Abstract
We evaluated the performance of a new chromogenic medium for detection of Clostridium difficile, chromID C. difficile agar (CDIF; bioMérieux, France), by comparison with BBL C. difficile Selective Agar (CDSA; Becton Dickinson and Company, USA). After heat pre-treatment (80℃, 5 min), 185 diarrheal stool samples were inoculated onto the two media types and incubated anaerobically for 24 hr and 48 hr for CDIF and for 48 hr and 72 hr for CDSA. All typical colonies on each medium were examined by Gram staining, and the gram-positive rods confirmed to contain the tpi gene by PCR were identified as C. difficile. C. difficile was recovered from 36 samples by using a combination of the two media. The sensitivity with CDIF 48 hr was highest (100%) and was significantly higher than that with CDIF 24 hr (58.3%; P<0.001), because samples with a low burden of C. difficile tended to require prolonged incubation up to 48 hr (P<0.001). The specificity of CDIF 24 hr and CDIF 48 hr (99.3% and 90.6%, respectively) was significantly higher than that of CDSA 48 hr and CDSA 72 hr (72.5% and 67.1%, respectively; P<0.001). CDIF was effective for detecting C. difficile in heat-pretreated stool specimens, thus reducing unnecessary testing for toxin production in non-C. difficile isolates and turnaround time.
Keywords: Clostridium difficile, Chromogenic agar, Performance, ChromID C. difficile agar, Toxigenic culture
Clostridium difficile infection (CDI) is increasingly recognized in both hospital and community settings [1, 2, 3]. For culture of C. difficile, pre-reduced cycloserine-cefoxitin-fructose agar with a high cycloserine concentration (0.5 g/L) is the most widely used medium, but it requires an incubation of at least 48 hr. In addition, intense growth of stool flora is frequently observed, thereby making detection and isolation of C. difficile difficult. To limit the growth of contaminating flora, various treatments such as heat-shock and alcohol-shock may be applied to specimens for culture [4, 5, 6, 7, 8]. Recently, a new chromogenic medium for C. difficile, chromID C. difficile agar (CDIF; bioMérieux, Marcy l'Etoile, France), which can detect C. difficile within 24 hr, was introduced to the market. This study aimed to evaluate the performance of CDIF by comparing it with BBL C. difficile Selective Agar (CDSA; Becton Dickinson and Company, Sparks, MD, USA).
This prospective study was conducted at Seoul St. Mary's hospital, a 1,300-bed teaching hospital in Seoul, Korea. In total, 185 consecutive diarrheal stool samples from patients suspected of having CDI were included from April to May 2012. Culture was performed within 72 hr of collection, and stool specimens were stored at -4℃ until processing. Each specimen (1 mL) was mixed with 1 mL of thioglycollate broth and, after vortexing, placed in an 80℃ water bath for 5 min [6, 7, 8].
A 100-µL aliquot of this suspension was inoculated onto CDIF and CDSA and spread to obtain isolated bacterial colonies. All media were incubated in a Bactron anaerobic/environmental chamber (Shel Lab, Cornelius, OR, USA) at 37℃, either for 24 hr (CDIF) or for 48-72 hr (CDSA) as recommended by the manufacturer and then removed into air for a maximum of 15 min for counting colonies. CDIF plates were reincubated anaerobically for a further 24 hr and further counts were performed.
The colonies were read by two investigators and the presence of C. difficile was evaluated semiquantitatively as follows: 1+ for 1-10 colonies, 2+ for 11-50 colonies, 3+ for 51-100 colonies, and 4+ for more than 100 colonies. Growth of typical colonies of C. difficile (gray to black color with an irregular or smooth border on CDIF and flat, yellow colonies with a ground glass-like appearance and a slightly filamentous edge on CDSA) were regarded as positive on each medium. All the colonies were examined by Gram staining, and the gram-positive rods were subjected to PCR to detect the tpi gene, a housekeeping gene of C. difficile [9]. If a colony harbored the tpi gene, it was determined to be C. difficile.
