Abstract
Background
The standard therapy regimen of conventional osteosarcoma includes neoadjuvant chemotherapy followed by surgical resection and postoperative chemotherapy. The percentage of necrotic tissue following induction chemotherapy is assessed by using the Huvos grading system, which classifies patients as “poor responders” (PR) and “good responders” (GR). The aim of this study was to identify molecular markers expressed differentially between good and poor responders to neoadjuvant chemotherapy in order to predict the response to chemotherapy in conventional osteosarcomas before beginning treatment.
Methods
Suppression Substractive Hybridization (SSH) was performed by using cDNA from frozen biopsy specimens. Expression of selected relevant genes identified by SSH was validated by using QRT-PCR. Immunohistochemistry (IHC) on tissue microarray (TMA) sections of 52 biopsies was performed to investigate protein expression in an independent cohort.
Results
ERK1 and STAT3 mRNA level were significantly different between PR and GR in an independent cohort. Phosphorylated STAT3 and ERK1 expressions by IHC on TMA were correlated with poor response to chemotherapy.
Conclusions
Our results suggest that ERK1 and STAT3 expression are good predictive markers for chemotherapy response and that inhibitors might be used in combination with common chemotherapeutic drugs in conventional osteosarcomas.
Keywords: Conventional osteosarcomas, Predictive factors, Chemotherapy response, STAT3, ERK1
Background
Osteosarcoma, the most common type of primary bone cancer, is a rare disease. Approximately 900 new cases of osteosarcoma are diagnosed each year in the United States (http://www.cancer.org/docroot/home/index.asp) and 200 in France, including 150 in children (http://www.fnclcc.fr/sor/SSP/CancersEnfant/PeauTissusSoutien/Osteosarcome). Adjuvant and neoadjuvant chemotherapy have significantly improved the long-term survival rate for patients with osteosarcoma [1–3]. Nevertheless, recurrent disease still occurs in about 30–40% of patients and more than 70% of them die of their tumor, despite second-line treatment. The standard therapy regimen of high-grade osteosarcoma includes induction by multiagent chemotherapy followed by surgical resection and postoperative chemotherapy [4]. The percentage of necrotic tissue following induction chemotherapy is classified with the Huvos grading system [5]. Patients with <90% tumor necrosis following induction therapy are classified as “poor responders” (PR) or Huvos grade I/II [6], while more than 90% necrosis corresponds to Huvos grade III and complete necrosis to Huvos grade IV. Good responders (GR) correspond to Huvos grade III/IV. The degree of necrosis remains the only reliable prognostic factor for the patients presenting with localized disease and is used to guide the choice of postoperative chemotherapy. Numerous clinical trials have attempted to increase the disease-free survival rate for poorly responding patients with intensified postoperative therapy. No survival benefit has been convincingly shown through the administration of more intensified therapy to poor responders [3, 7–10]. This suggests that there may be an innate biological difference between good responsive and poor responsive tumors. Previous transcriptomic studies have shown that in chemoresistant tumors, the genes involved in osteoclastogenesis, extracellular matrix remodeling, bone development, tumor progression, drug resistance and angiogenesis are up-regulated [11–17]. However, none of these molecular predictive factors can be used routinely. Therefore, there is a need to establish reliable predictive biomarkers for the response to chemotherapy at the time of diagnosis. The aim of this study was to identify molecular markers expressed differentially between “good” and “poor” responders to neoadjuvant chemotherapy in order to predict the response to chemotherapy in conventional osteosarcoma before beginning treatment, and to elucidate the mechanisms involved in this response. We identified several subsets of novel potential candidate genes. In particular, our data suggest that ERK1 and STAT3 expression are involved in the response to chemotherapy and that they could be therapeutic targets.
Methods
Patients and tumor specimens
The response to preoperative chemotherapy was assessed on resected specimens according to Rosen’s protocol [4]. To identify differentially expressed genes between “good” and “poor” responders (GR and PR) to chemotherapy, Suppression Subtractive Hybridization (SSH) was performed by using cDNA from frozen biopsy specimens taken for diagnosis prior to treatment. SSH was performed by using 5 samples of GR patients (three males and two females, mean age 14 years) and 4 samples of PR patients (two males and two females, mean age 13.5 years). All patients received preoperative and postoperative chemotherapy derived from the SFOP OS 94 regimen [18]. Clinicopathological characteristics of the patients studied by SSH are presented in Table 1. The two groups were similar in tumor volume, tumor location and histological subtype. Expression of selected relevant genes identified by SSH was validated by using real-time quantitative RT-PCR (QRT-PCR). For QRT-PCR, the same specimens and additional specimens of 22 patients were obtained. The whole cohort consisted of 13 GR and 18 PR. Immunohistochemistry (IHC) was performed on Tissue Microarray (TMA) sections consisting of 52 biopsies of patients with a conventional osteosarcoma. Six of 9 samples used for SSH were used for TMA. 18 of the 31 samples used in QRT-PCR were used for TMA. In total, among the 52 patients in the TMA validation cohort, only 5 received chemotherapy without high-dose MTX. The vast majority of patients (40) were those treated according to protocol OS94 or by neoadjuvant chemotherapy with methotrexate, vepeside and ifosfamide. All samples were obtained after informed consent from patients or their parents when the patients were under the age of 18.
Table 1.
Clinicopathological characteristics of the 9 patients studied by SSH
| Good responders | Poor responders | |
|---|---|---|
| Number of patients | 5 | 4 |
| Mean age at diagnosis [95% IC] (years) | 14 [5-17] | 13.5 [13-16] |
| Sex | ||
| Male | 3 | 2 |
| Female | 2 | 2 |
| Tumor location | ||
| Upper limb | 2 | 1 |
| Lower limb | 3 | 3 |
| Histological subtype | ||
| Osteoblastic | 4 | 3 |
| Osteoblastic and chondroblastic | 1 | 1 |
| Mean tumor size [95% IC] (cm) | 12.5 [8-34] | 9 [6.7-25] |
| Mean viable residual tumor cells [95% IC] (%) | 2.5 [1-4.5] | 25 [17-37] |
Research involving the patients have been performed with the approval of Protection of the Person Center: CPP sud Méditerranée 1 ethics committee (authorization number: DC-2008-309) in compliance with the Helsinki Declaration. Samples were from a tumor bank that respects the ethical charter of the French National Cancer Institute (AP-HM Biobank authorization number 2013-1786).
RNApreparation
Total RNA extraction was performed from frozen tumor specimens by using the acid guanidinium isothiocyanate/phenol/chloroform procedure. Before use, RNA samples were treated with 10U ribonuclease-free deoxyribonuclease (Promega, France) at 37°C for 15 min. Tests for purity and quality were performed on a nanodrop spectrophotometer and the Agilent 2100 Bioanalyser RNA LabChip kit (Agilent Technologies, Palo Alto, CA, USA) [19, 20]. Only samples with RNA integrity Number (RIN) > 7 and no evidence of ribosomal degradation were included.
