Abstract
Background
In past, a number of peptides have been reported to possess highly diverse properties ranging from cell penetrating, tumor homing, anticancer, anti-hypertensive, antiviral to antimicrobials. Owing to their excellent specificity, low-toxicity, rich chemical diversity and availability from natural sources, FDA has successfully approved a number of peptide-based drugs and several are in various stages of drug development. Though peptides are proven good drug candidates, their usage is still hindered mainly because of their high susceptibility towards proteases degradation. We have developed an in silico method to predict the half-life of peptides in intestine-like environment and to design better peptides having optimized physicochemical properties and half-life.
Results
In this study, we have used 10mer (HL10) and 16mer (HL16) peptides dataset to develop prediction models for peptide half-life in intestine-like environment. First, SVM based models were developed on HL10 dataset which achieved maximum correlation R/R2 of 0.57/0.32, 0.68/0.46, and 0.69/0.47 using amino acid, dipeptide and tripeptide composition, respectively. Secondly, models developed on HL16 dataset showed maximum R/R2 of 0.91/0.82, 0.90/0.39, and 0.90/0.31 using amino acid, dipeptide and tripeptide composition, respectively. Furthermore, models that were developed on selected features, achieved a correlation (R) of 0.70 and 0.98 on HL10 and HL16 dataset, respectively. Preliminary analysis suggests the role of charged residue and amino acid size in peptide half-life/stability. Based on above models, we have developed a web server named HLP (Half Life Prediction), for predicting and designing peptides with desired half-life. The web server provides three facilities; i) half-life prediction, ii) physicochemical properties calculation and iii) designing mutant peptides.
Conclusion
In summary, this study describes a web server ‘HLP’ that has been developed for assisting scientific community for predicting intestinal half-life of peptides and to design mutant peptides with better half-life and physicochemical properties. HLP models were trained using a dataset of peptides whose half-lives have been determined experimentally in crude intestinal proteases preparation. Thus, HLP server will help in designing peptides possessing the potential to be administered via oral route (http://www.imtech.res.in/raghava/hlp/).
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2105-15-282) contains supplementary material, which is available to authorized users.
Background
Due to rapid advancement in peptide and peptidomimetic techniques, pharmaceutical companies are focusing towards peptides-based therapeutics [1]. A large number of peptides like insulin [2], cyclosporine [3], corticotropin [4] are used for treating various diseases like diabetes and immunoregulatory disorders [5–7]. Owing to their immense therapeutic importance, peptides have been curated from literature and stored in form of databases such as Hemolytik [8], PhytAMP [9], APD2 [10], DAMPD [11], CAMP [12], YADAMP [13], CPPsite [14], TumorHoPe [15], Quorumpeps [16], Brainpeps [17], MilkAMP [18], DADP [19], LAMP [20] and AVPdb [21] to name a few. Also, a number of computation tools such as CellPPD [22], TumorHPD [23], AntiCP [24], AVPpred [25] and ToxinPred [26] have been developed to predict and design cell penetrating, tumor homing, anticancer, antiviral and toxic peptides, respectively. Presently, most of the therapeutic peptides are used in the form of injection via subcutaneous, intravenous/intramuscular route. In addition, alternate routes such as pulmonary [27], oral [28, 29], intranasal [30], buccal [31], transdermal [32], ocular [33] and rectal [34] have been tested. Among all routes of drug delivery, oral is the most preferred route because oral formulations are less expensive and less prone to infection caused by inappropriate use/reuse of needles. Additionally, orally available peptides are highly accepted by patients, which increases the therapeutic value of the drug. However, designing and formulating an oral peptide has been considered as a challenging job. This is because of their undesirable physicochemical properties like large molecular size, high susceptibility to enzymatic degradation (proteases), hepatic and renal clearance, etc.
Amongst the above factors, peptide stability is one of the most difficult tasks to maintain. Their high susceptibility to proteases in the gut and serum (especially for cationic peptides) and fast degradation rate due to their arginine and lysine content makes them low orally bioavailable [5, 35, 36]. A major concern associated with the use of peptide therapeutics is improving their stability by protection against degrading proteases [37]. Although, a number of experimental techniques like peptide modification are available in order to improve peptide stability with high accuracy [5, 38], yet these experimental procedures are costly and time consuming. In view of its importance, there is a need to develop an in silico method for predicting 1) half-life of peptides as well as 2) prediction of mutations required in a peptide, in order to increase its intestinal half-life. In past, computational methods for predicting half-life of proteins/peptides in blood and kidney cells/cell lines have been developed. For example, ProtParam is a tool that helps to estimate half-life of a protein stored in Swiss-Prot/TrEMBL or a user entered protein. The estimation of half-life is done for three experimental models namely: yeast in vivo, mammalian reticulocytes (immature red blood cells) in vitro and Escherichia coli in vivo [39]. Stability Prediction tool predicts the stability of HIV-derived peptides in cytosolic extracts from human peripheral blood mononuclear cells [40]. SProtP is a web server to recognize the short-lived proteins (half-life < 30 minutes) in 293 T cells (a variant of human embryonic kidney cell line) [41, 42]. The N-end rule [43, 44] based ‘ProtLifePred server’ [http://protein-n-end-rule.leadhoster.com/] takes into account the ubiquitination (a process that involves post-translational modification of proteins) process of proteins [45–47] and predicts their stability in S. cerevisiae, E. coli and mammalian cells. Best of author’s knowledge there is no computational method developed for designing/predicting stability/half-life of peptides in intestine-like environment.
