the two sexes in humans reveals that males die at a faster rate than females, at nearly every life stage beginning before birth (Migeon, 2007). Without a direct comparison of the sexes, it would not occur to us to seek to explain what protects females, and/or what makes males vulnerable. If a protective factor can be found, then it might be manipulated to prevent deaths in both sexes. A second example, of the advantages of direct comparison of the sexes, is that sometimes understanding the physiology of one sex requires the comparison to the other. For example, comparison of the sexes is required to understand the evolution and function of X-inactivation (transcriptional silencing of one X chromosome in XX cells), a process that occurs in nearly every XX female somatic cell, but never in XY male somatic cells. X-inactivation solves problems that arise when the ratio of expression of X to autosomal genes is different in one sex than the other, because such sexual imbalance would mean that the ratio would be non-optimal in at least one of the sexes (Charlesworth, 1996). The main effect of X inactivation is to reduce the sexual disparity in X to autosome dose, so that X genes are not inherently expressed higher in females (except for some exceptions discussed below)(Itoh et al., 2007; Arnold et al., 2008). If one tried to study X-inactivation only in females, it would be impossible to understand its function. The conceptual importance of the X to autosome ratio becomes quite relevant when attempting to understand the sex-biased impact of the genes that escape X-inactivation, which is discussed below.
2. A General Theory of Sex Determination and Sexual Differentiation
The goal of basic biomedical science is to explain the causal pathways that control physiology and disease. Thus, we envision the function of cells, tissues, and individuals to be controlled by complex intersecting causal pathways, in which specific physical events cause changes in other events. Genes (and their products, RNA and protein) form networks of interactions as they control and are controlled by each other. The gene networks can be thought to be composed of nodes (gene products) that are connected to limited number of other nodes (van Nas A. et al., 2009; Arnold et al., 2009; Arnold and Lusis, 2011). In this analogy, functional gene networks pulsate with activity, with specific nodes increasing and decreasing in their activity, stimulating and inhibiting each other, creating a dynamic net of interactions that lead to emergent phenotypes (such as heart rate, fat and energy metabolism, etc.). Sex differences in gene networks. and in the phenotypes that they control, are created when the activity of some nodes is greater in one sex than in the other; the sex differences in network functions are caused by sex-specific factors acting in the network. The totality of sex-biased factors in the network comprised the sexome (Arnold and Lusis, 2011). A major goal is to identify these sex-biasing factors together with their downstream effects on specific parts of gene networks. These factors, and the downstream gene products that they bias sexually, are candidates for manipulation to mimic sex-specific protection from disease.
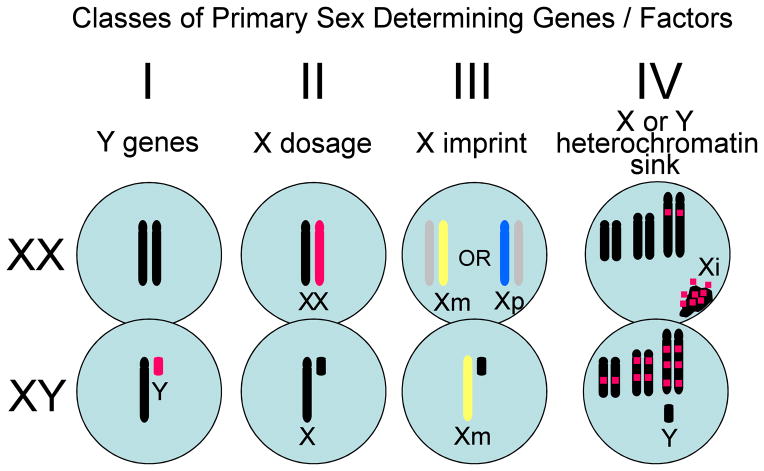
We can distinguish primary sex-determining factors, and secondary factors that are downstream from the primary factors (Arnold, 2009b; Arnold, 2011). The primary factors are encoded by the sex chromosomes, because all sex differences start with the sex chromosomes at some point in life. The sex chromosomes are the only factors that differ in the male and female zygote, and thus they are the factors that give rise to all downstream sex differences thereafter. Four classes of X and Y factors are postulated to comprise the primary sex determining genes (De Vries et al., 2002; Arnold, 2011; see figure 1). Class I are Y genes, which can only have effects in males. Among the Y genes known to be required to make a complete male are the testis-determining gene Sry (Goodfellow and Lovell-Badge, 1993), and several Y genes required for spermatogenesis (Burgoyne and Mitchell, 2007). Class II are X genes that escape X-inactivation and are expressed from both X chromosomes, resulting in constitutively higher expression in XX cells than XY cells. Because X inactivation appears to vary across tissues and age, the number of such X escapees is likely to depend on species, developmental stage, and tissue, but is greater in humans than in mice (Berletch et al., 2010; Carrel and Willard, 2005). Class III are X genes that are expressed at a higher or lower level in XX than XY cells because of a parental imprint on the gene from the mother or father. Parental imprints on X genes are inherently unequal in the two sexes, because XY cells can only express a maternal imprint on imprinted X genes, whereas XX cells can show the effects of a maternal or paternal X imprint depending on which X chromosome is active in a specific cell. The presence of the paternal imprint in about half of the XX cells (when the active X chromosome is from the father) could make XX individuals different from XY. Although some X genes are imprinted (Raefski and O’Neill, 2005; Davies et al., 2005; Gregg et al., 2010), and XO mice and humans differ in their cognitive or social behavior depending on the parent of origin of their X chromosome (Davies et al., 2005; Skuse et al., 1997), there are no established cases yet of a sex difference caused by Class III genes. Class IV is a newly proposed and speculative class, not of specific genes, but of non-coding regions of the sex chromosomes. These are sex chromosome regions that are heterochromatic in one sex more than the other, and which may alter the availability of heterochromatizing factors that regulate gene expression on all chromosomes. The best evidence for sex-specific heterochromatizing effects is in Drosophila, in which the large heterochromatic Y chromosome alters the expression of autosomal genes, not because of any expression of genes from the Y chromosome, but by its effects on the epigenetic status of other chromosomes (Jiang et al., 2010; Lemos et al., 2010). The Y chromosome is also largely heterochromatic but is much smaller in mammals than in Drosophila, but it could theoretically have a male-specific effect of this type, although evidence is lacking at present. In addition, however, XX mammalian cells each possess a heterochromatic inactive X chromosome that is absent in XY cells. It is unknown if these chromosomal regions bias expression from the autosomes, but some evidence argues in favor of this idea (Wijchers and Festenstein, 2011; Wijchers et al., 2010).
Figure 1.
Four classes of primary sex-determining factors that are encoded by the sex chromosomes. Class I are Y genes found only in males. Class II are X genes that escape inactivation and are inherently expressed higher in females than males. Class III are X genes that are imprinted and have a sex-biasing effect because of expression of the paternal imprint only in XX cells. Class IV are putatitve heterochromatic regions on the sex chromosomes (the X chromosome is illustrated here), which act as sinks to sequester heterochromatizing factors from other chromosomes and alter the epigenetic status of autosomes. Reprinted from Arnold, 2011, Trends in Genetics.