SMART-Suppression Subtractive Hybridization (SMART: “switching mechanism at 5’ end of the RNA transcript”)
Poly(A) + mRNA were isolated from GR and PR total RNA using an Oligotex mRNA isolation kit (Qiagen, France) and gene expression between these two mRNA populations was compared by SMART-SSH using a Super PCR cDNA Synthesis Kit for cDNA synthesis (Clontech) and a PCR-Select cDNA subtraction kit (Clontech), a principle previously described by Diatchenko et al. [21].
Cloning and analysis of subtracted clones
Products from the final PCR amplification were cloned into a Topo TA cloning vector (Invitrogen Life Technologies, France) and electro-transferred into One Shot E. coli. Differential screening was performed to eliminate false positives. Hybridizations were performed in duplicate according to standard procedures. Specific clones were prepared by using a Qiagen plasmid mini-kit and sequenced (QIAGEN France SAS, Coutaboeuf, France). Nucleic acid homology searches were carried out with the BLAST program at the NCBI, USA.
Quantitative Reverse Transcription Polymerase Chain Reaction (QRT-PCR)
QRT-PCR was used to accurately detect the changes of expression of selected relevant genes: ERK1 and STAT3 gene expression levels and ribosomal 18S RNA as reference sequence. Total RNA (1 μg) DNA-free was reverse-transcribed into cDNA using hexamers (Pharmacia Biotech, Orsay, France) and Superscript II Reverse Transcriptase (Invitrogen Life Technologies, France). Genes of interest and 18S rRNA were amplified, detected and quantified in real-time by using the Light Cycler Real-Time PCR (Roche Applied Science, Meylan, France). QRT-PCR was performed by using the oligonucleotides and sequence parameters described in Table 2 in a medium containing 1X LightCycler 480 SYBR Green I master mix, 0.25 μM of each primer and 20 ng of cDNA. Each PCR reaction was preceded by one activation cycle of 95°C for 5 min and ended by establishing a melting curve 5 degrees above the oligonucleotide melting temperature.
Table 2.
Description of oligonucleotides and sequence parameters for QRT-PCR
| Name gene | Oligo direct | Oligo reverse | PCR conditions | Cycle number | GeneInfo identifier |
|---|---|---|---|---|---|
| 18S | CTACCACATCCAAGGAAGGCA | TTTTTCGTCACTACCTCCCCG | 95°C 15 sec | 35 | 124517659 |
| 67°C 30 sec | |||||
| ERK1 | CTAAAGCCCTCCAACCTGCT | CAGCCCACAGACCAGATGT | 95°C 15 sec | 45 | 158138506 |
| 60°C 30 sec | |||||
| STAT3 | AAAGTCAGGTTGCTGGTCAAA | TGCCGTTGTTGGATTCTTC | 95°C 15 sec | 45 | 76253927 |
| 60°C 30 sec |
Immunohistochemistry (IHC) on tissue microarray sections (TMA)
Automated immunohistochemistry was performed on slides of TMA paraffin blocks. The 52 tumor specimens were all fixed in 4% formalin. Fleshy tissue was separated from calcified areas to avoid unnecessary decalcification. When necessary, tumor specimens were decalcified in a solution of 22% formic acid. TMA were prepared as previously described [20]. For each sample, three representative sample areas were carefully selected from a hematoxylin–eosin-stained section of a donor block. Core cylinders with a diameter of 1 mm each were punched from three representative areas and deposited onto two separate recipient paraffin blocks by using a specific arraying device (Alphelys). To determine the expression of activated forms of STAT3 and ERK1 proteins, we used anti-phospho-STAT3 (Tyr705) (polyclonal, 9131 from Cell Signaling Technology, dilution 1/20) and anti-phospho-ERK1 (polyclonal, clone 20G11 from Cell Signaling Technology, dilution: 1/100) antibodies. Automated IHC was performed with a Ventana automate (Benchmark XT, Ventana Medical Systems SA, Illkirch, France). Positive external control was a glioblastoma for both pSTAT3 and pERK1. Negative controls were also included and corresponded to omission of primary antibody or irrelevant antibodies of the same isotype. IHC was scored positive when nuclear staining was observed. A semi-quantitative analysis was done for positive specimens without knowledge of clinical data. Percentage of stained cells and staining intensity (weak, moderate, high) were taken into account to obtain the score. Score 0 was attributed to tumors with absence of staining. Score 1 was attributed to tumors with low intensity of staining whatever the number of stained nuclei or to tumors with no more than 25% of nuclei immunostained with moderate intensity. Score 2 corresponded to stained nuclei numbering between 25% and 50% with moderate intensity or to fewer than 25% of stained nuclei with high intensity. Score 3 was defined as either more than 50% of stained nuclei with moderate intensity or more than 25% of stained nuclei with high staining intensity. A mean score was proposed for the three areas of each tumor. Three independent observers evaluated the IHC results blind to clinical data. A consensus score was reached and statistical analysis was performed from the consensus score.
Data analysis
Relationships between response to chemotherapy (GR vs. PR) and other parameters used were obtained by using non-parametric tests, the Fisher exact test and the Mann-Whitney test when qualitative and continuous respectively. All tests were two-sided. P-value was considered significant when ≤ 5%. SAS System version 9.2 (SAS Institute Inc., Cary, NC) and Stata software (version 10.1 Special Edition, StataCorp, College Station, Texas) were used to perform data analyses.
Results
Patients
Clinicopathological characteristics of the patients studied are presented in Table 3.
Table 3.
Clinicopathological characteristics of the 52 patients studied by IHC including those studied by SSH and QRT-PCR
| Whole cohort | |
|---|---|
| Number of patients | 52 |
| Age | |
| Mean age at diagnosis [95% IC]a (years) | 17.4 [5;80] |
| Sex | |
| Male (%) | 34 (65.4) |
| Female (%) | 18 (34.6) |
| Histologic response | |
| Good responders | 24 |
| Poor responders | 28 |
| Histological diagnosis and subtype | |
| High-grade osteosarcomas of central “conventional” type | 52 |
| Osteoblastic (%) | 38 (73) |
| Chondroblastic (%) | 5 (9.5) |
| Telangiectasic (%) | 3 (6) |
| Fibroblastic (%) | 2 (4) |
| Mixed subtypeb (%) | 4 (7.5) |
aConfidence Interval, for the whole cohort, to ascertain that the screening cohort is a representative subset of the whole.
bOsteoblastic and chondroblastic or fibroblastic.
