Abstract
Background
The SUV3 (suppressor of Var 3) gene encodes a DNA and RNA helicase, which is localized in the mitochondria. Plant SUV3 has not yet been characterized in detail. However, the Arabidopsis ortholog of SUV3 (AT4G14790) has been shown to be involved in embryo sac development. Previously, we have reported that rice SUV3 functions as DNA and RNA helicase and provides salinity stress tolerance by maintaining photosynthesis and antioxidant machinery. Here, we report further analysis of the transgenic OsSUV3 rice plants under salt stress.
Findings
The transgenic OsSUV3 overexpressing rice T1 lines showed significantly higher endogenous content of plant hormones viz., gibberellic acid (GA3), zeatin (Z) and indole-3-acetic acid (IAA) in leaf, stem and root as compared to wild-type (WT), vector control (VC) and antisense (AS) plants under salt (200 mM NaCl) stress condition. A similar trend of endogenous plant hormones profile was also reflected in the T2 generation of OsSUV3 transgenic rice under defined parameters and stress condition.
Conclusions
In response to stress, OsSUV3 rice plants maintained plant hormone levels that regulate the expression of several stress-induced genes and reduce adverse effects of salt on plant growth and development and therefore sustains crop productivity.
Keywords: Helicases, Oryza sativa, Plant hormones, Plant tolerance, Salinity stress
Findings
Abiotic stresses (such as high salinity, drought, flood and high and low temperatures) cause the greatest constraint on crop productivity. These affect the growth and productivity and trigger a series of biochemical and molecular changes in the plants, which appear in the form of morphological and physiological variations in the crops. Globally, it has been estimated that approximately 70% of yield reduction is the direct result of the negative effects of abiotic stresses on various crops (Acquaah [2007]). High salinity causes the hyper-ionic and hyper-osmotic stress to plant cells leading to reduction in plant growth and productivity, which in severe cases may lead to plant death. Plants may cope with the adverse effects of high salinity by improving their photosynthesis and antioxidant machinery (Tuteja [2007]; Tuteja et al. [2013]). The rice SUV3 helicase has been shown to be involved in salinity, cadmium and zinc stress tolerance (Tuteja et al. [2013]; Sahoo and Tuteja [2014]). The endogenous content of plant hormones directs the molecular and biochemical mechanisms to confer increased stress tolerance to improve plant growth and development and thereby better survival (Osakabe et al. [2013]).
Plant hormones regulate several aspects of plant growth and developmental processes in response to the multiple abiotic and biotic stresses. The molecular events in plant hormone responses and environmental stress adjustment have been reported (Harrison [2012]). Gibberellic acid (GA3) has been reported to be involved in plant response to abiotic stress and GA3 mediated growth variability in plants relieves the adverse effects of salt, oxidative, and heat stresses (Qin et al. [2011]). Stress-induced production of cytokinin in plants facilitates sink strengthening via a cytokinin-dependent co-ordinated regulation of carbon and nitrogen metabolism that provides an improved tolerance to salinity and drought stress (Ha et al. [2012]). In response to stress, plants synchronize their development by exogenous signals and signal transduction cascades. The essential roles of auxin in various growth and developmental processes have been well documented. Additionally, the role of auxin in the regulation of biotic and abiotic stresses has been suggested (Kazan et al. [2013]).
The OsSUV3 gene (1.74 kb) (accession number: GQ982584; locus ID NM_001057785 on chromosome 3) was cloned previously (Tuteja et al. [2013]). The OsSUV3 gene shows high homology with previously existing sequences (accession numbers AK101069.1; XM_006650512.1) in the GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The IR64 rice transgenic plants overexpressing OsSUV3 in sense and antisense orientations were developed as described previously (Tuteja et al. [2013]). A competent strain of Agrobacterium tumefaciens (LBA4404) transformed with empty vector (pCAMBIA1301) construct was used as vector control to raise control plants of rice via standard protocol as described earlier (Tuteja et al. [2013]). The plants viz., T1OsSUV3 rice, T2OsSUV3 rice, wild-type (WT), vector control (VC), and antisense (AS) were allowed to grow under 200 mM salt (NaCl). After 24 h, the plant samples such as leaves, stem and root were collected for extraction and purification of the endogenous plant hormones (GA3, zeatin and IAA) by adopting method of Chen et al. ([1996]).