Which of the primary sex-determining factors is most important for causing sex differences in downstream pathways and diseases? The prize would have to go to Sry, the Class I Y gene that is turned on in the undifferentiated embryonic gonad, and causes differentiation of testes in males, including the activation of genes that inhibit ovarian differentiation (Koopman, 2010). In the absence of Sry, autosomal and/or X chromosome genes initiate ovarian differentiation in the XX gonad, activating genes that also block testicular differentiation (Chassot et al., 2008; Dinapoli and Capel, 2008). Thus, the molecular switches controlled initially by Sry represent the choice between testicular and ovarian development, and therefore set up a lifelong difference in the secretion of gonadal hormones such as testosterone in males vs. estradiol and progesterone in females. These gonadal hormones act on gene networks and are probably the cause of the large majority of known sex differences in function and disease. The molecular effects of gonadal hormones are diverse and beyond the scope of this review.
The effects of the hormones have historically been lumped into two broad classes, activational and organizational. The acute or activational effects of gonadal hormones are those that are reversible. In animal models, sex differences that are erased by gonadectomy are attributed to the ongoing activational effects of either testicular or ovarian secretions that were removed by gonadectomy. To do the experiment properly in animals, one has to remove the gonads of both sexes to determine if the sex difference is caused entirely by gonadal secretions. In one study, for example, thousands of genes were found to be expressed consistently at different levels in livers from male or female mice. After removing the gonads, virtually all of these differences disappeared, suggesting the most sex differences in adult mouse liver are caused by activational effects (van Nas A. et al., 2009). Sometimes, however, sex differences persist when comparing gonadectomized males and females. Males castrated in adulthood, for example, continue to have male genitalia that differ from those of the female, and structural sex differences in the brain. In many cases, these differences are caused by the long-lasting or permanent organizational effects of gonadal hormones (Arnold and Gorski, 1984). Masculine differentiation of the genitalia and brain is caused largely by the effects of testosterone on the fetus and neonate, which last for the rest of the male’s life and differentiates the male from the female. Although the dichotomy between activational and organizational effects has a long history (Phoenix et al., 1959; Arnold and Breedlove, 1985; Arnold, 2009b), the effects more likely lie along a continuum, with some steroid hormone effects lasting longer than others, after the level of hormone declines. Exposure of male hamsters to androgens at the time of puberty, like the fetal and neonatal exposure to testosterone, also has long-lasting effects that can be classified as organizational (Schulz et al., 2009).
Although gonadal steroids (downstream of Sry’s effect on the gonads) are the dominant factors causing sex differences in physiology and disease, the Class I–IV sex chromosome factors also act, via molecular pathways that are not mediated by gonadal hormones, to bias the function of XX and XY cells. These “sex chromosome effects” have been uncovered predominantly in the last 25 years. In a few cases, sex differences have been discovered that occur before the differentiation of gonads, which are therefore not explained by sex differences in gonadal hormones (Renfree and Short, 1988; Burgoyne et al., 1995; Bermejo-Alvarez et al., 2011; Dewing et al., 2003). Particularly intriguing is the finding that sex differences in the size and other traits of mammalian embryos exist well before the differentiation of the gonads, even before implantation of the embryo. In bovine blastocysts, nearly a third of all genes measured show sex differences in the level of expression, which probably is the result of the higher expression of X genes in females (Bermejo-Alvarez et al., 2010). The generally higher expression of X genes in females than males is probably due the incomplete inactivation of one X chromosome each XX cell of the inner cell mass (the precursor of the embryo proper), so the X genes as a group show higher expression in females (mostly Class II effects, figure 1). In turn, the sex difference in expression of X genes causes sex differences in expression in some autosomal genes. Thus, the mammalian embryo appears to pass through a stage of considerable sexual dimorphism in gene expression, prior to random X-inactivation. It is not known if long term sex-specific effects might be caused by this sexual inequality. Once X inactivation has occurred, the X genes show about the same amount of sex difference globally as shown by the autosomal genes (Itoh et al., 2007) (Figure 2).
Figure 2.
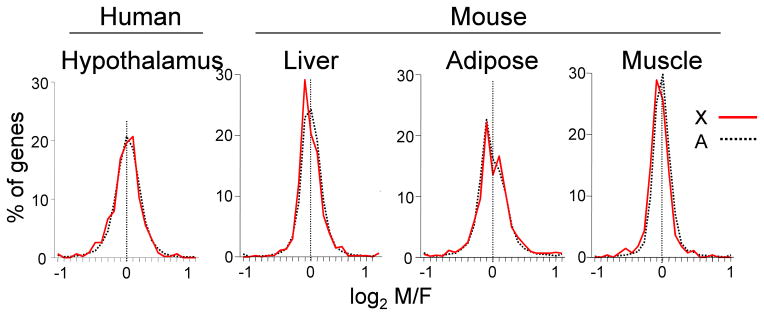
Sex differences in the mammalian transcriptome. Data from microarray profiling are illustrated. Histograms show the distribution of M?F ratios of expression of all genes measured, including autosomal genes (black, dotted line) and X chromosome genes (red). In each tissue, about the same number of genes are expressed higher in males than females, and most sex differences are well below two-fold. X inactivation is effective in preventing higher expression of most X genes in females. Although the amount of sexual dimorphism (width of the histograms) differs across tissues, the degree of sexual bias in X genes is matched, tissue for tissue, to the sexual bias of autosomal genes, presumably because they interact with each other in gene networks. Reprinted from Itoh et al., (2007).
