Abstract
Mouse cancer models have provided critical insights into tumor biology; however, clinical translation of these findings has been challenging. This perspective posits that factors impacting on successful translation start with limitations in capturing human cancer pathophysiology and end with challenges in generating robust translatable preclinical end points. A comprehensive approach that considers clinically relevant mouse models with both an integrated biomarker strategy and a complementary modeling and simulation effort will strengthen the current oncology drug development paradigm.
Successful submission and regulatory approval of innovative drugs is growing ever more challenging. Each drug candidate must navigate the so-called drug discovery and development “funnel,” characterized by a series of steps and associated probabilities of success. The relatively low historical success rate for registration of cancer drugs1 suggests that the funnel can be prohibitively narrow for this medication class. This can be attributed in part to discovery and development constraints that are either specific to, or more pronounced in, oncology relative to other therapeutic areas. Difficulties in proper interpretation and quantitative translation of oncology animal cancer model data, commonly originating from mice, are frequently cited as important factors that act to narrow the funnel.
Preclinical mouse cancer models have provided a wealth of information regarding our understanding of tumor biology. However, extrapolation from bench to bedside has been problematic, leading to a vigorous scientific debate regarding the predictability of antitumor efficacy from mouse experiments. Even with this uncertainty, mouse cancer models occupy a central role, especially in early drug development and the translational space. The need to increase drug development efficiency remains a major priority for grievous diseases such as cancer. Thus, the robust clinical translation of mouse data is of correspondingly high importance.
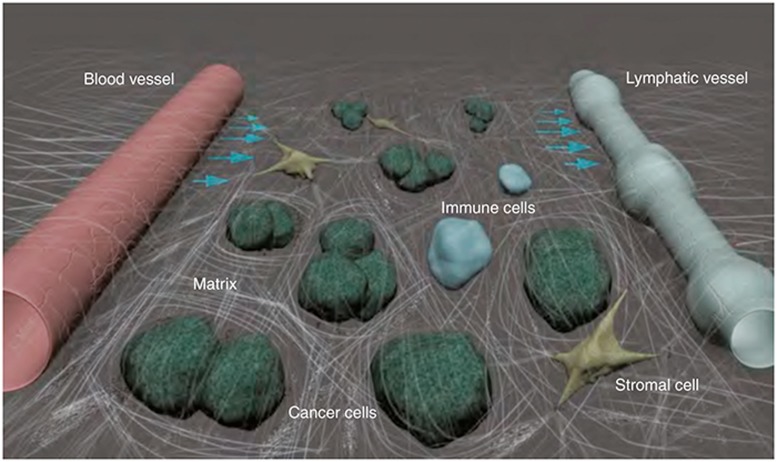
Our understanding of the factors that influence extrapolation of mouse findings to the clinic is evolving and suggests a high degree of complexity. Though cancer is characterized by uncontrolled cell proliferation caused by oncogenic driver mutations, the tumor microenvironment—comprised of blood and lymphatic vessels, nonmalignant host cells, and extracellular matrix—is critical to cancer progression and treatment (Figure 1). Indeed, 10 drugs targeting the tumor vasculature are currently approved by the US Food and Drug Administration.2 Others targeting the immunosuppressive or desmoplastic tumor microenvironment are in clinical development. Accordingly, our ability to reasonably approximate the human disease in mice should not be determined solely by selection of a relevant cancer cell line or model, but also by local interactions with the stroma and parenchyma, as well as systemic interactions with the host. Our current understanding of “cancer as an organ system” suggests that such interactions are influential, at both individual tumor compartment (i.e., tumor vessels, perivascular cells, extracellular matrix, etc.) and molecular levels. For example, at the compartmental level, intravital imaging provided direct evidence that tumor vessel morphology is vastly different following transplantation of the same cancer cell line at multiple sites in mice.2 These morphological changes in vessel characteristics bring important consequences for drug delivery to the tumor. Accordingly, extrapolation of preclinical tumor drug delivery findings to the clinic may be especially sensitive to this kind of cancer cell–stromal cell interaction. In addition, we see evidence of cancer cell–stromal cell interactions at the molecular level. A first example comes from medulloblastoma, the most frequent pediatric brain malignancy. A recent study suggests that medulloblastoma growth depends upon PlGF secretion from the cerebellar stroma irrespective of its genetic subtype and that PlGF expression is mediated by tumor-derived Sonic hedgehog.3 As with the previous example at compartmental level, this molecular level cancer cell–stromal cell interaction finding has potentially important ramifications for clinical translation; selection of a relevant preclinical model was essential in revealing this tissue-specific interaction. Similarly, at molecular level, a growing body of evidence implicates activation of the chemokine CXCL12 (stromal cell-derived factor-1α) pathway in tumor resistance to various treatments.4 The resistance mechanism involves direct promotion of cancer cell survival and invasion, as well as indirect effects on tumor recurrence and metastasis via the stroma. Accordingly, the clinical translation of anti-CXCL12 agents for use as sensitizers to existing cancer therapies should rely upon the development and use of preclinical models capable of capturing the response to these same therapies in patients.