In order to facilitate researchers working in the field of therapeutic peptides, we have developed in silico models for predicting half-life of peptides. These models were developed using a set of peptides whose half-lives have been experimentally determined in crude intestinal proteases preparation [37]. Server developed in this study can be used to identify minimum mutations in a peptide required to optimize their half-life.
Results
Analysis of half-life data
We computed and analyzed physicochemical properties of peptides in both HL10 and HL16 datasets to understand relation between property of amino acids and their half-life. Based on the half-life value, peptides were classified into two categories peptides having long half-life (highly stable) and peptides having short half-life (poorly stable or unstable). Each category have top 20 peptides, it means 20 peptides having longest half-life were classified as stable and 20 peptides having shortest half-life were classified as unstable. We observed that negatively charged, neutral and tiny types of residues are more prominent in highly stable peptides (Figure 1A and B). We have also computed amino acid composition of peptides in highly and lowest/poorly stable peptide datasets. As shown in Figure 1 (C and D), residues Ala, Asp, Glu, Gly, Gln, Ser and Thr are abundant in peptide dataset with longer half-life. As evident from Additional file 1: Table S3, residues D (Asp), F (Phe), G (Gly), L (Leu), Q (Gln), R (Arg) and Y (Tyr) show significant differences (p < 0.05) in amino acid composition in 10mer dataset. Similarly, residues D (Asp), F (Phe), G (Gly), K (Lys), M (Met), N (Asn), R (Arg) and Y (Tyr) shows statistically significant differences (p < 0.05) in amino acid composition for 16mer dataset (Additional file 1: Table S4).
Figure 1.
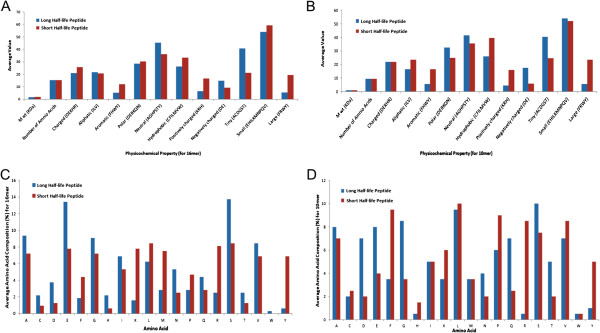
Physicochemical properties and amino acid composition of top 20 peptides having longest half-life (stable peptides) and top 20 peptides having shortest half-life (unstable peptides). (A) physicochemical properties of 16mer peptides having short and long half-life; (B) physicochemical properties of 10mer peptides having short and long half-life; (C) average amino acid composition 16mer peptides having short and long half-life; (D) shows average amino acid composition of 10mer peptides having short and long half-life.
Prediction of Half-Life
SVM based models on HL10 dataset
The model was developed on HL10 dataset using amino acid composition-based and achieved maximum R/R2 of 0.57/0.32 with mean absolute error (MAE) 1.87. Similarly models were developed using dipeptide and tripeptide composition resulted in R/R2 of 0.68/0.46 and 0.69/0.47 with MAE 1.44 and 1.38 respectively (Table 1). As shown in Table 1, performance of binary pattern based model is poor as compared to composition-based model with R/R2 of 0.22/0.02 and MAE 1.87. In order to understand role of residues at terminus, models were also developed using 5-residues from N and C terminal called 5 N-terminal and 5 C-terminal residues. We achieved maximum R/R2 of 0.39/0.12 and 0.32/0.09 using amino acid composition of 5 N-terminal and 5 C-terminal residues, respectively (Table 1). It is evident from the above results that composition-based models perform better than models based on binary profile/pattern.
Table 1.