3. Mouse Models for Uncovering the Origins of Sex Differences
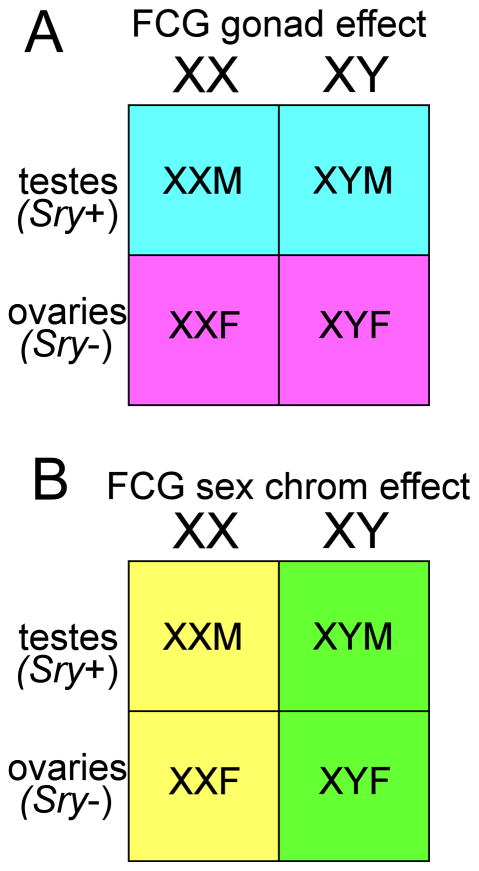
The appreciation of the role of sex chromosome effects has grown steadily in the last 10 years because of the development of mouse models that allow one to separate them from the effects of the gonads (Arnold, 2009a). These models are of three types. (1) One approach has been is to manipulate the expression of Sry directly in non-gonadal tissues, to demonstrate or suggest a male-specific effect of this gene that is independent of Sry’s differentiating effect in the testes (Dewing et al., 2006; Turner et al., 2011). Similarly, the manipulation of other Y genes in the germ line has demonstrated the importance of several Y genes for specific stages of spermatogenesis (Mazeyrat et al., 2001; Vernet et al., 2011; Burgoyne and Mitchell, 2007). The same approach could also work for other class I–IV genes or factors but these experiments have not yet been performed. (2) A second approach is to study mice that lack gonads, to find sex differences that occur in the complete absence of gonadal secretions (but not necessarily in the absence of sex steroid hormones synthesized outside of the gonads). So far this approach has been reported for mice with a null mutation of Sf1 (steroid factor 1). Sf1 is required for gonadal and adrenal differentiation, and thus Sf1 knock out mice are born also without both tissues. The neonates are kept alive by treating them with glucocorticoids to reverse the adrenal insufficiency and then transplanting an adrenal. Such mice show some sex differences in the hypothalamus, which cannot have been caused by gonadal secretions (Majdic and Tobet, 2011; Budefeld et al., 2008). (3) The third approach is the use of Four Core Genotypes (FCG) mice and related models (Arnold and Chen, 2009). In FCG mice, the Y chromosome lacks Sry because of a small deletion of a portion of the Y chromosome thought to disrupt only the Sry locus (Lovell-Badge and Robertson, 1990). This Y chromosome is designated Yminus (Y−). The loss of Sry is complemented by the insertion of an Sry transgene onto an autosome (Mahadevaiah et al., 1998). Thus, XY− mice with the autosomal Sry transgene (called XYM here) are functional gonadal males, whereas XY− mice lacking the Sry transgene are gonadal females (called XYF here). Mating XYM to normal XX gonadal females (XXF) gives four core genotypes, XYM, XYF, XXF, XXM (figure 3). The four genotypes are a 2 X 2 comparison that varies sex chromosome complement (XX vs. XY) independently of presence / absence of Sry (Figure 3). This model is useful for dissecting differences between XX and XY mice caused by gonadal hormones and sex chromosome complement. For example, when a phenotype is measured in the four groups, and the two groups with Sry (gonadal males) are similar to each other but both differ from the two groups without Sry (gonadal females), which are also similar to each other, then we conclude that the difference is caused by Sry. This Sry effect is most likely the result of different effects of gonadal hormones (i.e., different effects of testicular hormones in mice with Sry vs. ovarian hormones in mice without Sry), but could also be caused by a direct effect of Sry on some other tissue. If, on the other hand, the two XX groups are similar to each other, but both are different from the XY groups that are also similar to each other, then we conclude that the sex chromosomes have contributed to the sex difference. In the second example, because the two XX groups are similar to each other, and the two XY groups are similar to each other, there is no apparent effect of gonadal type. Because the FCG model shows effects of gonadal type and also of sex chromosome complement, it offers the possibility of measuring interactions between the two variables, for example when a gonadal effect is different in XX than XY, or when the difference between XX and XY differs in gonadal males and females.
Figure 3.
The Four Core Genotypes Model. When XY gonadal males (XYM) are mated to XX gonadal females, the offspring comprise four genotypes: XX gonadal females and males (XXF, XXM both without Sry), and XY gonadal males and females (XYF, XYM, both with Sry). The four genotypes represent a 2 X 2 comparison of the effects of gonadal sex (comparing mice with testes vs. mice with ovaries) and the effects of sex chromosome complement (XX vs. XY). A, When a sex difference is caused by gonadal hormones, the two groups of mice with testes differ from the two groups with ovaries, irrespective of their sex chromosome complement. B, When the sex difference is caused by sex chromosome complement, then the two groups of XX mice differ from the two XY groups, irrespective of their type of gonad. Not diagramed are cases in which the two factors interact.
By the late 20th Century, the dominant theory of sexual differentiation stated that all sex differences in mammals, outside of the gonad, were caused by gonadal secretions (Arnold and Gorski, 1984). At the time of the emergence of the FCG model, some evidence argued that sex chromosome complement could also produce sex differences (Arnold, 1996; Arnold and Burgoyne, 2004). When the FCG model was used to study non-gonadal tissues such as brain and behavior, the results confirmed that several classic sex differences in the brain and behavior were differentiated by organizational effects of gonadal secretions (figure 4). For example, male patterns of sexual behavior were found in mice that developed with testes, not mice with ovaries, irrespective of whether they were XX or XY. Thus, the FCG model underscored the dominant effects of gonadal hormones. In addition, study of FCG mice has uncovered a growing number of cases in which XX mice differ from XY mice, irrespective of their gonadal sex. In these studies, the FCG mice have been gonadectomized as adults, and treated with equal amounts of testosterone or with nothing. The point of gonadectomizing the mice is that at the time of testing, the levels of gonadal hormones are equivalent across groups, and group differences in phenotype cannot be attributed to activational effects of hormones. Under these conditions, when effects of Sry are found (differences between mice that previously had ovaries vs. testes), they are most likely caused by effects of gonadal secretions that persisted after gonadectomy, for example organizational effects of gonadal hormones. Because of the novelty of the sex chromosome effects, which were not acknowledged by most 20th century theories of sexual differentiation, the published studies using FCG mice have emphasized cases in which XX and XY mice have a different phenotype or different susceptibility to disease.
Figure 4.
Representative differences among FCG mice. FCG mice were gonadectomized as adults, and then implanted with equal amounts of testosterone (left) or nothing (right). On the left is graphed the number of neurons in the spinal nucleus of the bulbocavernosus (SNB). Gonadal males have more neurons than gonadal females, irresective of their sex chromosome complement, indicating that this sex difference is dominantly controlled by gonadal hormones. To measure nociception (right), the mice were place on a hot plate and the latency to lick the paws was measured. XX mice responded more slowly than XY mice, irrespective of their previous gonadal status, indicating that the complement of sex chromosomes causes the difference. SNB data from De Vries et al., 2002, and nociception data from Gioiosa, Chen, et al., (2008a).
4. Examples of sex chromosome effects on phenotype
Several mouse models of disease show sex differences that are controlled in part by sex chromosome complement. These include models of autoimmune disease, viral infections, neural tube closure defects, and hypertension.
4.1 Autoimmuine disease
In humans, autoimmune diseases, such as multiple sclerosis and systemic lupus erythematosus (SLE), affect females much more than males. A similar predominance of multiple sclerosis in females also occurs in Experimental Autoimmune Encephalomyelitis (EAE), a rodent model of multiple sclerosis. Although androgens are protective in EAE, and therefore account in part for the sex difference (Voskuhl, 2011), XX mice are affected by EAE much more than XY mice, irrespective of the type of gonad the mice have (Palaszynski et al., 2005; Smith-Bouvier et al., 2008). Similarly, in a mouse model of SLE, XX mice die sooner and show other signs of greater disease than XY mice. In both models, various immune system markers are influenced by the disease differently in XX vs. XY mice. Differences in sex chromosome number and X chromosome dose have also been suggested to influence the incidence of SLE in humans (Scofield et al., 2008).
4.2 Viral infection
One study examined sex differences in two types of viral infections in mice, cocksackie virus and influenza A (Robinson et al., 2011a). Both showed sex differences, caused at least in part by gonadal hormones (Robinson et al., 2011b). In addition, however, after gonadectomy XX mice showed greater myocarditis than XY mice in response to cocksackie infection, irrespective of their gonadal type, and the protection in XY mice correlated with increased activation of regulatory T cells and expression of CD4+ forkhead box P3 mRNA.