Identification of differentially expressed genes by SSH in PR
A subtractive cDNA library of PR was generated. 126 selected clones were sequenced (Table 4). The following genes were selected on the basis of their known roles in tumorigenesis or chemoresistance: ACTN1, AKT2, ANXA2, CADM1, CDKN2C(P18), FN1, GAL1, HRAS, IGFBP3, LMNA, ERK1 and STAT3. Particularly, STAT3 is a key factor for chemosensitivity in human epithelial ovarian cancer cells and thyroid cancer-derived CD133+ cells [22–24]. Recent studies show that ERKs may also be activated in response to chemotherapeutic drugs, and pERK1/2 played critical roles in drug resistance [25–28]. Thus, these selected genes were tested by QRT-PCR.
Table 4.
Identification of genes differentially expressed by SSH in PR
| Gene title | Gene symbol | Chromosomal location |
|---|---|---|
| Actin, alpha 1, skeletal muscle | ACTA1 | chr1q42.13-q42.2 |
| Actin, beta | ACTB | chr7p15-p12 |
| Actin, gamma 1 | ACTG1 | chr17q25 |
| Actinin, alpha 1 | ACTN1 | chr14q24.1-q24.2|14q24|14q22-q24 |
| ADAM metallopeptidase with thrombospondin type 1 motif, 20 | ADAMTS20 | chr12q12 |
| v-akt murine thymoma viral oncogene homolog 2 | AKT2 | chr19q13.1-q13.2 |
| Ankyrin repeat domain 11 | ANKRD11 | chr16q24.3 |
| Annexin A2 | ANXA2 | chr15q21-q22 |
| AT rich interactive domain 4B (RBP1-like) | ARID4B | chr1q42.1-q43 |
| Actin-related protein 2/3 complex, subunit 2, 34 kDa | ARPC2 | chr2q36.1 |
| ATPase family, AAA domain containing 3A | ATAD3A | chr1p36.33 |
| ATP synthase, H + transporting, mitochondrial F0 complex, subunit E///major facilitator superfamily domain containing 7 | ATP5I///MFSD7 | chr4p16.3 |
| Bromo adjacent homology domain containing 1 | BAHD1 | chr15q15.1 |
| Breast carcinoma amplified sequence 3 | BCAS3 | chr17q23 |
| Branched chain aminotransferase 2, mitochondrial | BCAT2 | chr19q13 |
| Chromosome 14 open reading frame 112 | C14orf112 | chr14q24.2 |
| Chromosome 14 open reading frame 2 | C14orf2 | chr14q32.33 |
| Chromosome 20 open reading frame 194 | C20orf194 | chr20p13 |
| Cell adhesion molecule 1 | CADM1 | chr11q23.2 |
| Coiled-coil domain containing 28B | CCDC28B | chr1p35.1 |
| Chaperonin containing TCP1, subunit 8 (theta) | CCT8 | chr21q22.11 |
| Cell division cycle 34 homolog (S. cerevisiae) | CDC34 | chr19p13.3 |
| Cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) | CDKN2C | chr1p32 |
| Carbohydrate (chondroitin 4) sulfotransferase 11 | CHST11 | chr12q |
| Creatine kinase, brain | CKB | chr14q32 |
| CDC28 protein kinase regulatory subunit 1B | CKS1B | chr1q21.2 |
| CLPTM1-like | CLPTM1L | chr5pter-p15.3 |
| Cornifelin | CNFN | chr19q13.2 |
| Collagen, type V, alpha 1 | COL5A1 | chr9q34.2-q34.3 |
| Catechol-O-methyltransferase | COMT | chr22q11.21-q11.23|22q11.21 |
| Cytochrome c oxidase subunit VIa polypeptide 1 | COX6A1 | chr12q24.2|12q24.2 |
| Cytokine receptor-like factor 1 | CRLF1 | chr19p12 |
| Chondroitin sulfate glucuronyltransferase | CSGlcA-T | chr7q36.1 |
| Casein kinase 2, alpha prime polypeptide | CSNK2A2 | chr16q21 |
| cutA divalent cation tolerance homolog (E. coli) | CUTA | chr6pter-p21.31 |
| dodecenoyl-Coenzyme A delta isomerase (3,2 trans-enoyl-Coenzyme A isomerase) | DCI | chr16p13.3 |
| Dicarbonyl/L-xylulose reductase | DCXR | chr17q25.3 |
| DEAD (Asp-Glu-Ala-As) box polypeptide 19A | DDX19A | chr16q22.1 |
| DEAD (Asp-Glu-Ala-As) box polypeptide 19B///DEAD (Asp-Glu-Ala-As) box polypeptide 19A | DDX19A///DDX19B | chr16q22.1 |
| DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 | DDX39 | chr19p13.12 |
| Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | EEF1D | chr8q24.3 |
| Eukaryotic elongation factor-2 kinase | EEF2K | chr16p12.1 |
| Eukaryotic translation initiation factor 3, subunit H | EIF3H | chr8q24.11 |
| Eukaryotic translation initiation factor 4 gamma, 3 | EIF4G3 | chr1p36.12 |
| Fas apoptotic inhibitory molecule 3 | FAIM3 | chr1q32.1 |
| FK506 binding protein 7 | FKBP7 | chr2q31.2 |
| Kappa-actin | FKSG30 | chr2q21.1 |
| Flavin containing monooxygenase 5 | FMO5 | chr1q21.1 |
| Fibronectin 1 | FN1 | chr2q34 |
| FERM domain containing 5 | FRMD5 | chr15q15.3 |
| Golgi SNAP receptor complex member 2 | GOSR2 | chr17q21 |
| Glypican 1 | GPC1 | chr2q35-q37 |
| G protein-coupled receptor 108 | GPR108 | chr19p13.3 |
| Ribosomal protein L23a///similar to ribosomal protein L23A///ribosomal protein L23a-like | hCG_16001///hCG_2001000///RPL23A | chr17q11///chr17q23.2///chr3q26.1 |
| v-Ha-ras Harvey rat sarcoma viral oncogene homolog | HRAS | chr11p15.5 |
| Heparan sulfate proteoglycan 2 | HSPG2 | chr1p36.1-p34 |
| Insulin-like growth factor 2 mRNA binding protein 3 | IGF2BP3 | chr7p11 |
| Inositol(myo)-1(or 4)-monophosphatase 2 | IMPA2 | chr18p11.2 |
| Integrator complex subunit 1 | INTS1 | chr7p22.3 |
| Importin 11 | IPO11 | chr5q12.1 |
| Jumonji domain containing 2C | JMJD2C | chr9p24.1 |
| KIAA0999 protein | KIAA0999 | chr11q23.3 |
| Laminin, alpha 4 | LAMA4 | chr6q21 |
| Lectin, galactoside-binding, soluble, 1 (galectin 1) | LGALS1 | chr22q13.1 |
| Lamin A/C | LMNA | chr1q21.2-q21.3 |