The performance of SVM based models developed on HL10 dataset using different features of peptide sequences
| Input feature | Residues in peptides | Total attributes | R | R 2 | MAE |
|---|---|---|---|---|---|
| Amino acid composition | All residues | 20 | 0.57 | 0.32 | 1.87 |
| 5 N-terminal | 0.39 | 0.12 | 2.12 | ||
| 5 C-terminal | 0.32 | 0.09 | 1.99 | ||
| Binary pattern | All residues | 200 | 0.22 | 0.02 | 1.87 |
| 5 N-terminal | 100 | 0.06 | -0.06 | 1.27 | |
| 5 C-terminal | 100 | 0.34 | 0.12 | 2.01 | |
| Dipeptide composition | All residues | 400 | 0.68 | 0.46 | 1.44 |
| Tripeptide composition | All residues | 8000 | 0.69 | 0.47 | 1.38 |
Models developed on selected features
After removing useless features, we developed models for predicting half-life using following techniques SMOreg, IBk, KStar and DecisionTable (implemented in Weka package). We developed composition-based models using best-selected features/compositions (four type of residues) and got maximum R/R2 of 0.61/0.35 with MAE 1.42 (Table 2, Additional file 1: Table S1). Similarly, we developed models using best-selected dipeptides (eight type of dipeptides) and achieved maximum R/R2 of 0.70/0.46 along with MAE 1.22. We also developed model using selected 38 tripeptides and obtained maximum R/R2 of 0.73/0.35 with MAE 1.39 (Table 2, Additional file 1: Table S1).
Table 2.
The performance of Weka models developed using selected features on HL10 dataset
| Total attributes | Techniques | Selected attributes | Method | R | R 2 | MAE |
|---|---|---|---|---|---|---|
| 20 | KStar | D, G, P, R | Amino acid composition | 0.61 | 0.35 | 1.42 |
| 400 | IBk | EK, EL, GD, GF, IE, KP, PG, YL | Dipeptide composition | 0.70 | 0.46 | 1.22 |
| 8000 | IBk | AAH, AGR, AMP, ARE, ASV, DSI, EEK, ELY, ESK, FCI, FGD, FSL, FSS, FYC, GDS, GFG, GLF, GSI, GTS, ILP, INF, INK, IRN, ITK, KIL, KIS, KLP, LVL, MVL, PGF, PVQ, SGL, SIE, SLR, SVL, VFK, VLF, VYL | Tripeptide composition | 0.73 | 0.35 | 1.39 |
SVM based models on HL16 dataset
Similarly, SVM based models were developed on HL16 dataset. The amino acid composition-based model showed best performance with R/R2 of 0.91/0.82 with MAE 0.18 (Table 3). As shown in Table 3, the dipeptide and tripeptide composition based models showed an R/R2 of 0.90/0.39, and 0.90/0.31 with MAE 0.23 and 0.24, respectively. As compared to composition-based model, the binary pattern based model again showed poor performance with R/R2 of 0.13/0.01. Next, models were developed using 5 and 10 residues from N and C terminals, respectively. An increase in model performance with correlation value from 0.57 to 0.77 has been observed for N-terminal 5 to 10 residues based model developed using amino acid composition (Table 3). Similarly, the C-terminal based composition based model shows an increase in model performance with correlation (R) value of 0.37 to 0.81 and R2 of 0.13 to 0.65. The binary pattern based models performed poor than composition-based model; developed using N and C terminal residues (Table 3).
Table 3.
The performance SVM based models developed on HL16 dataset using composition and binary pattern
| Input features | Residues in peptides | Total attributes | R | R 2 | MAE |
|---|---|---|---|---|---|
| Amino acid composition | All residues | 20 | 0.91 | 0.82 | 0.18 |
| 5 N-terminal | 0.57 | 0.32 | 0.23 | ||
| 10 N-terminal | 0.77 | 0.60 | 0.24 | ||
| 5 C-terminal | 0.37 | 0.13 | 0.27 | ||
| 10 C-terminal | 0.81 | 0.65 | 0.24 | ||
| Binary pattern | All residues | 320 | 0.13 | 0.01 | 0.18 |
| 5 N-terminal | 100 | 0.17 | 0.02 | 0.20 | |
| 5 C-terminal | 100 | 0.03 | -0.002 | 0.18 | |
| 10 N-terminal | 200 | 0.22 | 0.02 | 0.18 | |
| 10 C-terminal | 200 | 0.09 | -0.002 | 0.18 | |
| Dipeptide composition | All residues | 400 | 0.90 | 0.39 | 0.23 |
| Tripeptide composition | All residues | 8000 | 0.90 | 0.31 | 0.24 |
Models developed on selected features
We identified/selected best features (residues or dipeptides or tripeptides) having high correlation with half-life of peptides. The first model developed using four important features from the amino acid composition and achieved maximum R/R2 of 0.97/0.93 along with MAE 0.07 (Table 4, Additional file 1: Table S2). As shown in Table 4, the three selected dipeptide (CG, GD, GF) results in R/R2 value of 0.98/0.96 with MAE 0.06. Further model was developed on selected five attributes from tripeptide composition shows equal performance with correlation coefficient value of 0.98 (Table 4, Additional file 1: Table S2).