4.3 Neural tube closure defects
In human populations, anterior neural tube closure defects influence females more than males. Numerous mouse knockouts produce problems of neural tube closure, presumably because this closure is a tightly regulated and complex process involving many gene pathways (Harris and Juriloff, 2007). Sex differences are found in some mouse models, including knockout of the tumor suppressor gene Trp53. Mice lacking functional Trp53 at birth are almost all males, because of the pre- or neonatal mortality of females that lack this gene, associated with severe neural tube closure defects. In FCG mice, the sex difference in mortality and neural tube closure is an effect of sex chromosome complement -- XX mice are affected more than XY mice irrespective of their gonadal type (Chen et al., 2008). Comparison of mice with two X chromosomes to mice with one X chromosome shows that the sex difference is caused by X chromosome number, not by the presence / absence of Y genes. That finding indicates that Class II–IV primary sex-determining factors are implicated in the sex differences (Figure 1).
4.4 Hypertension
In humans, hypertension is more prevalent in males than females. In rodent models, the sex difference is explained in part by effects of gonadal hormones, but recent evidence suggests that direct effects of sex chromosome genes also play a role. When FCG mice are treated with angiotensin II, either chronically or acutely, the changes in blood pressure and heart rate are different in XX and XY mice irrespective of their type of gonad (Ji et al., 2010; Caeiro et al., 2011). At this point, it is not clear which of the four sex chromosome factors (figure 1) are responsible for the sex chromosome effects, but in the FCG model the XX vs. XY difference cannot be attributed to the direct effects of Sry. In other studies, however, Sry is proposed as a regulator of hypertension because of its effects on the renin-angiotensin system and sympathetic nervous system (Turner et al., 2011; Ely et al., 2010). Sry is expressed in numerous non-gonadal tissues (Turner et al., 2011), and overexpression of Sry in adrenal or kidney causes an increase in blood pressure. Molecular studies in vitro indicate that expression of Sry leads to changes in promoter activity of tyrosine hydroxylase and genes in the renin-angiotensin related pathways. These intriguing studies suggest a role for male-specific expression of Sry as one mechanism leading to sex differences in regulation of blood pressure, but further studies are needed to down-regulate Sry expression in vivo to show that Sry normally plays this role.
4.5 Behavioral and brain phenotypes
FCG and Sf1 KO mice have also contributed significantly to the conclusion that sex chromosome complement contributes to sex differences in normal physiology and behavior of mice. Many studies have been done on the brain, and show that gene expression is different in XX and XY mice irrespective of gonadal type (or despite the complete absence of gonads). Genes that have been studied include vasopressin in the lateral septum (De Vries et al., 2002; Gatewood et al., 2006), and expression of nitric oxide synthase and calbindin in the hypothalamus, and prodynorphin in the striatum (Abel et al., 2011; Chen et al., 2008; Xu et al., 2005). In addition, neural expression of several Class II X genes is higher in XX than XY mice, including two histone demethylases (Xu et al., 2008a; Xu et al., 2008b; Chen et al., 2009). In at least one case, prodynorphin expression in the striatum, subsequent studies showed that the expression is regulated by X genes rather than Y genes (Classes II–IV) (Chen et al., 2009). Numerous studies of behavior show differences between XX and XY mice: in formation of habits related to models of alcohol abuse and drug addiction (Quinn et al., 2007; Barker et al., 2010), in aggressive and parenting behaviors (Gatewood et al., 2006), behavioral measures of nociception (Gioiosa, Chen et al., 2008a b), and investigative and social behaviors (McPhie-Lalmansingh et al., 2008; Cox and Rissman, 2011). In studies not using FCG mice, Sry has been found to be expressed in the substantia nigra of the midbrain, in neurons that project to the striatum (Dewing et al., 2006). These neurons die in Parkinson’s Disease, which affects males more than females. When Sry expression is experimentally reduced in the brain of adult male mice or rats, the motor behavior of the mice is degraded, and tyrosine hydroxylase is reduced. The deficits are reversed once Sry expression is restored. Thus, evidence suggests that there is a direct male-specific effect of Sry on the brain that is not mediated by its effects on gonadal differentiation.
5. Practical Approaches to the Study of Sex Differences in Physiology and Disease
As discussed above, the current theory of sexual differentiation suggests that the X and Y chromosomes harbor numerous genes that are the primary factors causing sexual differentiation, because these factors are inherently unequally represented in XX vs. XY zygotes (Arnold, 2011). These primary factors act to cause numerous sex differences in downstream genes and pathways that they regulate. Ultimately these downstream pathways intersect and interact with each other. In some cases, two sex-biased factors might inhibit each other, which would tend to make the sexes more similar rather than different. The general goal of research on sex differences is to identify the sex-biased factors that explain sex differences in physiology and disease, which involves studying the primary and downstream pathways.
Becker et al. (Becker et al., 2005) have suggested a practical experimental approach to finding the sex-biased factors and their downstream products in animal models. Because most sex differences may be caused by activational effects of gonadal hormones, a logical first experiment is to gonadectomize adult animals to determine if the sex difference is abolished. If it is, then one would investigate which gonadal hormones in adulthood were responsible for the sex difference, and investigate downstream pathways modulated by those hormones. If sex differences are still found in gonadectomized animals that have equivalent levels of gonadal hormones, then it is likely that organizational effects of gonadal hormones, or differences in sex chromosome complement, cause the residual sex difference. Organizational effects of gonadal hormones can be tested by manipulating the levels of gonadal hormones at early developmental stages before and/or after birth. In studies of the brain, one would likely start with manipulations of testosterone (or its metabolite estradiol), which is the main hormone causing masculinizing organizational effects in rodent model (McCarthy and Arnold, 2011). FCG mice are a good choice for screening for direct effects of sex chromosome complement, except that this model does not test for a direct effect of Sry that is independent of its effect on the gonads. If sex chromosome effects are found, then one independently manipulates the number of X and Y chromosomes to determine if the sex chromosome effect is due to X or Y genes (e.g., Chen et al., 2008; 2009)(Classes I–IV, figure 1). Ultimately the goal is to find the individual X or Y genes, and understand their physiological effects within specific tissues.
6. The role of epigenetics in sexual differentiation
The recent explosion in study of epigenetics has several important effects on the study of sex differences. Historically the genetic control of phenotype has concentrated on variations in phenotype caused by variations in the primary genetic sequence. Variation is also induced by transient and long-lasting epigenetic changes that alter the compaction and loosening of DNA and chromatin, which include processes such as methylation of cytosines in the primary DNA sequence, or changes in the chromatin because of various modifications (acetylation, methylation, ubiquitination, sumoylation, etc.) of specific amino acids in the histone tails comprising the histone octamer around which DNA is wrapped. The epigenetic modifications are fundamental to any biological process, so it is not surprising that they are increasingly studied in the context of sexual differentiation. Particularly intriguing is the finding that epigenetic modifications can last a long time, such that changes early in development can alter the phenotype of the animal much later in life (Zhang and Meaney, 2010). Some epigenetic modifications last across generations, and can influence subsequent generations (Guerrero-Bosagna and Skinner, 2011; Morgan and Bale, 2011). The persistence of epigenetic modifications makes this mechanism an attractive candidate mechanism for explaining some long-lasting effects of gonadal hormones, for example organizational effects exerted early in development. Accordingly, several groups have begun to analyze epigenetic parameters using research designs that compare the sexes or manipulate hormones at different times of life to determine if steroid hormones have short- or long-lasting influences on the epigenome (McCarthy et al., 2009; Auger and Auger, 2011).