| Ribosomal protein S16///similar to 40S ribosomal protein S16 | LOC441876///RPS16 | chr19q13.1///chr1p36.21 |
| Leucine-rich repeat containing 28 | LRRC28 | chr15q26.3 |
| Microtubule-associated protein 1S | MAP1S | chr19p13.11 |
| Mitogen-activated protein kinase 3 | MAPK3 (ERK1) | chr16p11.2 |
| Major facilitator superfamily domain containing 5 | MFSD5 | chr12q13.13 |
| Mitochondrial ribosomal protein S7 | MRPS7 | chr17q25 |
| NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9 kDa | NDUFA4 | chr7p21.3 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20 kDa (NADH-coenzyme Q reductase) | NDUFS7 | chr19p13.3 |
| NADH dehydrogenase (ubiquinone) flavoprotein 1, 51 kDa | NDUFV1 | chr11q13 |
| Nuclear factor I/C (CCAAT-binding transcription factor) | NFIC | chr19p13.3 |
| NOL1/NOP2/Sun domain family, member 5 | NSUN5 | chr7q11.23 |
| NOL1/NOP2/Sun domain family, member 5B | NSUN5B | chr7q11.23 |
| NOL1/NOP2/Sun domain family, member 5C | NSUN5C | chr7q11.23 |
| Nucleoporin 214 kDa | NUP214 | chr9q34.1 |
| Nucleoporin 85 kDa | NUP85 | chr17q25.1 |
| PDZ domain containing 2 | PDZD2 | chr5p13.3 |
| Periplakin | PPL | chr16p13.3 |
| Protein phosphatase 1, regulatory (inhibitor) subunit 12B | PPP1R12B | chr1q32.1 |
| Protein phosphatase 2 (formerly 2A), regulatory subunit A, alpha isoform | PPP2R1A | chr19q13.33 |
| Protein kinase C substrate 80 K-H | PRKCSH | chr19p13.2 |
| Protein arginine methyltransferase 2 | PRMT2 | chr21q22.3 |
| RNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) | RALY | chr20q11.21-q11.23 |
| RNA binding motif protein 4 | RBM4 | chr11q13 |
| RNA binding motif protein 4B | RBM4B | chr11q13 |
| RNA binding motif protein 8A | RBM8A | chr1q12 |
| Ribosomal protein L13 | RPL13 | chr16q24.3|17p11.2 |
| Ribosomal protein L13a | RPL13A | chr19q13.3 |
| Ribosomal protein L19 | RPL19 | chr17q11.2-q12 |
| Ribosomal protein L23a | RPL23A | chr17q11 |
| Ribosomal protein L31 | RPL31 | chr2q11.2 |
| Ribosomal protein, large, P1 | RPLP1 | chr15q22 |
| Ribosomal protein S12 | RPS12 | chr6q23.2 |
| Ribosomal protein S14 | RPS14 | chr5q31-q33 |
| Ribosomal protein S17 | RPS17 | chr15q |
| Ribosomal protein S21 | RPS21 | chr20q13.3 |
| Ribosomal protein S27 (metallopanstimulin 1) | RPS27 | chr1q21 |
| Ribosomal protein S6 | RPS6 | chr9p21 |
| RNA pseudouridylate synthase domain containing 4 | RPUSD4 | chr11q24.2 |
| Ribosomal RNA processing 1 homolog B (S. cerevisiae) | RRP1B | chr21q22.3 |
| Retinoid X receptor, alpha | RXRA | chr9q34.3 |
| Synaptonemal complex protein SC65 | SC65 | chr17q21.2 |
| Splicing factor, arginine/serine-rich 3 | SFRS3 | chr6p21 |
| Serine hydroxymethyltransferase 2 (mitochondrial) | SHMT2 | chr12q12-q14 |
| SIVA1, apoptosis-inducing factor | SIVA1 | chr14q32.33 |
| SIVA1, apoptosis-inducing factor | SIVA1 | chr14q32.33 |
| Solute carrier family 16, member 8 (monocarboxylic acid transporter 3) | SLC16A8 | chr22q12.3-q13.2 |
| Solute carrier family 20 (phosphate transporter), member 2 | SLC20A2 | chr8p12-p11 |
| Small nuclear ribonucleoprotein D3 polypeptide 18 kDa | SNRPD3 | chr22q11.23 |
| Signal transducer and activator of transcription 3 (acute-phase response factor) | STAT3 | chr17q21.31 |
| Serine/threonine kinase 24 (STE20 homolog, yeast) | STK24 | chr13q31.2-q32.3 |
| T-cell, immune regulator 1, ATPase, H + transporting, lysosomal V0 subunit A3 | TCIRG1 | chr11q13.2 |
| Testis-specific kinase 1 | TESK1 | chr9p13 |
| Thymosin, beta 10 | TMSB10 | chr2p11.2 |
| Transportin 3 | TNPO3 | chr7q32.1 |
| Tetraspanin 9 | TSPAN9 | chr12p13.33-p13.32 |
| Ubiquitin A-52 residue ribosomal protein fusion product 1 | UBA52 | chr19p13.1-p12 |
| Vacuolar protein sorting 28 homolog (S. cerevisiae) | VPS28 | chr8q24.3 |
| Williams-Beuren syndrome chromosome region 16 | WBSCR16 | chr7q11.23 |
| WW domain containing oxidoreductase | WWOX | chr16q23.3-q24.1 |
| X antigen family, member 1D///X antigen family, member 1C///X antigen family, member 1E///X antigen family, member 1///X antigen family, member 1B | XAGE1///XAGE1B///XAGE1C///XAGE1D///XAGE1E | chrXp11.22 |
| Zinc finger protein 449 | ZNF449 | chrXq26.3 |
QRT-PCR validation of selected genes expressed in PR versus GR by SSH
Only STAT3 mRNA level and ERK1 mRNA level were significantly different between PR and GR. Quantification of STAT3 and ERK1 mRNA transcripts revealed higher mRNA levels in PR compared to GR samples (p = 0.019 and p = 0.046 respectively). The mean level of STAT3 mRNA was 0.820 [0.280-13.970] in PR versus 0.310 [0.230-2.370] in GR samples (Figure 1A) and the mean level of ERK1 was 0.270 [0.110-4.340] in PR versus 0.150 [0.088-0.710] in GR samples (Figure 1B).
Figure 1.
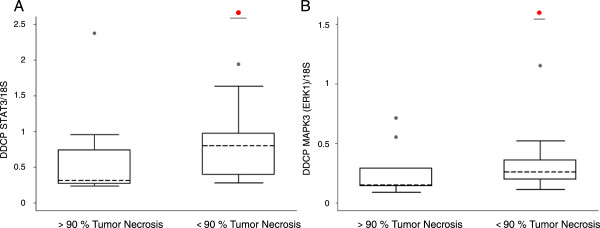
RTQ-PCR analysis of STAT3 and ERK1 genes. A: Quantification of STAT3 mRNA with 18S rRNA reference gene transcript confirmed higher STAT3 mRNA levels in poor responder (PR) samples compared with good responder (GR) samples (p = 0.019). B: Quantification of MAPK3 (ERK1) mRNA with 18S rRNA reference gene transcript confirmed higher MAPK3 (ERK1) mRNA levels in PR samples compared with GR samples (p = 0.046).