By using this criterion, growth of C. difficile confirmed to contain the tpi gene in any medium was defined as true positive. False positive was defined as growth of an isolate showing typical colony morphology of C. difficile but not identified as C. difficile. The four combinations (CDIF 24 hr, CDIF 48 hr, CDSA 48 hr, and CDSA 72 hr) were compared statistically by using the exact McNemar test with Bonferroni adjustment for proportions and the Mantel-Haenszel chi-squared test for trend analysis. P<0.05 was considered statistically significant. The Institutional Review Board of Seoul St. Mary's Hospital approved this study (Protocol No. KC11SISI0655).
C. difficile was recovered from 36 stool samples (19.5%) by using a combination of two media. The number of stool samples positive for C. difficile was 21 (58.3%) and 36 (100%) on CDIF at 24 hr and 48 hr, respectively, and 30 (83.3%) and 31 (86.1%) on CDSA at 48 hr and 72 hr, respectively. The sensitivity of CDIF at 48 hr was significantly higher than that of CDIF 24 hr (P<0.001, exact McNemar test using Bonferroni adjustment), but it was not significantly higher than that of CDSA 48 hr or CDSA 72 hr (Table 1). Of the 36 CDIF-positive samples, the distribution of colony-count-grades was as follows; 1+ in 10 samples, 2+ in 5 samples, 3+ in one sample, and 4+ in 20 samples. The proportion of samples that presented growth at 24 hr according to the colony-count-grade was 0% in grade 1+, 40% in grade 2+, 0% in grade 3+, and 95% in grade 4+ (P for trend <0.001, Mantel-Haenszel chi-squared test; Table 2).
Table 1.
Comparison of four combinations (CDIF 24 hr, CDIF 48 hr, CDSA 48 hr, and CDSA 72 hr)
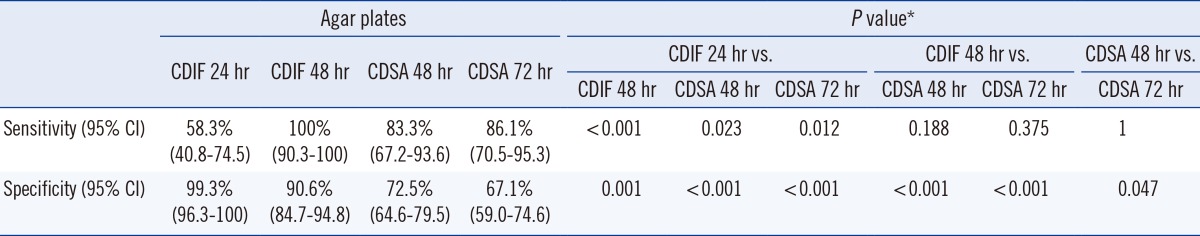
*Obtained by exact McNemar test using Bonferroni adjustment.
Abbreviations: CDIF 24 hr, growth of C. difficile colonies on chromID C. difficile agar (CDIF) at 24 hr incubation; CDIF 48 hr, growth of C. difficile colonies on CDIF agar at 48 hr incubation; CDSA 48 hr, growth of C. difficile colonies on BBL C. difficile Selective Agar (CDSA) at 48 hr incubation; CDSA 72 hr, growth of C. difficile colonies on CDSA at 72 hr incubation; CI, confidence intervals.
Table 2.