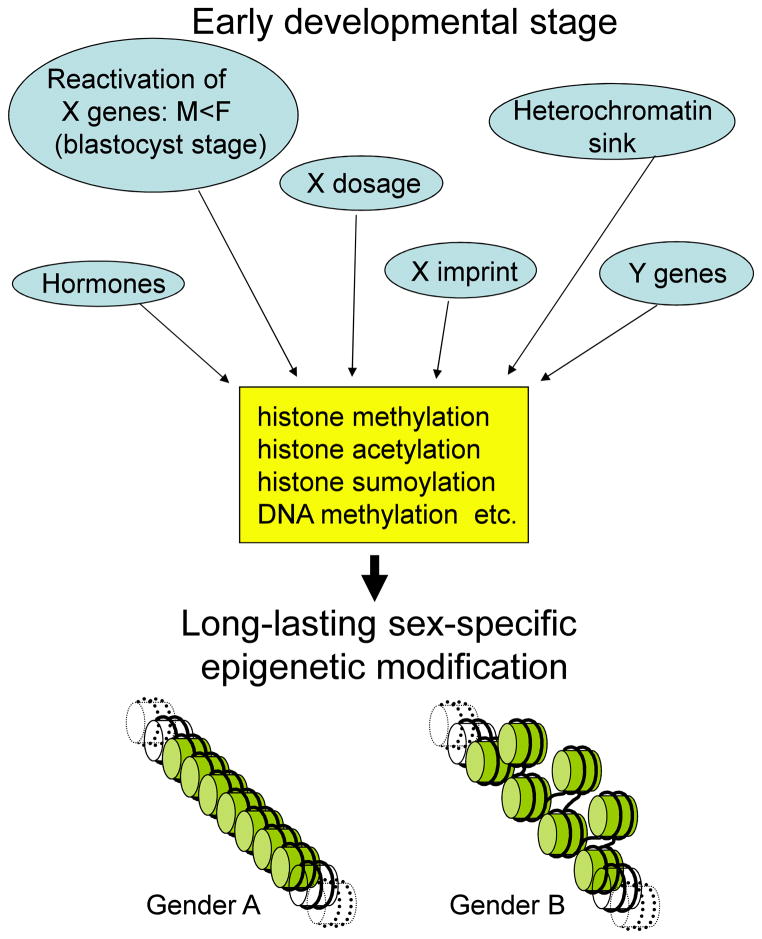
Several considerations support the importance of these epigenetic modifications (figure 5).
Figure 5.
Summary of possible sex-specific epigenetic modifications that could influence chromatin status and gene expression in a gender-specific manner.
The mechanism of action of gonadal hormones involves modification of histones. Sex steroid hormones bind to nuclear receptors (androgen or estrogen receptors, for example), which bind to hormone response elements in the DNA and attract various cofactors that have inherent histone acetyltransferase or methyltransferase activity. The histone-modifying enzymes alter the epigenetic state of gene promoters to which the nuclear receptors bind, and change gene expression (Fu et al., 2004; Leader et al., 2006; Green and Carroll, 2007; Auger et al., 2011). Nevertheless, more information is needed to understand where in the genome these changes occur, when in life, and how long they persist,
The list of sex chromosome signals that are inherently unequal in most male and female cells (figure 1) includes genes that are histone demethylases, Kdm5c and Kdm6a (Xu et al., 2008a b). These X-linked genes escape X-inactivation, are expressed widely in many cell types, and are often expressed higher in XX cells than XY cells. They are members of Class II of putative sex-biasing factors (figure 1). Because of their histone demethylase activity, these genes could have widespread sex-biasing roles in different tissues or stages of development, but the phenotypes influenced by these genes are just beginning to be described. Kdm5c is implicated in tumor suppression and mental retardation (Santos-Reboucas et al., 2011; Niu et al., 2011), whereas Kdm6a is involved in oncogenesis (Kristensen et al., 2011). However, neither gene has yet to be implicated in the sex bias of an emergent phenotype.
In some brain regions, the two sexes differ in patterns of acetylation or methylation of histones by several days before birth, indicating that sex-biased signals have already impacted the brain epigenome by that stage (Tsai et al., 2009; Matsuda et al., 2011). The sex differences are dynamic in this period, with some sex differences appearing and disappearing in the course of a few days (Matsuda et al., 2011). In one study, treatment of female embryos with testosterone masculinized the pattern of acetylation of histone 3 measured at birth, but did not sex-reverse the pattern of methylation of histone 3 (Matsuda et al., 2011). Thus, diverse sex-biased signals, including testosterone secreted by the male, appear to sexually differentiate histone modifications during the perinatal period. Administration of valproic acid, an inhibitor of histone deactylase, at the time of birth, alters acetylation of histone 3 and blocks testosterone-dependent masculine development of the bed nucleus of stria terminalis (Murray et al., 2009; 2011). Knock-down of histone deacetylases in the mating-control regions of the preoptic area in rats, inhibited the adult expression of a sexually differentiated behavior, male copulatory behavior (Matsuda et al., 2011).
Several studies have examined the patterns of methylation of promoters of interesting relevant genes in several brain regions that are known to be sexually differentiated (masculinized) by testosterone (or estradiol, which is a natural active metabolite of testosterone mediating masculinization of the neonatal male’s brain) (McCarthy et al., 2009; Nugent and McCarthy, 2011). These studies have found sex differences in the patterns of methylation of cytosines during the critical period for brain masculinization (Edelmann and Auger, 2011), and have shown that some but not all of these sex differences can be reversed by treating females with estradiol. That result indicates that neonatal levels of estradiol, which are known to have permanent masculinizing effects, also alter the epigenetic status of some genes. Because some of the methylation patterns are not sex-reversed by treating females with estradiol, it is possible that other sex-biasing signals (e.g., sex chromosome complement) may cause the observed sex differences. The sex differences in pattern of methylation are complex in that they differ according to gene, brain region, age of development, and sometimes according to the lab performing the study. Most of the gene-specific patterns that are masculinized by estradiol are not found to persist into adulthood, so that it is not yet clear how the sex-biased alterations of the neonatal epigenome contribute to the long-term development of permanent sex differences in brain function (Schwarz et al., 2010; Nugent et al., 2011). Even when permanent sex differences are found, an important question is whether sex-biased signals such as estradiol cause specific changes in the neonatal epigenome which then change brain function for much of the animal’s life, or if the estradiol causes structural and functional differences (e.g., modifying the distribution of cell types in a tissue) which brings with them long-term changes in the epigenome. Many of the sex differences and estradiol-induced changes, in methylation of specific gene promoters, are measured as relatively small differences in methylation. When a specific cytosine is found to be methylated a few percent more often in one sex than the other, it is not clear how large a functional impact would be expected on gene function. All in all, this field is in its infancy. Only a tiny part of the epigenome has been studied in the context of sex differences, in a few brain regions, so we can expect a great deal of work to clarify these issues in the future (McCarthy et al., 2009; Nugent and McCarthy, 2011).