Validation at protein level using immunohistochemistry for pSTAT3 and pERK1 (Tables 5 and 6)
Table 5.
Correlation between phosphorylated STAT3 and ERK1 IHC expression to poor response to chemotherapy for a percentage of viable residual cells ≤10%
| IHC score | Good responders | Poor responders | p-value | |
|---|---|---|---|---|
| Phosphorylated STAT3 | 0 or 1 | 16 | 9 | 0.036 |
| 2 or 3 | 6 | 14 | ||
| VPP(PR) = 14/20 = 70%/VPN(GR) = 16/25 = 64% | ||||
| Phosphorylated ERK1 | 0 or 1 | 17 | 8 | 0.007 |
| 2 or 3 | 5 | 15 | ||
| VPP(PR) = 15/20 = 75%/VPN(GR) = 17/25 = 68% | ||||
| Phosphorylated STAT3 and ERK1 | Both 0-1 | 12 | 4 | 0.003 |
| Intermediate | 8 | 7 | ||
| Both 2-3 | 1 | 10 | ||
| VPP(both/PR) = 10/11 = 91%/VPN(both/GR) = 12/16 = 75% | ||||
Table 6.
Correlation between phosphorylated STAT3 and ERK1 IHC expression to poor response to chemotherapy for a percentage of viable residual cells lower than 5%
| IHC score | Good responders | Poor responders | p-value | |
|---|---|---|---|---|
| Phosphorylated STAT3 | 0 or 1 | 13 | 12 | 0.013 |
| 2 or 3 | 3 | 17 | ||
| VPP(PR) = 17/20 = 85%/VPN(GR) = 13/25 = 52% | ||||
| Phosphorylated ERK1 | 0 or 1 | 13 | 12 | 0.035 |
| 2 or 3 | 4 | 16 | ||
| VPP(PR) = 16/20 = 80%/VPN(GR) = 13/25 = 52% | ||||
| Phosphorylated STAT3 and ERK1 | Both 0-1 | 11 | 5 | 0.007 |
| Intermediate | 5 | 10 | ||
| Both 2-3 | 1 | 10 | ||
| VPP(both/PR) = 10/11 = 91%/VPN(both/GR) = 11/16 = 69% | ||||
pSTAT3 nuclear expression was examined in 45 cases out of 52 and a high score was observed in 20 cases (score 2 and 3). pERK1 expression was examined in 45 cases out of 52 (low score in 25 cases and high score in 20 cases) (Figure 2). pSTAT3 protein expression was correlated to poor response to chemotherapy for a percentage of viable residual cells ≤10%, with the higher scores in the PR group (p = 0.036). A statistically significant correlation was also found between pERK1 protein expression and response to chemotherapy when comparing low scores (0-1) versus high scores (2-3) (p = 0.007). Moreover, the correlation between the expression of pSTAT3 and pERK1 in IHC and the response to chemotherapy remained statistically significant for patients under 25 years (p = 0.024 and p = 0.010 respectively). For a percentage of viable residual cells lower than 5%, a statistically significant correlation was still found between pSTAT3 or pERK1 protein expression and response to chemotherapy (p = 0.013 and p = 0.035 respectively). Whatever the threshold (5 or 10%), positive predictive value (probability of belonging to the group of PR in case of high score) of both pSTAT3 and pERK1 in combination was 91%. Negative predictive value (probability of belonging to the group of GR in case of low score) of both pSTAT3 and pERK1 in combination for a 5 and 10% threshold were 69% and 75% respectively
Figure 2.
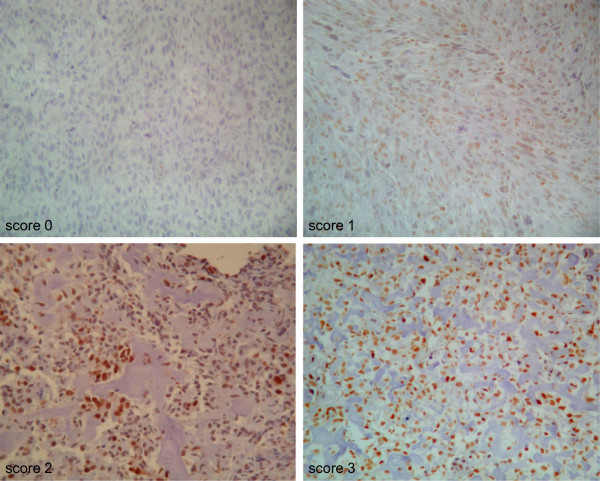
IHC pSTAT3 scores. Score 0: negative staining (X200). Score 1: >50% of nuclei are labeled with low staining intensity (X200). Score 2: <50% cells with moderate staining intensity and <25% of cells are highly stained (X200). Score 3: more than 50% of nuclei are stained with moderate staining intensity (X200).
Discussion
SSH is a molecular biology technique that enables the identification of differentially expressed genes between two groups with high sensitivity. By comparing PR to GR prior to chemotherapy among patients with an osteosarcoma, we found 126 clones. ERK1 and STAT3, the genes selected on the basis of their roles in tumorigenesis or chemoresistance, were further studied by QRT-PCR in an independent cohort. ERK1 and STAT3 expressions assessed by QRT-PCR and IHC were significantly linked to the response to chemotherapy. The protein encoded by ERK1 is a member of the MAP kinase family and acts in a signalling cascade that regulates various cellular processes such as proliferation, differentiation, and cell cycle progression in response to a variety of extracellular signals. We found ERK1/2 positivity score by IHC and ERK1/2 IHC high score (score 2 and 3) in 78% and 51% of our cohort, respectively. These results suggested that ERK1/2 pathway could be involved in osteosarcoma as it has supported by Pignochino et al. study that showed activated ERK 1/2 pathway in 66.6% of osteosarcoma samples. Moreover, the same team also showed that Sorafenib, a tyrosine kinase inhibitor, blocks tumor growth, angiogenesis and metastatic potential in preclinical models of osteosarcoma through a mechanism potentially involving the inhibition of ERK1/2 [29]. No attempt to investigate the link between ERK1 expression and response to chemotherapy was made in vivo. However, our work suggested that ERK1 could be involved in drug resistance as reported recently by Si et al. with an approach by RNAi-mediated knockdown of ERK1/2 inhibiting cell proliferation and invasion and increasing chemosensitivity to cisplatin in human osteosarcoma U2-OS cells in vitro [30].