Table 4.
The performance of various models (Weka) using selected features on HL16 dataset
| Total attributes | Techniques | Selected attributes | Method | R | R 2 | MAE |
|---|---|---|---|---|---|---|
| 20 | DecisionTable | C, D, G, R | Amino acid composition | 0.97 | 0.93 | 0.07 |
| 400 | DecisionTable | CG, GD, GF | Dipeptide composition | 0.98 | 0.96 | 0.06 |
| 8000 | DecisionTable | AQC, EAQ, FGD, GFG, QCG | Tripeptide composition | 0.98 | 0.96 | 0.06 |
Development of web server/software
Based on above study, we developed a web server for predicting intestinal half-life of peptides to design stable peptides through mutant generation and whole protein scanning. HLP server has three main modules called Peptide, Protein and Batch module. First module i.e., Peptide, allows users to submit single peptide at a time to the web server for predicting its intestinal half-life and designing mutants which may have better half-life and physicochemical properties than original peptide. Protein module allows a user to submit a protein for scanning to find peptide having high intestinal half-life. Furthermore, it allow to generate mutant peptides corresponding to user-selected peptide. Third module i.e., Batch, designed for high throughput scanning where a user can submit a large number of peptides for filtering best peptides (having high intestinal half-life and desired physicochemical properties). Our HLP server implement total six type of models; these can be divided in two classes: SVM based models (implemented using SVMlight package) and Weka based models (implemented using WEKA package). Our SVM based models implemented using following type of information of peptides; i) dipeptide composition in case of 10mer peptides, ii) amino acid composition for 16mer peptides and iii) type of composition depend on peptide length in case of user-selected sequence length. In case of Weka based models, following techniques and peptide information is used; i) IBK model using eight dipeptides (EK, EL, GD, GF, IE, KP, PG, YL) for 10mer peptides, ii) DecisionTable using three dipeptides (CG, GD, GF) for 16mer peptides and iii) model selection depends on peptide length in case of user-selected sequence length. The HLP web server was developed using Perl, CGI and HTML languages; it is available freely for public use from URL http://www.imtech.res.in/raghava/hlp/.
Discussion
Although a number of peptides have entered in market against various indications (such as diabetes, hypertension, cancer, HIV, etc.) still one of the major challenges in success of peptide-based therapy is their low half-life in serum and gut. There are number of factors responsible for degrading or shortening half-life of peptides [5]; where in silico designing of peptides seems to be a promising field in the area of peptide based therapy. The presence of intestinal proteases is one of the major factors behind degradation of peptides. In vitro peptide stability screening is considered to be the most difficult and time-consuming task. In order to assist scientific community to screen stable peptide in intestine-like environment, we have made a systematic attempt in this direction. In this study, models have been developed for predicting half-life of peptides on dataset derived from Gorris et al., 2009, where half-lives of peptides have been experimentally determined in the presence of crude intestinal proteases (high concentration). Firstly, they calculated half-life of peptides in undiluted proteolytic solution and then it has been subsequently extrapolated from results obtained from diluted solutions. The analysis based on these datasets suggest that charge and size of an amino acid are the two major factors governing the peptide stability. As reported in previous studies on bioactive peptides [22–24], positively charge amino acids Arg and Lys are preferred among majority of these kind of peptides while the present study reports these residues responsible to decrease peptide’s intestinal half-life. Therefore, the residues affecting peptide’s half-life and bioactivity should be optimized simultaneously while designing highly stable and potent bioactive peptides. The presence of large size amino acids such as Phe, Arg, Tyr and Trp will increase protease susceptibility and tiny amino acids (such as Gly, Ala, Ser, Thr) may help in their stabilization. Among these large-size amino acids, three (Phe, Tyr, Trp) belong to aromatic class and are also responsible for the decreased half-life. HLP models, developed on a small dataset shows good correlation (R) of 0.70 and 0.98 between predicted and actual half-life value on HL10 and HL16 dataset, respectively. One of two major limitations of our method is that it is not suitable for predicting the stability of modified peptides like N-terminal/C-terminal modification or effect of disulphide bond in a peptide. Therefore, our model is only suitable for predicting the stability of peptide composed of natural amino acids without any chemical modifications. Another limitation of the present study is the presence of small dataset in training and testing the half-life models due to unavailability of large dataset on peptide half-life in intestine from literature. The actual half-lives have been calculated in highly concentrated environment. Gorris et al. assume that the half-life values would be very high in real life. Unfortunately, they haven’t mentioned quantitatively, how high these half-lives values would be in real life. Thus, the half-life dataset helps in estimating half-lives of peptides relatively rather than in absolute terms. Just for our satisfaction and to test how good the models will work on peptides having very high half-lives, we developed new models after multiplying peptide’s actual half-life with 10 and 1000. As evident from Additional file 2: Tables S1-S6, half-life prediction models performs equally well for higher half-life containing peptides, irrespective of very high/low half-life. Hence, HLP can prove to be a extremely good resource to relatively estimate the half-life of peptides in intestine like environment. Additionally, this can help in improving half-life of peptide(s) half-life by mutating residue. We hope that our study will be useful to scientific community for designing/prediction of stable peptides and thus helpful in solving the major barrier in peptide based drug development.