6. Conclusions: Understanding the Sexome
To understand the sexome, the aggregate sex-biasing actions that change cellular systems, the following steps are important: (1) Identify the inherent primary genetic sex-biasing factors, encoded by the sex chromosomes, that initiate the process of sexual differentiation. Figure 1 summarizes four possible categories of primary sex-biasing factors, which act in parallel to cause sexual differentiation. (2) Identify the secondary proximate factors, often downstream of the action of primary factors, that act on specific molecular networks to cause the networks to function differently in males and females. (3) Identify which molecular network components are sexually dimorphic, and how the influence of sex-biasing factors is propagated throughout the network. Once the sex-biasing process is understood, it may well be possible to find sex-biased factors that protect from disease, and target those factors to develop new therapies
The study of sex differences in reproductive tissues in the last 100 years has given rise to a general theory of sexual differentiation, which provides a conceptual framework for approaching the study of sex differences, as well as experimental strategies and animal models for recognizing and deconstructing the sexome. The vast majority of studies on sexual differentiation of non-gonadal tissues has involved the manipulation of gonadal hormones, which are the most potent proximate factors controlling sexual differentiation. In the past decade, however, animal models have been investigated that allow the study of sex chromosome effects (differential effects of an XX vs. XY genome) independent of the action of gonadal hormones. The Four Core Genotypes mouse model, for example, produces XX and XY mice, each with testes or ovaries. Under some conditions, XX and XY mice differ from each other, not because of differences in gonadal secretions. Those results indicate that the constitutive sexual bias in X and Y genes contributes to sexual differentiation of cells. The imbalance of X and Y genes are both important, indicating that multiple primary sex-biasing factors are encoded in the sex chromosomes, and these act in parallel to cause sex differences. The modern theory of sexual differentiation, therefore, envisions multiple sex-biasing signals that act in parallel but also interact with each other, such that multiple factors can sum with each other, or counteract each other to buffer and reduce the individual effect of any one sex-biasing factor. Thus, understanding the sexome involves unraveling numerous downstream pathways and figuring out where and how cellular systems are impacted.
Take home messages.
Multiple primary sex-determining genes are found on the sex chromosomes, including Sry.
The proximate factors causing sex differences in physiology and disease are primary acute (activational) effects of gonadal hormones, organizational (long-lasting) effects of gonadal hormones, and sex chromosome effects not mediated by gonadal hormones.
The Four Core Genotypes mouse model is useful for dissecting the sex-biasing effects of gonadal hormones and of sex chromosome complement.
Numerous sex differences in physiology and disease can now be traced to the different effect of XX vs. XY sex chromosomes.
Numerous hormonal and non-hormonal mechanisms likely alter the epigenome and regulate sex differences via epigenetic mechanisms.
Reference List
- Abel JM, Witt DM, Rissman EF. Sex Differences in the Cerebellum and Frontal Cortex: Roles of Estrogen Receptor Alpha and Sex Chromosome Genes. Neuroendocrinol. 2011;93:230–40. doi: 10.1159/000324402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. Genetically triggered sexual differentiation of brain and behavior. Horm Behav. 1996;30:495–505. doi: 10.1006/hbeh.1996.0053. [DOI] [PubMed] [Google Scholar]
- Arnold AP. Mouse models for evaluating sex chromosome effects that cause sex differences in non-gonadal tissues. J Neuroendocrinol. 2009a;21:377–386. doi: 10.1111/j.1365-2826.2009.01831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm Behav. 2009b;55:570–578. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. Promoting the understanding of sex differences to enhance equity and excellence in biomedical science. Biol Sex Differ. 2010;1:1. doi: 10.1186/2042-6410-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP. The end of gonad-centric sex determination in mammals. Trends Genet. 2011 doi: 10.1016/j.tig.2011.10.004. Epub Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Breedlove SM. Organizational and activational effects of sex steroid hormones on vertebrate brain and behavior: a re-analysis. Horm Behav. 1985;19:469–498. doi: 10.1016/0018-506x(85)90042-x. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Burgoyne PS. Are XX and XY brain cells intrinsically different? Trends Endocrinol Metab. 2004;15:6–11. doi: 10.1016/j.tem.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Chen X. What does the “four core genotypes” mouse model tell us about sex differences in the brain and other tissues? Front Neuroendocrinol. 2009;30:1–9. doi: 10.1016/j.yfrne.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Gorski RA. Gonadal steroid induction of structural sex differences in the CNS. Annu Rev Neurosci. 1984;7:413–442. doi: 10.1146/annurev.ne.07.030184.002213. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Itoh Y, Melamed E. A bird’s-eye view of sex chromosome dosage compensation. Annu Rev Genomics Hum Genet. 2008;9:109–127. doi: 10.1146/annurev.genom.9.081307.164220. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Lusis AJ. Understanding the sexome. Endocrinol. 2011 doi: 10.1210/en.2011-2134. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, van NA, Lusis AJ. Systems biology asks new questions about sex differences. Trends Endocrinol Metab. 2009;20:471–476. doi: 10.1016/j.tem.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger AP, Auger CJ. Epigenetic turn ons and turn offs: chromatin reorganization and brain differentiation. Endocrinol. 2011;152:349–353. doi: 10.1210/en.2010-0793. [DOI] [PubMed] [Google Scholar]
- Auger CJ, Coss D, Auger AP, Forbes-Lorman RM. Epigenetic control of vasopressin expression is maintained by steroid hormones in the adult male rat brain. Proc Natl Acad Sci U S A. 2011;108:4242–4247. doi: 10.1073/pnas.1100314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JM, Torregrossa MM, Arnold AP, Taylor JR. Dissociation of genetic and hormonal influences on sex differences in alcoholism-related behaviors. J Neurosci. 2010;30:9140–9144. doi: 10.1523/JNEUROSCI.0548-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JB, Arnold AP, Berkley KJ, Blaustein JD, Eckel LA, Hampson E, Herman JP, Marts S, Sadee W, Steiner M, Taylor J, Young E. Strategies and methods for research on sex differences in brain and behavior. Endocrinol. 2005;146:1650–1673. doi: 10.1210/en.2004-1142. [DOI] [PubMed] [Google Scholar]
- Beery AK, Zucker I. Sex bias in neuroscience and biomedical research. Neurosci Biobehav Rev. 2011;35:565–572. doi: 10.1016/j.neubiorev.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Alvarez P, Rizos D, Lonergan P, Gutierrez-Adan A. Transcriptional sexual dimorphism during preimplantation embryo development and its consequences for developmental competence and adult health and disease. Reproduction. 2011;141:563–70. doi: 10.1530/REP-10-0482. [DOI] [PubMed] [Google Scholar]