STAT3 is one of the transcription factors reported to play an important role in tumor survival, proliferation, angiogenesis and metastasis. In normal cells, STAT3 is activated transiently to maintain homeostasis. However, if STAT3 continues to be activated, the abnormal level of expression can trigger oncogenic pathways. Aberrant active STAT3 promotes uncontrolled growth and survival through dysregulation of expression of downstream targeted genes including survivin, Bcl-xL, Bcl-2, Mcl-1, c-Myc and cyclin D1. Constitutive activation of the STAT3 pathway has recently been shown in several malignancies, especially osteosarcoma [31]. It has recently been implicated in resistance to chemotherapy-induced apoptosis [32]. Furthermore, activation of STAT3 in several cancers has been found to be correlated with clinical outcome especially in osteosarcoma. A high level of expression of STAT3 by IHC in 76 biopsies of patients with an osteosarcoma was a poor prognostic factor for both overall survival and disease-free survival in univariate and multivariate analysis [33]. High staining with pSTAT3 was also of prognostic value in another series of 51 conventional osteosarcomas [34]. In addition, inhibition of STAT3 plays a role in proliferation, apoptosis and migration in osteosarcoma cells in vitro. The down-regulation of STAT3 by miR-125b suppresses in vitro proliferation and migration of osteosarcoma cells [35]. STAT3 inhibition by RNA interference induces inhibition of proliferation and apoptosis enhancement in osteosarcoma cells [33]. The novel curcumin analog FLLL32 decreases STAT3 DNA binding activity and expression, and induces apoptosis in osteosarcoma cell lines [36]. The small molecules, LLL12 and FLLL32, inhibit STAT3 phosphorylation and exhibit potent growth suppressive activity in osteosarcoma cells and tumor growth in mice [37]. In contrast, oncostatin M promotes STAT3 activation, VEGF production, and invasion in osteosarcoma cell lines [38]. Finally, STAT3 is involved in drug resistance in osteosarcoma cell lines. Ryu et al. recently showed that the STAT3 pathway was overexpressed in MDR osteosarcoma cells and that inhibitors of STAT3 such as CDDO-Me could reduce resistance to doxorubicin in these cell lines [31]. In our study, we have showed an expression of pSTAT3 in 58% of cases. This activated STAT3 pathway was correlated to poor response to chemotherapy. Thus, our results are consistent with the results of the literature in vitro through the analysis of patient samples.
The effects of EGFR are mediated by activation of downstream signal transduction cascades that include Janus tyrosine kinases (Jak), Signal Transducers and Activators of Transcription (STAT), Phophatidyl Inositol 3 Kinase (PI3K)/Akt and Ras/RAf/MAP kinase (ERK). The prognostic value of EGFR and its downstream signaling molecules such as STAT3 and ERK1 have been studied in many tumor types. Only one study [39] simultaneously examined the status of EGFR and four downstream molecules - pSTAT3, pERK1, pAkt, survivin - by IHC in 47 samples of conventional osteosarcomas. ERK1 and survivin expression were statistically correlated with survival. A high expression was negatively correlated with prognosis. Furthermore, EGFR expression was correlated with expression of ERK1 and it was observed a significant association of survivin expression with STAT3 and ERK activation. These results and ours support the idea that ERK is a downstream signaling molecule of EGFR and also suggest a link between the EGFR signaling pathway and drug resistance through ERK1 and STAT3 expression in conventional osteosarcoma.
Conclusions
We have shown that high pSTAT3 and pERK1 expression in the biopsies are suggestive of poor response to chemotherapy. The elevated positive predictive value of high score of both pSTAT3 and pERK1 in combination (91%) highly suggests that IHC test could be used at the time of diagnosis to stratifying patients enrolled in randomized trials. Our results also suggest that STAT3 and ERK1 inhibitors might be used in combination with common chemotherapeutic drugs in osteosarcoma in order to increase the response to chemotherapy and to improve the prognosis. Finally, other genes identified by SSH remain to be explored and a prospective validation phase on a larger cohort is still needed before these biomarkers could be used in clinical practice.
Acknowledgements
We are grateful to R. Cooke for editing the English. Samples were collected and stored in the Assistance Publique des Hôpitaux de Marseille tumor bank (Tumor Biobank authorized nb AC-2013-1786.). This study was supported by grants from “projet libre INCA recherche Biomédicale 2010.
Footnotes
Competing interests
The author(s) declare that they have no competing interests.
Authors’ contributions
Conceived and designed the experiments: SS, CJJ, CB. Performed the experiments: CJJ, CB, CB, FF, AP, FF. Analyzed the data: SS, LC, PJ. Contributed reagents/materials/analysis tools: SS, JLD, JCG. Wrote the manuscript: SS, CJJ. All authors read and approved the final manuscript.
Contributor Information
Sébastien Salas, Email: sebastien.salas@ap-hm.fr.
Carine Jiguet-Jiglaire, Email: Carine.JIGUET-JIGLAIRE@univ-amu.fr.
Loic Campion, Email: Loic.Campion@ico.unicancer.fr.
Catherine Bartoli, Email: catherine.bartoli@univ-amu.fr.
Frédéric Frassineti, Email: frederic.frassineti@univ-amu.fr.
Jean-Laurent Deville, Email: Jean-laurent.DEVILLE@ap-hm.fr.
André Maues De Paula, Email: Andre.MAUESDEPAULA@ap-hm.fr.
Fabien Forest, Email: fabienforest@gmail.com.
Pascal Jézéquel, Email: Pascal.Jezequel@ico.unicancer.fr.
Jean-Claude Gentet, Email: Jean-claude.GENTET@ap-hm.fr.
Corinne Bouvier, Email: corinne.bouvier@univ-amu.fr.