Figure 1.
The tumor microenvironment. The tumor microenvironment is comprised of blood and lymphatic vessels, nonmalignant host cells, and extracellular matrix, all of which shape tumor progression and response or resistance to therapy. Courtesy of Lance L. Munn, Massachusetts General Hospital and Harvard Medical School, Boston, MA.
Given the challenges in fully capturing the cancer disease process in the mouse model, a translational program including quantitative assessment of continuous pharmacodynamic biomarkers (complementary to the existing discovery framework) promises to augment the cumulative database available for clinical translation and subsequent development. This is especially true given continuing increasing trends in developing molecularly targeted (vs. cytotoxic) agents in cancer, where mechanistic biomarkers may provide important insight into the intended mechanism of action. Despite this promise, biomarker approaches to inform clinical development in oncology have thus far been limited. A review of 2,483 phase I trials submitted to the American Society of Clinical Oncology suggested that 20% of trials incorporated biomarker assessments.5 Use of these biomarker data in clinical decision-making (i.e., proof of mechanism) was relatively infrequent, with biomarker-based support for phase II dose selection reported only in 13% of cases.
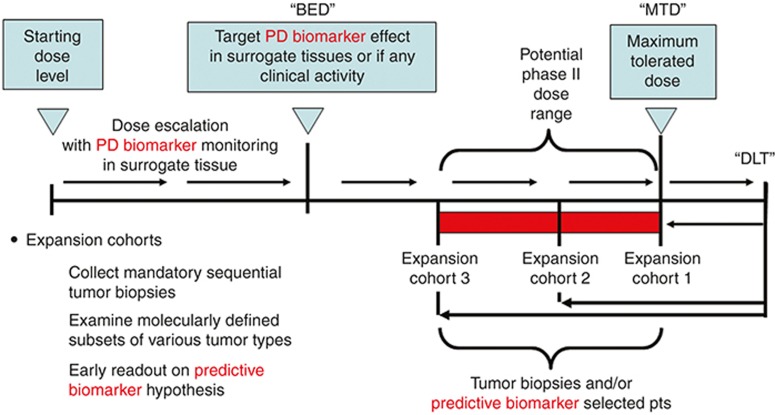
To increase the impact of biomarkers in oncology, it is important to have a robust framework for biomarker implementation into clinical development. The pharmacological audit trail (PhAT) can be especially useful.6 The PhAT partitions the determinants of drug action into a series of multilevel connected questions, progressing from target molecule to clinical response. Measurements at each of these levels permit a more holistic understanding of the drug's performance and provide context both for interpretation of biomarker readout and for broader use in dose selection. The emphasis upon interconnectivity between drug and effect enables identification of a biologically effective dose; this is in contrast to the more common selection of maximally tolerated dose that in turn focuses only on one aspect of the PhAT (i.e., tolerability). Accordingly, the PhAT naturally lends itself to defense of biologically effective dose as an alternative to dose selection based upon maximally tolerated dose. Figure 2 provides an example of how we can incorporate this approach into early development clinical designs. Examples of studies using biologically effective dose as a primary end point include trials with agents such as PARP inhibitors and antiangiogenic agents.7,8
Figure 2.