- Bermejo-Alvarez P, Rizos D, Rath D, Lonergan P, Gutierrez-Adan A. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci U S A. 2010;107:3394–3399. doi: 10.1073/pnas.0913843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budefeld T, Grgurevic N, Tobet SA, Majdic G. Sex differences in brain developing in the presence or absence of gonads. Dev Neurobiol. 2008;68:981–995. doi: 10.1002/dneu.20638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Mitchell MJ. The role of mouse Y chromosome genes in spermatogenesis. In: Lau YFC, Chan WY, editors. Y Chromosome and Male Germ Cell Biology. Hackensack NJ: World Scientific Publishers; 2007. pp. 27–45. [Google Scholar]
- Burgoyne PS, Thornhill AR, Boudrean SK, Darling SM, Bishop CE, Evans EP. The genetic basis of XX-XY differences present before gonadal sex differentiation in the mouse. Philos Trans R Soc Lond B Biol Sci. 1995;350:253–260. doi: 10.1098/rstb.1995.0159. [DOI] [PubMed] [Google Scholar]
- Caeiro XE, Mir FR, Vivas LM, Carrer HF, Cambiasso MJ. Sex Chromosome Complement Contributes to Sex Differences in Bradycardic Baroreflex Response. Hypertension. 2011;58:505–11. doi: 10.1161/HYPERTENSIONAHA.111.175661. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Chassot AA, Gregoire EP, Magliano M, Lavery R, Chaboissier MC. Genetics of ovarian differentiation: Rspo1, a major player. Sex Dev. 2008;2:219–227. doi: 10.1159/000152038. [DOI] [PubMed] [Google Scholar]
- Chen X, Grisham W, Arnold AP. X chromosome number causes sex differences in gene expression in adult mouse striatum. Eur J Neurosci. 2009;29:768–776. doi: 10.1111/j.1460-9568.2009.06610.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Watkins R, Delot E, Reliene R, Schiestl RH, Burgoyne PS, Arnold AP. Sex difference in neural tube defects in p53-null mice is caused by differences in the complement of X not Y genes. Dev Neurobiol. 2008;68:265–273. doi: 10.1002/dneu.20581. [DOI] [PubMed] [Google Scholar]
- Cox KH, Rissman EF. Sex differences in juvenile mouse social behavior are influenced by sex chromosomes and social context. Genes Brain Behav. 2011;10:465–472. doi: 10.1111/j.1601-183X.2011.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37:625–629. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci. 2002;22:9005–9014. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewing P, Chiang CWK, Sinchak K, Sim H, Fernagut PO, Kelly S, Chesselet MF, Micevych PE, Albrecht KH, Harley VR, Vilain E. Direct regulation of adult brain function by the male-specific factor SRY. Curr Biol. 2006;16:415–420. doi: 10.1016/j.cub.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Dewing P, Shi T, Horvath S, Vilain E. Sexually dimorphic gene expression in mouse brain precedes gonadal differentiation. Brain Res Mol Brain Res. 2003;118:82–90. doi: 10.1016/s0169-328x(03)00339-5. [DOI] [PubMed] [Google Scholar]
- Dinapoli L, Capel B. SRY and the standoff in sex determination. Mol Endocrinol. 2008;22:1–9. doi: 10.1210/me.2007-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann MN, Auger AP. Epigenetic impact of simulated maternal grooming on estrogen receptor alpha within the developing amygdala. Brain Behav Immun. 2011;25:1299–1304. doi: 10.1016/j.bbi.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely D, Underwood A, Dunphy G, Boehme S, Turner M, Milsted A. Review of the Y chromosome, Sry and hypertension. Steroids. 2010;75:747–753. doi: 10.1016/j.steroids.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Wang C, Zhang X, Pestell RG. Acetylation of nuclear receptors in cellular growth and apoptosis. Biochem Pharmacol. 2004;68:1199–1208. doi: 10.1016/j.bcp.2004.05.037. [DOI] [PubMed] [Google Scholar]
- Gatewood JD, Wills A, Shetty S, Xu J, Arnold AP, Burgoyne PS, Rissman EF. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J Neurosci. 2006;26:2335–2342. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioiosa L, Chen X, Watkins R, Klanfer N, Bryant CD, Evans CJ, Arnold AP. Sex chromosome complement affects nociception in tests of acute and chronic exposure to morphine in mice. Horm Behav. 2008a;53:124–130. doi: 10.1016/j.yhbeh.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioiosa L, Chen X, Watkins R, Umeda EA, Arnold AP. Sex chromosome complement affects nociception and analgesia in newborn mice. J Pain. 2008b;9:962–969. doi: 10.1016/j.jpain.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow PN, Lovell-Badge R. SRY and sex determination in mammals. Annu Rev Genet. 1993;27:71–92. doi: 10.1146/annurev.ge.27.120193.000443. [DOI] [PubMed] [Google Scholar]
- Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of cofactors and chromatin state. Nat Rev Cancer. 2007;7:713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of phenotype and disease. Mol Cell Endocrinol. 2011 doi: 10.1016/j.mce.2011.10.004. Epub Oct 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MJ, Juriloff DM. Mouse mutants with neural tube closure defects and their role in understanding human neural tube defects. Birth Defects Research Part A-Clinical and Molecular Teratology. 2007;79:187–210. doi: 10.1002/bdra.20333. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Zheng W, Wu X, Liu J, Ecelbarger CM, Watkins R, Arnold AP, Sandberg K. Sex Chromosome Effects Unmasked in Angiotensin II-Induced Hypertension. Hypertension. 2010;55:1275–82. doi: 10.1161/HYPERTENSIONAHA.109.144949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang PP, Hartl DL, Lemos B. Y not a dead end: epistatic interactions between Y-linked regulatory polymorphisms and genetic background affect global gene expression in Drosophila melanogaster. Genetics. 2010;186:109–118. doi: 10.1534/genetics.110.118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P. The delicate balance between male and female sex determining pathways: potential for disruption of early steps in sexual development. Int J Androl. 2010;33:252–258. doi: 10.1111/j.1365-2605.2009.01001.x. [DOI] [PubMed] [Google Scholar]
- Kristensen JB, Nielsen AL, Jorgensen L, Kristensen LH, Helgstrand C, Juknaite L, Kristensen JL, Kastrup JS, Clausen RP, Olsen L, Gajhede M. Enzyme kinetic studies of histone demethylases KDM4C and KDM6A: towards understanding selectivity of inhibitors targeting oncogenic histone demethylases. FEBS Lett. 2011;585:1951–1956. doi: 10.1016/j.febslet.2011.05.023. [DOI] [PubMed] [Google Scholar]
- Leader JE, Wang C, Popov VM, Fu M, Pestell RG. Epigenetics and the estrogen receptor. Ann N Y Acad Sci. 2006;1089:73–87. doi: 10.1196/annals.1386.047. [DOI] [PubMed] [Google Scholar]
- Lemos B, Branco AT, Hartl DL. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc Natl Acad Sci U S A. 2010;107:15826–15831. doi: 10.1073/pnas.1010383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene. Tdy Dev. 1990;109:635–646. doi: 10.1242/dev.109.3.635. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, Washburn lL, McCarrey JR, Cattanach BM, Lovell-Badge R, Burgoyne PS. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet. 1998;7:715–727. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- Majdic G, Tobet S. Cooperation of sex chromosomal genes and endocrine influences for hypothalamic sexual differentiation. Front Neuroendocrinol. 2011;32:137–145. doi: 10.1016/j.yfrne.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda KI, Mori H, Nugent BM, Pfaff DW, McCarthy MM, Kawata M. Histone deacetylation during brain development is essential for permanent masculinization of sexual behavior. Endocrinol. 2011;152:2760–2767. doi: 10.1210/en.2011-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazeyrat S, Saut N, Grigoriev V, Mahadevaiah SK, Ojarikre OA, Rattigan A, Bishop C, Eicher EM, Mitchell MJ, Burgoyne PS. A Y-encoded subunit of the translation initiation factor Eif2 is essential for mouse spermatogenesis. Nat Genet. 2001;29:49–53. doi: 10.1038/ng717. [DOI] [PubMed] [Google Scholar]