Conclusion
There is a growing demand of peptide-based drugs but the major bottleneck in their development is their short half-life. The present study can provide an efficient method to design therapeutic peptides having better half-life and physicochemical properties. We hope that our method would promote the usage of various types of therapeutic peptides in drug discovery and development.
Methods
Datasets
In this study, we have used two datasets called HL10 and HL16 for developing prediction models. The datasets containing peptides with their half-life, were obtained from Gorris et al., 2009 [37]. HL10 dataset consists of 189 peptides with their half-life value varies from 0.0008 - 40.1296 seconds. HL16 dataset consists of 186 peptides with their half-life value varies from 0.0008 - 6.4211 seconds. Additionally, the distribution of half-lives (for both 10mer and 16mer) can be clearly seen from Figure five by Gorris et al., 2009.
Input features
Physicochemical properties
It is well established that activity or function of a peptide or protein depends on its amino acid sequence. Each amino acid has unique set of physicochemical properties like hydrophobicity, polarity, charge, size. Thus each peptide has different physicochemical properties depending on type of residue it contains. Thus, physicochemical properties of peptide play a significant role in determining the activity of peptides; therefore we had calculated more than 15 different properties of peptides like hydrophobicity, volume, isoelectric point, flexibility [48–51].
Residue composition as input features
In the past, it has been observed that amino acids composition encapsulates the protein information. Based on this approach, numbers of methods have been developed like protein folding rate prediction, sub-cellular localization prediction [51, 52]. In this study, we computed amino acid, dipeptide and tripeptide composition and used as input feature for developing prediction models. These compositions are represented by a vector of dimension 20, 400, and 8000 for residue/amino acid, dipeptide and tripeptide composition respectively [53].
Binary pattern
Each amino acid of a peptide is represented by binary pattern of 20, where 1′s represents the presence of concerned amino acid at that position and 0′s for the absence of other 19 amino acids (e.g. Ala is represented by 1,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0,0) [22]. Thus, a vector of dimension N X 20 is used to represent the peptide of length N (size of peptide).
Models based on machine learning techniques
In this study various machine-learning techniques have been used for developing regression models. In order to implement support vector machine for developing models we used commonly used highly efficient software called SVMlight. This package allows to implement various kernels (e.g., linear, polynomial, RBF) as well as it allow to tune various parameters.
In addition to SVMlight, we also used Weka, a Java based software package having various machine learning and feature selection algorithms [54]. In this study, we have used SMOreg (Sequential Minimization Optimization) module of Weka for building model for predicting half-life/stability of peptides. We also used IBK, KStar and DecisionTable based classifier, implemented in Weka for model building and optimization.
Feature selection
Previously, studies have shown that all input features are not equally important during the construction of an in silico model [55]. Thus, feature selection seems to be more relevant in order to find out the most important and contributing input features. In this study for feature selection, we have used CfsSubsetEval attribute evaluator with BestFirst (with parameters: -D 1 -N 5) search method (using full training set as attribute selection mode) implemented in Weka software for both HL10 and HL16 dataset.
Evaluation of performance
The performance of the models was evaluated by employing a five-fold cross-validation technique. The whole dataset is divided into five sets in such a way that every time four sets are used for training and one set for testing. This process is repeated five times in such a way that each set is used for testing. Once the model is constructed, fitness has been accessed using the following statistical parameters.
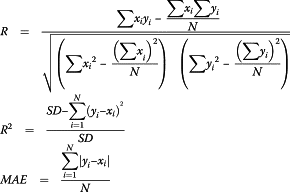 |
Where yi and xi represent predicted and actual half-life values for ith peptide. N is total number of peptides. SD is the sum of the squared deviations between the activities of the test set and mean activities of the training peptides. The value of correlation coefficient (R) and coefficient of determination (R2) is used to measure the quality of model. The value of R varies from -1 to +1 while R2 is from 0 to 1. The negative value of R shows the negative correlation with a particular property or feature. Thus, higher the value of R and R2, better will be the quality of model in term of prediction of half-life of peptides.
Availability and requirements
We have developed a web server HLP, freely available for predicting half-life of peptides in intestine-like environment. This web server was developed using PERL, CGI and HTML programming languages.