Translational phase I study with biomarker-defined end points. Phase I oncology studies classically treat advanced cancer patients in cohorts of three at progressively increasing starting dose levels until drug-related toxicities are observed. When the incidence and severity of these toxicities exceed levels predefined as dose limiting in the study protocol, dose escalation is halted. Additional patients are then treated at a lower dose to determine the maximum tolerated dose level. In a modern translational phase I study design, these classical phase I end points are augmented by pharmacodynamic biomarker monitoring to define the minimum exposures associated with biological activity. At the end of dose escalation, patient expansion cohorts are often added to collect additional biomarker, safety, and pharmacokinetic data. Thus, these early clinical studies mimic the comprehensive preclinical studies used to define pharmacologically active drug exposures. Modeling and simulation can be used at every step along the sequence to integrate exposure, biomarker, and other data collected during the trials. BED, biologically effective dose; DLT, dose-limiting toxicity; MTD, maximum tolerated dose.
Modeling, encompassing the “fit for purpose” continuum from mechanistic systems pharmacology to pharmacometrics through semi-mechanistic pharmacokinetics/pharmacodynamics (PK-PD), provides a quantitative approach to invoke and implement PhAT principles in drug development and to address the significant challenge of scaling results to the clinic. Both mechanistic and semi-mechanistic models can be amenable to scaling. For example, systems approaches have been used to capture the pathophysiological determinants of tumor drug delivery to the intended target and to successfully project the impact of elevated interstitial fluid pressure on fluid flow rates exiting the tumor, both in rodent models and in human cancers.2 The semi-mechanistic tumor growth inhibition (TGI) model also lends itself to scaling. With the TGI model, the observed tumor size is usually expressed as a competition between tumor growth in the absence of drug (system property) and kill rate (drug property); the TGI model may be further modified to allow for resistance and decreased drug effect in time. This conceptual partition provides the framework to scale findings from mice to the clinic, either directly or in conjunction with biomarker data, which can also be integrated in the modeling process.9
The case example of crizotinib (PF02341066; Xalkori), a dual inhibitor of anaplastic lymphoma kinase (ALK) and mesenchymal-epithelial transition factor (MET), illustrates the use of TGI modeling together with biomarker data to infer minimally efficacious levels of biomarker modulation (i.e., ALK and MET inhibition).10 In this example, a modeling and simulation approach was used to describe two exposure–response relationships, i.e., between crizotinib levels and the extent of inhibition of biomarker and tumor growth, respectively. This approach provides a quantitative framework to benchmark biomarker readout against TGI; the potency of crizotinib was captured as the minimal efficacious concentration for 50% of full effect (EC50) on TGI. Comparing the EC50 estimate for TGI and biomarker inhibition, the authors postulated a minimal required biomarker inhibition in the clinic. To optimally inform both TGI and biomarker inhibition relationships, MET and ALK inhibition data were obtained in biologically appropriate settings: GTL16 gastric and H3122 non–small-cell lung carcinoma mouse models, respectively. The EC50 for TGI was found to be similar to the EC50 for ALK inhibition and the EC90 for MET inhibition; accordingly, minimal required ALK and MET biomarker inhibition were predicted to be 50 and 90%, respectively. Subsequently, the crizotinib PK-PD relationship was simulated using the predicted (pre-first-in-human) and observed (post-first-in-human) pharmacokinetic parameters in patients. In these simulations, the crizotinib pharmacodynamics and unbound concentration (relationships the latter consistently with the “free drug hypothesis”) from nonclinical models were assumed to be analogous to those in cancer patients. The PK-PD prediction at the recommended phase II dose was especially important for decision making. That is, at 250 mg twice daily crizotinib, the model-predicted inhibition for both ALK and MET were higher (specifically, greater than 75% for ALK and 95% for MET) than their minimal required inhibition proposed from biomarker and TGI modeling, suggesting that target modulation in patients at the approved dosing regimen would be sufficient to attain desired antitumor efficacy.