- McCarthy MM, Arnold AP. Reframing sexual differentiation of the brain. Nat Neurosci. 2011;14:677–683. doi: 10.1038/nn.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MM, Auger AP, Bale TL, De Vries GJ, Dunn GA, Forger NG, Murray EK, Nugent BM, Schwarz JM, Wilson ME. The epigenetics of sex differences in the brain. J Neurosci. 2009;29:12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhie-Lalmansingh AA, Tejada LD, Weaver JL, Rissman EF. Sex chromosome complement affects social interactions in mice. Horm Behav. 2008;54:565–570. doi: 10.1016/j.yhbeh.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migeon BR. Females are mosaic: X inactivation and sex differences in disease. Oxford: Oxford University Press; 2007. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Chanda ML. The case for the inclusion of female subjects in basic science studies of pain. Pain. 2005;117:1–5. doi: 10.1016/j.pain.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci. 2011;31:11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EK, Hien A, De Vries GJ, Forger NG. Epigenetic control of sexual differentiation of the bed nucleus of the stria terminalis. Endocrinol. 2009;150:4241–4247. doi: 10.1210/en.2009-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray EK, Varnum MM, Fernandez JL, De Vries GJ, Forger NG. Effects of neonatal treatment with valproic acid on vasopressin immunoreactivity and olfactory behaviour in mice. J Neuroendocrinol. 2011;23:906–914. doi: 10.1111/j.1365-2826.2011.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu X, Zhang T, Liao L, Zhou L, Lindner DJ, Zhou M, Rini B, Yan Q, Yang H. The von Hippel-Lindau tumor suppressor protein regulates gene expression and tumor growth through histone demethylase JARID1C. Oncogene. 2011 Jul 4; doi: 10.1038/onc.2011.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent BM, McCarthy MM. Epigenetic underpinnings of developmental sex differences in the brain. Neuroendocrinol. 2011;93:150–158. doi: 10.1159/000325264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent BM, Schwarz JM, McCarthy MM. Hormonally mediated epigenetic changes to steroid receptors in the developing brain: implications for sexual differentiation. Horm Behav. 2011;59:338–344. doi: 10.1016/j.yhbeh.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C, Loisel DA, Gilad Y. Sex-specific genetic architecture of human disease. Nat Rev Genet. 2008;9:911–922. doi: 10.1038/nrg2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaszynski KM, Smith DL, Kamrava S, Burgoyne PS, Arnold AP, Voskuhl RR. A Yin-Yang effect between sex chromosome complement and sex hormones on the immune response. Endocrinol. 2005;146:3280–3285. doi: 10.1210/en.2005-0284. [DOI] [PubMed] [Google Scholar]
- Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinol. 1959;65:369–382. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Hitchcott PK, Umeda EA, Arnold AP, Taylor JR. Sex chromosome complement regulates habit formation. Nat Neurosci. 2007;10:1398–1400. doi: 10.1038/nn1994. [DOI] [PubMed] [Google Scholar]
- Raefski AS, O’Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nature Genetics. 2005;37:620–624. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- Regitz-Zagrosek V, Lehmkuhl E, Weickert MO. Gender differences in the metabolic syndrome and their role for cardiovascular disease. Clin Res Cardiol. 2006;95:136–147. doi: 10.1007/s00392-006-0351-5. [DOI] [PubMed] [Google Scholar]
- Renfree MB, Short RV. Sex determination in marsupials: evidence for a marsupial-eutherian dichotomy. Phil Trans Roy Soc Lond B:Biol Sci. 1988;322:41–53. doi: 10.1098/rstb.1988.0112. [DOI] [PubMed] [Google Scholar]
- Robinson DP, Huber SA, Moussawi M, Roberts B, Teuscher C, Watkins R, Arnold AP, Klein SL. Sex chromosome complement contributes to sex differences in Coxsackievirus B3 but not Influenza A virus pathogenesis. Biol Sex Differ. 2011a;2:8. doi: 10.1186/2042-6410-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DP, Lorenzo ME, Jian W, Klein SL. Elevated 17beta-estradiol protects females from influenza A virus pathogenesis by suppressing inflammatory responses. PLoS Pathog. 2011b;7:e1002149. doi: 10.1371/journal.ppat.1002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Reboucas CB, Fintelman-Rodrigues N, Jensen LR, Kuss AW, Ribeiro MG, Campos M, Jr, Santos JM, Pimentel MM. A novel nonsense mutation in KDM5C/JARID1C gene causing intellectual disability, short stature and speech delay. Neurosci Lett. 2011;498:67–71. doi: 10.1016/j.neulet.2011.04.065. [DOI] [PubMed] [Google Scholar]
- Schulz KM, Molenda-Figueira HA, Sisk CL. Back to the future: The organizational-activational hypothesis adapted to puberty and adolescence. Horm Behav. 2009;55:597–604. doi: 10.1016/j.yhbeh.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JM, Nugent BM, McCarthy MM. Developmental and hormone-induced epigenetic changes to estrogen and progesterone receptor genes in brain are dynamic across the life span. Endocrinol. 2010;151:4871–4881. doi: 10.1210/en.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, Reveille JD, Alarcon GS, Vila LM, Reid J, Harris B, Li S, Kelly JA, Harley JB. Klinefelter’s syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58:2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skuse DH, James RS, Bishop DVM, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA. Evidence from Turner’s syndrome of an imprinted x-linked locus affecting cognitive function. Nature. 1997;387:705–708. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, King JK, Arnold AP, Singh RR, Voskuhl RR. A role for sex chromosome complement in the female bias in autoimmune disease. J Exp Med. 2008;205:1099–1108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KE, Vallejo-Giraldo C, Schaible NS, Zakeri R, Miller VM. Reporting of sex as a variable in cardiovascular studies using cultured cells. Biol Sex Differ. 2011;2:11. doi: 10.1186/2042-6410-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4:47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner ME, Ely DL, Prokop J, Milsted A. Sry, more than testis determination? Am J Physiol Regul Integr Comp Physiol. 2011;301:R561–71. doi: 10.1152/ajpregu.00645.2010. [DOI] [PubMed] [Google Scholar]
- van Nas A, GuhaThakurta D, Wang SS, Yehya N, Horvath S, Zhang B, Ingram-Drake L, Chaudhuri G, Schadt EE, Drake TA, Arnold AP, Lusis AJ. Elucidating the role of gonadal hormones in sexually dimorphic gene coexpression networks. Endocrinol. 2009;150:1235–1249. doi: 10.1210/en.2008-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet N, Mahadevaiah SK, Ojarikre OA, Longepied G, Prosser HM, Bradley A, Mitchell MJ, Burgoyne PS. The Y-encoded gene zfy2 acts to remove cells with unpaired chromosomes at the first meiotic metaphase in male mice. Curr Biol. 2011;21:787–793. doi: 10.1016/j.cub.2011.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuhl R. Sex differences in autoimmune diseases. Biol Sex Differ. 2011;2:1. doi: 10.1186/2042-6410-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJ, Festenstein RJ. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–40. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Yandim C, Panousopoulou E, Ahmad M, Harker N, Saveliev A, Burgoyne PS, Festenstein R. Sexual dimorphism in mammalian autosomal gene regulation is determined not only by Sry but by sex chromosome complement as well. Dev Cell. 2010;19:477–484. doi: 10.1016/j.devcel.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PLoS One. 2008a;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci. 2008b;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Taya S, Kaibuchi K, Arnold AP. Sexually dimorphic expression of Usp9x is related to sex chromosome complement in adult mouse brain. Eur J Neurosci. 2005;21:3017–3022. doi: 10.1111/j.1460-9568.2005.04134.x. [DOI] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Annu Rev Psychol. 2010;61:439–3. doi: 10.1146/annurev.psych.60.110707.163625. [DOI] [PubMed] [Google Scholar]