Electronic supplementary material
Additional file 1: Weka based results for both HL10 (Table S1) and HL16 (Table S2) using selected features. Differences in amino acid composition for longest half-life (stable) Vs shortest half-life (unstable) containing 10mer (Table S3) and 16mer peptides (Table S4). (XLS 35 KB)
Additional file 2: Performance of SVM and Weka based models (Tables S1-S6) after multiplying their actual half-life with 10 and 1000. Longest half-life (Stable) and shortest half-life containing 10mer (Table S7) and 16mer peptides (Table S8). (DOC 419 KB)
Acknowledgments
We are thankful to Mr. Gaurav for providing technical help. We are also thankful to Dr. Amit Arora for critically reviewing the manuscript. Authors are thankful to Council of Scientific and Industrial Research (CSIR), Open Source for Drug Discovery (OSDD) foundation and Department of Biotechnology (DBT), Govt. of India for financial support.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AS developed the prediction models for predicting half-life of peptide and assisted in developing the web server. DS and AS performed analysis on datasets. MR implemented modules for predicting stability of peptides and designed the web server with features like sorting of columns. GPSR conceived the project, coordinated it and refined the manuscript drafted by AS and DS. This manuscript has been seen and approved by all authors. All authors read and approved the final manuscript.
Contributor Information
Arun Sharma, Email: bioinfo.arun@gmail.com.
Deepak Singla, Email: deep_kumar@imtech.res.in.
Mamoon Rashid, Email: Mamoon.rashid@kaust.edu.sa.
Gajendra Pal Singh Raghava, Email: raghava@imtech.res.in.
References
- 1.Rubinstein M, Niv MY. Peptidic modulators of protein-protein interactions: progress and challenges in computational design. Biopolymers. 2009;91(7):505–513. doi: 10.1002/bip.21164. [DOI] [PubMed] [Google Scholar]
- 2.Morris AD, Boyle DI, McMahon AD, Greene SA, MacDonald TM, Newton RW. Adherence to insulin treatment, glycaemic control, and ketoacidosis in insulin-dependent diabetes mellitus. The DARTS/MEMO Collaboration. Diabetes Audit and Research in Tayside Scotland. Medicines Monitoring Unit. Lancet. 1997;350(9090):1505–1510. doi: 10.1016/S0140-6736(97)06234-X. [DOI] [PubMed] [Google Scholar]
- 3.Colombo D, Ammirati E. Cyclosporine in transplantation - a history of converging timelines. J Biol Regul Homeost Agents. 2011;25(4):493–504. [PubMed] [Google Scholar]
- 4.Carey LC, Su Y, Valego NK, Rose JC. Infusion of ACTH stimulates expression of adrenal ACTH receptor and steroidogenic acute regulatory protein mRNA in fetal sheep. Am J Physiol Endocrinol Metab. 2006;291(2):E214–E220. doi: 10.1152/ajpendo.00578.2005. [DOI] [PubMed] [Google Scholar]
- 5.Vlieghe P, Lisowski V, Martinez J, Khrestchatisky M. Synthetic therapeutic peptides: science and market. Drug Discov Today. 2010;15(1–2):40–56. doi: 10.1016/j.drudis.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Mason JM. Design and development of peptides and peptide mimetics as antagonists for therapeutic intervention. Future Med Chem. 2010;2(12):1813–1822. doi: 10.4155/fmc.10.259. [DOI] [PubMed] [Google Scholar]
- 7.Walter R, Yamanaka T, Sakakibara S. A neurohypophyseal hormone analog with selective oxytocin-like activities and resistance to enzymatic inactivation: an approach to the design of peptide drugs. Proc Natl Acad Sci U S A. 1974;71(5):1901–1905. doi: 10.1073/pnas.71.5.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautam A, Chaudhary K, Singh S, Joshi A, Anand P, Tuknait A, Mathur D, Varshney GC, Raghava GPS. Hemolytik: a database of experimentally determined hemolytic and non-hemolytic peptides. Nucleic Acids Res. 2014;42:D444–D449. doi: 10.1093/nar/gkt1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammami R, Ben Hamida J, Vergoten G, Fliss I. PhytAMP: a database dedicated to antimicrobial plant peptides. Nucleic Acids Res. 2009;37:D963–D968. doi: 10.1093/nar/gkn655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G, Li X, Wang Z. APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009;37:D933–D937. doi: 10.1093/nar/gkn823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seshadri Sundararajan V, Gabere MN, Pretorius A, Adam S, Christoffels A, Lehväslaiho M, Archer JA, Bajic VB. DAMPD: a manually curated antimicrobial peptide database. Nucleic Acids Res. 2012;40:D1108–D1112. doi: 10.1093/nar/gkr1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waghu FH, Gopi L, Barai RS, Ramteke P, Nizami B, Idicula-Thomas S. CAMP: Collection of sequences and structures of antimicrobial peptides. Nucleic Acids Res. 2014;42:D1154–D1158. doi: 10.1093/nar/gkt1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piotto SP, Sessa L, Concilio S, Iannelli P. YADAMP: yet another database of antimicrobial peptides. Int J Antimicrob Agents. 