The number of agar plates presenting growth according to the medium, time, and colony-count-grades
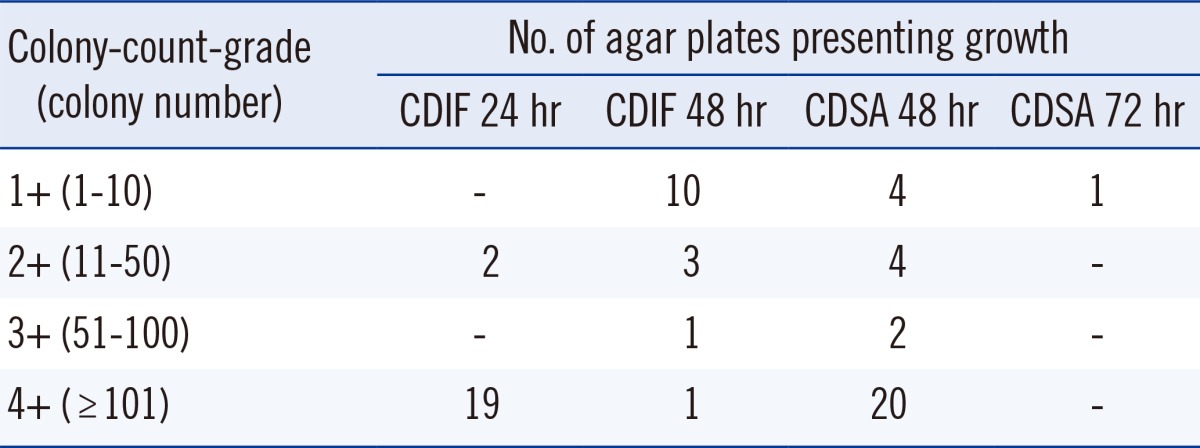
Abbreviations: CDIF 24 hr, growth of C. difficile colonies on chromID C. difficile agar (CDIF) at 24 hr incubation; CDIF 48 hr, growth of C. difficile colonies on CDIF agar at 48 hr incubation; CDSA 48 hr, growth of C. difficile colonies on BBL C. difficile Selective Agar (CDSA) at 48 hr incubation; CDSA 72 hr, growth of C. difficile colonies on CDSA at 72 hr incubation; CI, confidence intervals.
With CDIF, non-C. difficile isolates presenting gray to black colonies were recovered from approximately 9% of samples (16/185; 10 gram-positive bacillus [GPB], 1 gram-negative bacillus [GNB], 1 GPB/GNB, and 2 GPB/gram-positive coccus [GPC]/GNB), and they were much less common at 24 hr of incubation (two isolates). With CDSA, non-C. difficile isolates presenting typical, yellow colonies were recovered from approximately 30% (55/185; 53 GPB and 2 GPC) of samples, and 47 of them were recovered at 48 hr. Among them, the number of agar plates showing false-positive isolates from the 149 C. difficile-negative stool samples was one on CDIF at 24 hr, 14 on CDIF at 48 hr, 41 on CDSA at 48 hr, and 49 on CDSA at 72 hr, resulting in a specificity 99.3%, 90.6%, 72.5%, and 67.1%, respectively (Table 3). The specificity of CDIF 24 hr and CDIF 48 hr was significantly higher than that of CDSA 48 hr and 72 hr (P<0.001, exact McNemar test using Bonferroni adjustment; Table 1), and this low specificity of CDSA resulted from the frequent growth of non-C. difficile GPB (Table 3).
Table 3.
Non-C. difficile isolates presenting typical colonies recovered from 185 stool specimens
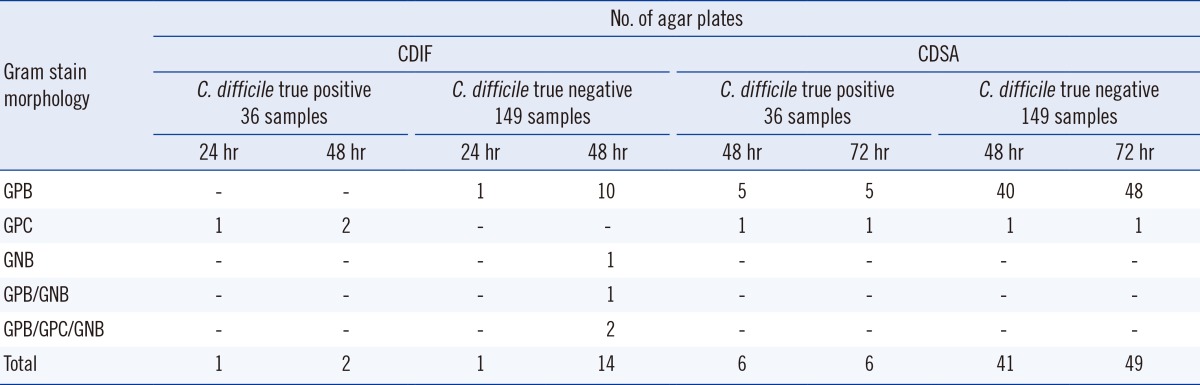
Abbreviations: CDIF 24 hr, growth of C. difficile colonies on chromID C. difficile agar (CDIF) at 24 hr incubation; CDIF 48 hr, growth of C. difficile colonies on CDIF agar at 48 hr incubation; CDSA 48 hr, growth of C. difficile colonies on BBL C. difficile Selective Agar (CDSA) at 48 hr incubation; CDSA 72 hr, growth of C. difficile colonies on CDSA at 72 hr incubation; CI, confidence intervals; GPB, gram-positive bacillus; GPC, gram-positive coccus; GNB, gram-negative bacillus.