References
- 1.Krailo M, Ertel I, Makley J, Fryer CJ, Baum E, Weetman R, Yunis E, Barnes L, Bleyer WA, Hammond GD. A randomized study comparing high-dose methotrexate with moderate-dose methotrexate as components of adjuvant chemotherapy in childhood nonmetastatic osteosarcoma: a report from the Childrens Cancer Study Group. Med Pediatr Oncol. 1987;15(2):69–77. doi: 10.1002/mpo.2950150205. [DOI] [PubMed] [Google Scholar]
- 2.Souhami RL, Craft AW, Van der Eijken JW, Nooij M, Spooner D, Bramwell VH, Wierzbicki R, Malcolm AJ, Kirkpatrick A, Uscinska BM, Van Glabbeke M, Machin D. Randomised trial of two regimens of chemotherapy in operable osteosarcoma: a study of the European Osteosarcoma Intergroup. Lancet. 1997;350(9082):911–917. doi: 10.1016/S0140-6736(97)02307-6. [DOI] [PubMed] [Google Scholar]
- 3.Bacci G, Picci P, Ruggieri P, Mercuri M, Avella M, Capanna R, Brach Del Prever A, Mancini A, Gherlinzoni F, Padovani G. Primary chemotherapy and delayed surgery (neoadjuvant chemotherapy) for osteosarcoma of the extremities. The Istituto Rizzoli Experience in 127 patients treated preoperatively with intravenous methotrexate (high versus moderate doses) and intraarterial cisplatin. Cancer. 1990;65(11):2539–2553. doi: 10.1002/1097-0142(19900601)65:11<2539::AID-CNCR2820651125>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 4.Rosen G, Caparros B, Huvos AG, Kosloff C, Nirenberg A, Cacavio A, Marcove RC, Lane JM, Mehta B, Urban C. Preoperative chemotherapy for osteogenic sarcoma: selection of postoperative adjuvant chemotherapy based on the response of the primary tumor to preoperative chemotherapy. Cancer. 1982;49(6):1221–1230. doi: 10.1002/1097-0142(19820315)49:6<1221::AID-CNCR2820490625>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 5.Rosen G, Marcove RC, Huvos AG, Caparros BI, Lane JM, Nirenberg A, Cacavio A, Groshen S. Primary osteogenic sarcoma: eight-year experience with adjuvant chemotherapy. J Cancer Res Clin Oncol. 1983;106(Suppl):55–67. doi: 10.1007/BF00625054. [DOI] [PubMed] [Google Scholar]
- 6.Meyers PA, Gorlick R. Osteosarcoma. Pediatr Clin North Am. 1997;44(4):973–989. doi: 10.1016/S0031-3955(05)70540-X. [DOI] [PubMed] [Google Scholar]
- 7.Kalifa C, Razafindrakoto H, Vassal G, Contesso G, Vanel D, Edeline V, Valteau D, Lemerle J. Chemotherapy in osteogenic sarcoma: the experience of the Pediatric Department of the Gustave Roussy Institute. Cancer Treat Res. 1993;62:347–349. doi: 10.1007/978-1-4615-3518-8_42. [DOI] [PubMed] [Google Scholar]
- 8.Winkler K, Beron G, Delling G, Heise U, Kabisch H, Purfurst C, Berger J, Ritter J, Jurgens H, Gerein V. Neoadjuvant chemotherapy of osteosarcoma: results of a randomized cooperative trial (COSS-82) with salvage chemotherapy based on histological tumor response. J Clin Oncol. 1988;6(2):329–337. doi: 10.1200/JCO.1988.6.2.329. [DOI] [PubMed] [Google Scholar]
- 9.Provisor AJ, Ettinger LJ, Nachman JB, Krailo MD, Makley JT, Yunis EJ, Huvos AG, Betcher DL, Baum ES, Kisker CT, Miser JS. Treatment of nonmetastatic osteosarcoma of the extremity with preoperative and postoperative chemotherapy: a report from the Children’s Cancer Group. J Clin Oncol. 1997;15(1):76–84. doi: 10.1200/JCO.1997.15.1.76. [DOI] [PubMed] [Google Scholar]
- 10.Bacci G, Ferrari S, Longhi A, Picci P, Mercuri M, Alvegard TA, Saeter G, Donati D, Manfrini M, Lari S, Briccoli A, Forni C. High dose ifosfamide in combination with high dose methotrexate, adriamycin and cisplatin in the neoadjuvant treatment of extremity osteosarcoma: preliminary results of an Italian Sarcoma Group/Scandinavian Sarcoma Group pilot study. J Chemother. 2002;14(2):198–206. doi: 10.1179/joc.2002.14.2.198. [DOI] [PubMed] [Google Scholar]
- 11.Man TK, Chintagumpala M, Visvanathan J, Shen J, Perlaky L, Hicks J, Johnson M, Davino N, Murray J, Helman L, Meyer W, Triche T, Wong KK, Lau CC. Expression profiles of osteosarcoma that can predict response to chemotherapy. Cancer Res. 2005;65(18):8142–8150. doi: 10.1158/0008-5472.CAN-05-0985. [DOI] [PubMed] [Google Scholar]
- 12.Mintz MB, Sowers R, Brown KM, Hilmer SC, Mazza B, Huvos AG, Meyers PA, Lafleur B, McDonough WS, Henry MM, Ramsey KE, Antonescu CR, Chen W, Healey JH, Daluski A, Berens ME, Macdonald TJ, Gorlick R, Stephan DA. An expression signature classifies chemotherapy-resistant pediatric osteosarcoma. Cancer Res. 2005;65(5):1748–1754. doi: 10.1158/0008-5472.CAN-04-2463. [DOI] [PubMed] [Google Scholar]
- 13.Ochi K, Daigo Y, Katagiri T, Nagayama S, Tsunoda T, Myoui A, Naka N, Araki N, Kudawara I, Ieguchi M, Toyama Y, Toguchida J, Yoshikawa H, Nakamura Y. Prediction of response to neoadjuvant chemotherapy for osteosarcoma by gene-expression profiles. Int J Oncol. 2004;24(3):647–655. [PubMed] [Google Scholar]
- 14.Dalla-Torre CA, de Toledo SR, Yoshimoto M, Petrilli AS, Andrade JA, Chilton-MacNeill S, Squire JA, Zielenska M. Expression of major vault protein gene in osteosarcoma patients. J Orthop Res. 2007;25(7):958–963. doi: 10.1002/jor.20371. [DOI] [PubMed] [Google Scholar]
- 15.Dalla-Torre CA, Yoshimoto M, Lee CH, Joshua AM, de Toledo SR, Petrilli AS, Andrade JA, Chilton-MacNeill S, Zielenska M, Squire JA. Effects of THBS3, SPARC and SPP1 expression on biological behavior and survival in patients with osteosarcoma. BMC Cancer. 2006;6:237. doi: 10.1186/1471-2407-6-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srivastava A, Fuchs B, Zhang K, Ruan M, Halder C, Mahlum E, Weber K, Bolander ME, Sarkar G. High WT1 expression is associated with very poor survival of patients with osteogenic sarcoma metastasis. Clin Cancer Res. 2006;12(14 Pt 1):4237–4243. doi: 10.1158/1078-0432.CCR-05-2307. [DOI] [PubMed] [Google Scholar]
- 17.Salas S, Jezequel P, Campion L, Deville JL, Chibon F, Bartoli C, Gentet JC, Charbonnel C, Gouraud W, Voutsinos-Porche B, Brouchet A, Duffaud F, Figarella-Branger D, Bouvier C. Molecular characterization of the response to chemotherapy in conventional osteosarcomas: predictive value of HSD17B10 and IFITM2. Int J Cancer. 2009;125(4):851–860. doi: 10.1002/ijc.24457. [DOI] [PubMed] [Google Scholar]