In summary, incomplete understanding of translation early in oncology drug development undermines the foundation for subsequent steps in the drug development process. The mouse model lies at the center of the translational effort in oncology, but it is frequently criticized as being poorly informative. Quantitative approaches have had significant impact in oncology drug development; their application to the interpretation of mechanistic nonclinical discovery data promises to continue providing significant value in the future. There is not a single factor limiting the value of the mouse model in informing early oncology development; thus, approaches to increase its value have to be multifaceted. Experiments in clinically relevant settings, coupled with an integrated mechanistically based biomarker program, provide the missing tiles in the current development paradigm mosaic. In addition, an equally active modeling and simulation effort can act to assemble these data pieces into a stronger foundation for bringing promising cancer candidates to the clinic more rapidly and efficiently.
Conflict of Interest
M.S. is an employee of Genentech, a member of the Roche Group, and a shareholder of Roche. D.G.D. has no conflicts of interest to disclose. C.H.T. is an employee and shareholder of Johnson & Johnson. S.Y. is an employee and shareholder of Pfizer. P.V. is an employee and shareholder of Pfizer. As an Associate Editor for CPT: Pharmacometrics & Systems Pharmacology, P.V. was not involved in the review or decision-making process for this paper.
Acknowledgments
The contents of this Perspective article were partially discussed at the 114th Annual Meeting of the American Society for Clinical Pharmacology and Therapeutics on 5–9 March in Indianapolis, during a workshop entitled “An Integrated Perspective on Translation of Mouse Model Data to the Clinic: From Tumor Biology to Mathematical Modeling.” The authors acknowledge the helpful input from numerous colleagues at Genentech, Massachusetts General Hospital, Janssen, and Pfizer into this Perspective.
References
- Kamb A., Wee S., Lengauer C. Why is cancer drug discovery so difficult. Nat. Rev. Drug Discov. 2007;6:115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- Jain R.K. Normalizing tumor microenvironment to treat cancer: bench to bedside to biomarkers. J. Clin. Oncol. 2013;31:2205–2218. doi: 10.1200/JCO.2012.46.3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snuderl M., et al. Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell. 2013;152:1065–1076. doi: 10.1016/j.cell.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda D.G., Kozin S.V., Kirkpatrick N.D., Xu L., Fukumura D., Jain R.K. CXCL12 (SDF1alpha)-CXCR4/CXCR7 pathway inhibition: an emerging sensitizer for anticancer therapies. Clin. Cancer Res. 2011;17:2074–2080. doi: 10.1158/1078-0432.CCR-10-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulart B.H., Clark J.W., Pien H.H., Roberts T.G., Finkelstein S.N., Chabner B.A. Trends in the use and role of biomarkers in phase I oncology trials. Clin. Cancer Res. 2007;13:6719–6726. doi: 10.1158/1078-0432.CCR-06-2860. [DOI] [PubMed] [Google Scholar]
- Workman P. Auditing the pharmacological accounts for Hsp90 molecular chaperone inhibitors: unfolding the relationship between pharmacokinetics and pharmacodynamics. Mol. Cancer Ther. 2003;2:131–138. [PubMed] [Google Scholar]
- Plummer R., et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin. Cancer Res. 2008;14:7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowlati A., et al. Novel Phase I dose de-escalation design trial to determine the biological modulatory dose of the antiangiogenic agent SU5416. Clin. Cancer Res. 2005;11:7938–7944. doi: 10.1158/1078-0432.CCR-04-2538. [DOI] [PubMed] [Google Scholar]
- Wang S., Guo P., Wang X., Zhou Q., Gallo J.M. Preclinical pharmacokinetic/pharmacodynamic models of gefitinib and the design of equivalent dosing regimens in EGFR wild-type and mutant tumor models. Mol. Cancer Ther. 2008;7:407–417. doi: 10.1158/1535-7163.MCT-07-2070. [DOI] [PubMed] [Google Scholar]
- Yamazaki S. Translational pharmacokinetic-pharmacodynamic modeling from nonclinical to clinical development: a case study of anticancer drug, crizotinib. AAPS J. 2013;15:354–366. doi: 10.1208/s12248-012-9436-4. [DOI] [PMC free article] [PubMed] [Google Scholar]