2012;39(4):346–351. doi: 10.1016/j.ijantimicag.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Gautam A, Singh H, Tyagi A, Chaudhary K, Kumar R, Kapoor P, Raghava GPS. CPPsite: a curated database of cell penetrating peptides. Database. 2012;2012:bas015. doi: 10.1093/database/bas015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapoor P, Singh H, Gautam A, Chaudhary K, Kumar R, Raghava GPS. TumorHoPe: a database of tumor homing peptides. PLoS ONE. 2012;7(4):e35187. doi: 10.1371/journal.pone.0035187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wynendaele E, Bronselaer A, Nielandt J, D’Hondt M, Stalmans S, Bracke N, Verbeke F, Van De Wiele C, De Tré G, De Spiegeleer B. Quorumpeps database: chemical space, microbial origin and functionality of quorum sensing peptides. Nucleic Acids Res. 2013;41:D655–D659. doi: 10.1093/nar/gks1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Dorpe S, Bronselaer A, Nielandt J, Stalmans S, Wynendaele E, Audenaert K, Van De Wiele C, Burvenich C, Peremans K, Hsuchou H, De Tré G, De Spiegeleer B. Brainpeps: the blood–brain barrier peptide database. Brain Struct Funct. 2012;217(3):687–718. doi: 10.1007/s00429-011-0375-0. [DOI] [PubMed] [Google Scholar]
- 18.Théolier J, Fliss I, Jean J, Hammami R. MilkAMP: a comprehensive database of antimicrobial peptides of dairy origin. Dairy Sci Technol. 2014;94(2):181–193. doi: 10.1007/s13594-013-0153-2. [DOI] [Google Scholar]
- 19.Novković M, Simunić J, Bojović V, Tossi A, Juretić D. DADP: the database of anuran defense peptides. Bioinformatics. 2012;28(10):1406–1407. doi: 10.1093/bioinformatics/bts141. [DOI] [PubMed] [Google Scholar]
- 20.Zhao X, Wu H, Lu H, Li G, Huang Q. LAMP: A Database Linking Antimicrobial Peptides. PLoS ONE. 2013;8(6):e66557. doi: 10.1371/journal.pone.0066557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qureshi A, Thakur N, Tandon H, Kumar M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014;42:D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gautam A, Chaudhary K, Kumar R, Sharma A, Kapoor P, Tyagi A. Open source drug discovery consortium. Raghava GPS: In silico approaches for designing highly effective cell penetrating peptides. J Transl Med. 2013;11:74. doi: 10.1186/1479-5876-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma A, Kapoor P, Gautam A, Chaudhary K, Kumar R, Chauhan JS, Tyagi A, Raghava GPS. Computational approach for designing tumor homing peptides. Sci Rep. 2013;3:1607. doi: 10.1038/srep01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tyagi A, Kapoor P, Kumar R, Chaudhary K, Gautam A, Raghava GPS. In silico models for designing and discovering novel anticancer peptides. Sci Rep. 2013;3:2984. doi: 10.1038/srep02984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thakur N, Qureshi A, Kumar M. AVPpred: collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012;40:W199–W204. doi: 10.1093/nar/gks450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta S, Kapoor P, Chaudhary K, Gautam A, Kumar R. Open Source Drug Discovery Consortium, Raghava GPS: In silico approach for predicting toxicity of peptides and proteins. PLoS ONE. 2013;8(9):e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Hagan DT, Illum L. Absorption of peptides and proteins from the respiratory tract and the potential for development of locally administered vaccine. Crit Rev Ther Drug Carrier Syst. 1990;7(1):35–97. [PubMed] [Google Scholar]
- 28.Hamman JH, Enslin GM, Kotzé AF. Oral delivery of peptide drugs: barriers and developments. BioDrugs. 2005;19(3):165–177. doi: 10.2165/00063030-200519030-00003. [DOI] [PubMed] [Google Scholar]
- 29.Shaji J, Patole V. Protein and Peptide drug delivery: oral approaches. Indian J Pharm Sci. 2008;70(3):269–277. doi: 10.4103/0250-474X.42967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torres-Lugo M, Peppas NA. Transmucosal delivery systems for calcitonin: a review. Biomaterials. 2000;21(12):1191–1196. doi: 10.1016/S0142-9612(00)00011-9. [DOI] [PubMed] [Google Scholar]
- 31.Sayani AP, Chien YW. Systemic delivery of peptides and proteins across absorptive mucosae. Crit Rev Ther Drug Carrier Syst. 1996;13(1–2):85–184. [PubMed] [Google Scholar]
- 32.Banga AK, Chien YW. Hydrogel-based iontotherapeutic delivery devices for transdermal delivery of peptide/protein drugs. Pharm Res. 1993;10(5):697–702. doi: 10.1023/A:1018955631835. [DOI] [PubMed] [Google Scholar]
- 33.Lee YC, Yalkowsky SH. Effect of formulation on the systemic absorption of insulin from enhancer-free ocular devices. Int J Pharm. 1999;185(2):199–204. doi: 10.1016/S0378-5173(99)00156-8. [DOI] [PubMed] [Google Scholar]