- 18.Le Deley MC, Guinebretiere JM, Gentet JC, Pacquement H, Pichon F, Marec-Berard P, Entz-Werle N, Schmitt C, Brugieres L, Vanel D, Dupoüy N, Tabone MD, Kalifa C. SFOP OS94: a randomised trial comparing preoperative high-dose methotrexate plus doxorubicin to high-dose methotrexate plus etoposide and ifosfamide in osteosarcoma patients. Eur J Cancer. 2007;43(4):752–761. doi: 10.1016/j.ejca.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 19.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162(1):156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- 20.Richter J, Wagner U, Kononen J, Fijan A, Bruderer J, Schmid U, Ackermann D, Maurer R, Alund G, Knonagel H, Rist M, Wilber K, Anabitarte M, Hering F, Hardmeier T, Schönenberger A, Flury R, Jäger P, Fehr JL, Schraml P, Moch H, Mihatsch MJ, Gasser T, Kallioniemi OP, Sauter G. High-throughput tissue microarray analysis of cyclin E gene amplification and overexpression in urinary bladder cancer. Am J Pathol. 2000;157(3):787–794. doi: 10.1016/S0002-9440(10)64592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93(12):6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page BD, Fletcher S, Yue P, Li Z, Zhang X, Sharmeen S, Datti A, Wrana JL, Trudel S, Schimmer AD, Turkson J, Gunning PT. Identification of a non-phosphorylated, cell permeable, small molecule ligand for the Stat3 SH2 domain. Bioorg Med Chem Lett. 2011;21(18):5605–5609. doi: 10.1016/j.bmcl.2011.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tseng LM, Huang PI, Chen YR, Chen YC, Chou YC, Chen YW, Chang YL, Hsu HS, Lan YT, Chen KH, Chi CW, Chiou SH, Yang DM, Lee CH. Targeting signal transducer and activator of transcription 3 pathway by cucurbitacin I diminishes self-renewing and radiochemoresistant abilities in thyroid cancer-derived CD133+ cells. J Pharmacol Exp Ther. 2012;341(2):410–423. doi: 10.1124/jpet.111.188730. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Liu P, Zhang B, Wang A, Yang M. Role of STAT3 decoy oligodeoxynucleotides on cell invasion and chemosensitivity in human epithelial ovarian cancer cells. Cancer Genet Cytogenet. 2010;197(1):46–53. doi: 10.1016/j.cancergencyto.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298(5600):1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 26.McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, Navolanic PM, Terrian DM, Franklin RA, D’Assoro AB, Salisbury JL, Mazzarino MC, Stivala F, Libra M. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Adv Enzym Regul. 2006;46:249–279. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 27.Chung LY, Tang SJ, Sun GH, Chou TY, Yeh TS, Yu SL, Sun KH. Galectin-1 promotes lung cancer progression and chemoresistance by upregulating p38 MAPK, ERK, and cyclooxygenase-2. Clin Cancer Res. 2012;18(15):4037–4047. doi: 10.1158/1078-0432.CCR-11-3348. [DOI] [PubMed] [Google Scholar]
- 28.Tang Y, Liu F, Zheng C, Sun S, Jiang Y. Knockdown of clusterin sensitizes pancreatic cancer cells to gemcitabine chemotherapy by ERK1/2 inactivation. J Exp Clin Cancer Res. 2012;31:73. doi: 10.1186/1756-9966-31-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pignochino Y, Grignani G, Cavalloni G, Motta M, Tapparo M, Bruno S, Bottos A, Gammaitoni L, Migliardi G, Camussi G, Alberghini M, Torchio B, Ferrari S, Bussolino F, Fagioli F, Picci P, Aglietta M. Sorafenib blocks tumour growth, angiogenesis and metastatic potential in preclinical models of osteosarcoma through a mechanism potentially involving the inhibition of ERK1/2, MCL-1 and ezrin pathways. Mol Cancer. 2009;8:118. doi: 10.1186/1476-4598-8-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Si H, Peng C, Li J, Wang X, Zhai L, Li X, Li J. RNAi-mediated knockdown of ERK1/2 inhibits cell proliferation and invasion osteosarcoma U2-OS cells in vitro. Int J Oncol. 2012;40(4):1291–1297. doi: 10.3892/ijo.2011.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryu K, Susa M, Choy E, Yang C, Hornicek FJ, Mankin HJ, Duan Z: Oleanane triterpenoid CDDO-Me induces apoptosis in multidrug resistant osteosarcoma cells through inhibition of Stat3 pathway.BMC Cancer10:187. [DOI] [PMC free article] [PubMed]
- 32.Duan Z, Ames RY, Ryan M, Hornicek FJ, Mankin H, Seiden MV. CDDO-Me, a synthetic triterpenoid, inhibits expression of IL-6 and Stat3 phosphorylation in multi-drug resistant ovarian cancer cells. Cancer Chemother Pharmacol. 2009;63(4):681–689. doi: 10.1007/s00280-008-0785-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YC, Zheng LH, Ma BA, Zhou Y, Zhang MH, Zhang DZ, Fan QY. Clinical value of signal transducers and activators of transcription 3 (STAT3) gene expression in human osteosarcoma. Acta Histochem. 2011;113(4):402–408. doi: 10.1016/j.acthis.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Ryu K, Choy E, Yang C, Susa M, Hornicek FJ, Mankin H, Duan Z. Activation of signal transducer and activator of transcription 3 (Stat3) pathway in osteosarcoma cells and overexpression of phosphorylated-Stat3 correlates with poor prognosis. J Orthop Res. 2010;28(7):971–978. doi: 10.1002/jor.21088. [DOI] [PubMed] [Google Scholar]
- 35.Liu LH, Li H, Li JP, Zhong H, Zhang HC, Chen J, Xiao T. miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem Biophys Res Commun. 2011;416(1-2):31–38. doi: 10.1016/j.bbrc.2011.10.117. [DOI] [PubMed] [Google Scholar]
- 36.Fossey SL, Bear MD, Lin J, Li C, Schwartz EB, Li PK, Fuchs JR, Fenger J, Kisseberth WC, London CA. The novel curcumin analog FLLL32 decreases STAT3 DNA binding activity and expression, and induces apoptosis in osteosarcoma cell lines. BMC Cancer. 2011;11:112. doi: 10.1186/1471-2407-11-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Onimoe GI, Liu A, Lin L, Wei CC, Schwartz EB, Bhasin D, Li C, Fuchs JR, Li PK, Houghton P, Termuhlen A, Gross T, Lin J. Small molecules, LLL12 and FLLL32, inhibit STAT3 and exhibit potent growth suppressive activity in osteosarcoma cells and tumor growth in mice. Investig New Drugs. 2012;30(3):916–926. doi: 10.1007/s10637-011-9645-1. [DOI] [PubMed] [Google Scholar]
- 38.Fossey SL, Bear MD, Kisseberth WC, Pennell M, London CA: Oncostatin M promotes STAT3 activation, VEGF production, and invasion in osteosarcoma cell lines.BMC Cancer11:125. [DOI] [PMC free article] [PubMed]
- 39.Do SI, Jung WW, Kim HS, Park YK. The expression of epidermal growth factor receptor and its downstream signaling molecules in osteosarcoma. Int J Oncol. 2009;34(3):797–803. doi: 10.3892/ijo_00000205. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/606/prepub