Toxigenic culture is a new reference method for diagnosis of CDI, as it is more sensitive than the fecal cytotoxin assay [10, 11, 12]. However, it is a two-step method that consists of isolating C. difficile strains on a selective medium and then testing the colonies for toxin production by PCR. To reduce unnecessary PCR tests and turnaround time, the occurrence of endogenous flora showing colonies of typical color should be low.
CDIF is a new chromogenic medium containing taurocholate and a chromogen mix that allows isolation of C. difficile in 24 hr. Although direct plating or alcohol pre-treatment is recommended by the manufacturer, we pre-treated stool samples to facilitate the plate reading and used heat-shock rather than alcohol-shock to shorten the turnaround time. The heat pre-treatment seemed to inhibit the growth of GPC and GNB effectively, as growth of GPC or GNB was only observed on rare occasions (one sample [0.5%] on CDIF at 24 hr, six samples [3.2%] on CDIF at 48 hr, two samples [1.1%] on CDSA at 48 hr, and two samples [1.1%] on CDSA at 72 hr among 185 stool samples). This finding is similar to that of Yim et al. [13], where GPC or GNB isolates were recovered from 3.1% (23/738) of samples on CDIF at 48 hr and superior to that of Perry et al. [14], where GPC or GNB isolates were recovered from 3.0% (11/368) and 9.0% (33/368) of samples on CDIF at 24 hr and 48 hr, respectively. In both studies [13, 14], alcohol pre-treatment was used.
Based on our results, the sensitivity for recovering CDIF at 24 hr was low (58.3%), but after 48 hr incubation, it increased to 100%. This finding agrees with that of Eckert et al. [15], who showed that prolonging incubation enhanced the recovery of C. difficile from 74.1% to 87%, but appears to contrast with the findings of Perry et al. [14], who showed that the sensitivity of CDIF at 24 hr incubation was 96.3%. However, looking into the study more closely, the recovery rate of C. difficile with CDIF at 24 hr incubation was different between the samples which were positive and negative for Vidas immunoassay; it was high (94.5%) in Vidas-positive samples, but low (68%) in Vidas-negative samples. In both studies, the recovery rate on CDIF at 24 hr was somewhat higher than that in our study. Further evaluation is needed to investigate whether this difference results from a difference in pre-treatment, because Eckert et al. [15] used the direct plating method and Perry et al. [14] used the alcohol-shock method.
The colony-count-grades in stool samples from which C. difficile was recovered on CDIF at 24 hr and 48 hr (3.8±0.6 and 1.5±0.9, respectively) indicate that a 24 hr incubation is sufficient for stool samples with a high burden (more than 50 colonies per agar plate), but the samples with a low burden (less than 50 colonies per agar plate) of C. difficile require prolonged incubation of up to 48 hr. This finding is similar to that of Shin et al. [16], where the no-growth-plates were 37.9% and 6.2% on day 1 and 2, respectively. For CDSA, of the 31 stool samples from which C. difficile was recovered, all but one were recovered after 48 hr of incubation, indicating that prolonged incubation of up to 72 hr was not necessary.
In conclusion, CDIF was highly effective for detecting C. difficile in heat-pretreated stool specimens, thus reducing unnecessary testing for toxin production on non-C. difficile isolates and turnaround time, and this medium exhibited the best sensitivity at 48 hr, especially with stool samples with low burden of C. difficile.
Acknowledgments
This work was supported by bioMérieux, Marcy l'Etoile, France.
Footnotes
No potential conflicts of interest relevant to this article were reported.