- 34.Jitendra, Sharma PK, Bansal S, Banik A. Noninvasive routes of proteins and peptides drug delivery. Indian J Pharm Sci. 2011;73(4):367–375. doi: 10.4103/0250-474X.95608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oyston PC, Fox MA, Richards SJ, Clark GC. Novel peptide therapeutics for treatment of infections. J Med Microbiol. 2009;58:977–987. doi: 10.1099/jmm.0.011122-0. [DOI] [PubMed] [Google Scholar]
- 36.Vaara M. New approaches in peptide antibiotics. Curr Opin Pharmacol. 2009;9(5):571–576. doi: 10.1016/j.coph.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Gorris HH, Bade S, Röckendorf N, Albers E, Schmidt MA, Fránek M, Frey A. Rapid profiling of peptide stability in proteolytic environments. Anal Chem. 2009;81(4):1580–1586. doi: 10.1021/ac802324f. [DOI] [PubMed] [Google Scholar]
- 38.Prabhakaran M. The distribution of physical, chemical and conformational properties in signal and nascent peptides. Biochem J. 1990;269(3):691–696. doi: 10.1042/bj2690691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, Hochstrasser DF. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. doi: 10.1385/1-59259-584-7:531. [DOI] [PubMed] [Google Scholar]
- 40.Lazaro E, Kadie C, Stamegna P, Zhang SC, Gourdain P, Lai NY, Zhang M, Martinez SA, Heckerman D, Gall SL. Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J Clin Invest. 2011;121(6):2480–2492. doi: 10.1172/JCI44932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song X, Zhou T, Jia H, Guo X, Zhang X, Han P, Sha J. SProtP: A Web Server to Recognize Those Short-Lived Proteins Based on Sequence-Derived Features in Human Cells. PLoS ONE. 2011;6(11):e27836. doi: 10.1371/journal.pone.0027836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global Protein Stability Profiling in Mammalian Cells. Science. 2008;322(5903):918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 43.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 44.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2(1):13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- 45.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 46.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315(5809):201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 47.Schnell JD, Hicke L. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J Biol Chem. 2003;278(38):35857–35860. doi: 10.1074/jbc.R300018200. [DOI] [PubMed] [Google Scholar]
- 48.Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25(19):5425–5432. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 49.Goldsack DE, Chalifoux RC. Contribution of the free energy of mixing of hydrophobic side chains to the stability of the tertiary structure of proteins. J Theor Biol. 1973;39(3):645–651. doi: 10.1016/0022-5193(73)90075-1. [DOI] [PubMed] [Google Scholar]
- 50.Zimmerman JM, Eliezer N, Simha R. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol. 1968;21(2):170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- 51.Gromiha MM, Thangakani AM, Selvaraj S. FOLD-RATE: prediction of protein folding rates from amino acid sequence. Nucleic Acids Res. 2006;34:W70–W74. doi: 10.1093/nar/gkl043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du P, Li Y. Prediction of protein submitochondria locations by hybridizing pseudo-amino acid composition with various physicochemical features of segmented sequence. BMC Bioinformatics. 2006;7:518. doi: 10.1186/1471-2105-7-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mishra NK, Kumar M, Raghava GPS. Support vector machine based prediction of glutathione S-transferase proteins. Protein Pept Lett. 2007;14(6):575–580. doi: 10.2174/092986607780990046. [DOI] [PubMed] [Google Scholar]
- 54.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20(15):2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 55.Wang P, Hu L, Liu G, Jiang N, Chen X, Xu J, Zheng W, Li L, Tan M, Chen Z, Song H, Cai Y, Chou K. Prediction of antimicrobial peptides based on sequence alignment and feature selection methods. PLoS ONE. 2011;6:e18476. doi: 10.1371/journal.pone.0018476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Weka based results for both HL10 (Table S1) and HL16 (Table S2) using selected features. Differences in amino acid composition for longest half-life (stable) Vs shortest half-life (unstable) containing 10mer (Table S3) and 16mer peptides (Table S4). (XLS 35 KB)
Additional file 2: Performance of SVM and Weka based models (Tables S1-S6) after multiplying their actual half-life with 10 and 1000. Longest half-life (Stable) and shortest half-life containing 10mer (Table S7) and 16mer peptides (Table S8). (DOC 419 KB)