References
- 1.Johnson S, Clabots CR, Linn FV, Olson MM, Peterson LR, Gerding DN. Nosocomial Clostridium difficile colonisation and disease. Lancet. 1990;336:97–100. doi: 10.1016/0140-6736(90)91605-a. [DOI] [PubMed] [Google Scholar]
- 2.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 3.Khanna S, Pardi DS, Aronson SL, Kammer PP, Orenstein R, St Sauver JL, et al. The epidemiology of community-acquired Clostridium difficile infection: a population-based study. Am J Gastroenterol. 2012;107:89–95. doi: 10.1038/ajg.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koransky JR, Allen SD, Dowell VR., Jr Use of ethanol for selective isolation of sporeforming microorganisms. Appl Environ Microbiol. 1978;35:762–765. doi: 10.1128/aem.35.4.762-765.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clabots CR, Gerding SJ, Olson MM, Peterson LR, Gerding DN. Detection of asymptomatic Clostridium difficile carriage by an alcohol shock procedure. J Clin Microbiol. 1989;27:2386–2387. doi: 10.1128/jcm.27.10.2386-2387.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura S, Yamakawa K, Izumi J, Nakashio S, Nishida S. Germinability and heat resistance of spores of Clostridium difficile strains. Microbiol Immunol. 1985;29:113–118. doi: 10.1111/j.1348-0421.1985.tb00809.x. [DOI] [PubMed] [Google Scholar]
- 7.Kamiya S, Yamakawa K, Ogura H, Nakamura S. Recovery of spores of Clostridium difficile altered by heat or alkali. J Med Microbiol. 1989;28:217–221. doi: 10.1099/00222615-28-3-217. [DOI] [PubMed] [Google Scholar]
- 8.Lahn M, Tyler G, Däubener W, Hadding U. Improvement of Clostridium difficile isolation by heat-shock and typing of the isolated strains by SDS-PAGE. Eur J Epidemiol. 1993;9:327–334. doi: 10.1007/BF00146272. [DOI] [PubMed] [Google Scholar]
- 9.Lemee L, Dhalluin A, Testelin S, Mattrat MA, Maillard K, Lemeland JF, et al. Multiplex PCR Targeting tpi (triose phosphate isomerase), tcdA (Toxin A), and tcdB (Toxin B) genes for toxigenic culture of Clostridium difficile. J Clin Microbiol. 2004;42:5710–5714. doi: 10.1128/JCM.42.12.5710-5714.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delmée M, Van Broeck J, Simon A, Janssens M, Avesani V. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: a plea for culture. J Med Microbiol. 2005;54:187–191. doi: 10.1099/jmm.0.45844-0. [DOI] [PubMed] [Google Scholar]
- 11.Reller ME, Lema CA, Perl TM, Cai M, Ross TL, Speck KA, et al. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J Clin Microbiol. 2007;45:3601–3605. doi: 10.1128/JCM.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jaksic AS, Nimmo GR, Dwyer BW. Laboratory diagnosis of Clostridium difficile-associated diarrhoea: microbiologist (should) do it with culture. Pathology. 2009;41:187–188. doi: 10.1080/00313020802579235. [DOI] [PubMed] [Google Scholar]
- 13.Yim JS, Hwang SM, Kim M, Lim HJ, Shin S, Chung HS, et al. Evaluation of a ChromID C. difficile Agar for the Isolation of Clostridium difficile. Korean J Clin Microbiol. 2012;15:88–91. [Google Scholar]
- 14.Perry JD, Asir K, Halimi D, Orenga S, Dale J, Payne M, et al. Evaluation ofa chromogenic culture medium for isolation of Clostridium difficile within 24 hours. J Clin Microbiol. 2010;48:3852–3858. doi: 10.1128/JCM.01288-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckert C, Burghoffer B, Lalande V, Barbut F. Evaluation of the chromogenic agar chromID C. difficile. J Clin Microbiol. 2013;51:1002–1004. doi: 10.1128/JCM.02601-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin BM. Comparison of ChromID agar and Clostridium difficile selective agar for effective isolation of C. difficile from stool specimens. Ann Lab Med. 2014;34:15–19. doi: 10.3343/alm.2014.34.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]