The OsSUV3 transgenic rice lines viz., Line 1 (L1), Line 2 (L2) and Line 3 (L3) showed higher endogenous content of plant hormones as compared to AS, VC and WT plants (Figure 1a-f). The plant hormones profile of transgenic OsSUV3 rice was assessed upto T2 generation. In T1 generation, GA3 content in leaves of OsSUV3 transgenic rice lines (L1, L2 and L3) was relatively higher (3.07, 3.1 and 3.04 μg/g fw, respectively) as compared to WT (1.54 μg/g fw), VC (1.44 μg/g fw) and AS (1.34 μg/g fw). GA3 content was 2.7, 2.72 and 2.7 μg/g fw in shoots and 1.9, 2.1 and 2.0 μg/g fw in roots of OsSUV3 rice lines (Figure 1a). Zeatin ranged between 0.7-1.27 μg/g fw, whereas control plants showed 0.2-0.52 of zeatin (μg/g fw) in leaf, shoot and roots (Figure 1b). The endogenous level of IAA in leaf, shoot and roots of transgenic rice lines ranged between 1.6-3.1 μg/g fw as opposed to control plants (1.4-1.6 μg/g fw) (Figure 1c). A similar trend of phytohormones (GA3, zeatin and IAA) was also reflected in T2 plants of OsSUV3 transgenic rice lines (L1, L2 and L3) (Figure 1d-f).
Figure 1.
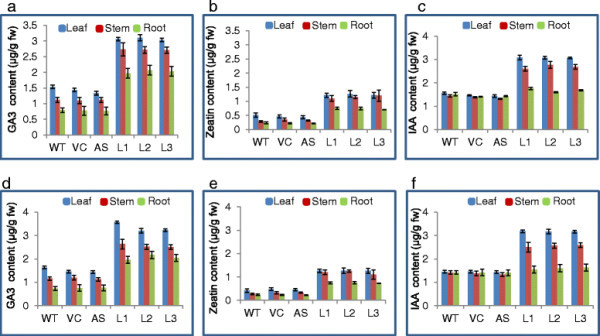
Plant hormones gibberellic acid (GA 3 ), zeatin and indole-3-acetic acid (IAA) profile in the transgenic OsSUV3 rice plants under salt stress. Endogenous content (μg/g fw) of GA3, zeatin and IAA in OsSUV3 transgenic rice lines (L1, L2 and L3) of T1 generation (a-c) and T2 generation (d-f) under 200 mM NaCl. The significant difference between the mean values (n = 3) of rice plants (WT, VC and AS) and OsSUV3 overexpressing transgenic rice lines (L1-L3) was determined by one-way analysis of variance (ANOVA) using SPSS 10.0 (SPSS, Inc., now IBM, http://www-01.ibm.com/software/analytics/spss). The WT, VC and transgenic lines at P < 0.05, P < 0.01 and P < 0.001 were considered statistically significant.
Multiple reports of effects of salinity on plants are available (Mahajan and Tuteja [2005]; Zolla et al. [2010]). To overcome the salinity stress-induced loss of crop productivity, investigations have focused more on the mechanisms of salt tolerance in plants (Tuteja [2007]). We recently reported that OsSUV3 over expression in rice functions in salt tolerance, it also improved growth performance in terms of plant height, number of tillers/plant, number of panicle/plant, number of filled grain/panicle, number of non-chaffy grains/panicle, straw dry weight, 100 grain weight, root length, root dry weight, leaf area, root and shoot lengths when compared to control plants (Tuteja et al. [2013]). Much attention is now focused on the plant hormones for their crucial roles in stress responses and adaptation (Kuppu et al. [2013]) and exploitation of different plant hormones for reducing the negative effects of salinity in growth parameters (Egamberdieva [2009]). In the present study, OsSUV3 rice transgenic T1 and T2 lines maintained higher endogenous content of GA3, zeatin and IAA under 200 mM NaCl as compared to the control plants. The findings of the present investigation suggest that plant hormones mitigate various adverse effects of salt to sustain plant productivity via modulating metabolism and signalling events in plants.
Competing interests
The authors declare no potential competing interests.
Authors’ contributions
NT conceived and designed the experiments. RKS screened the OsSUV3 rice transgenic lines and performed the experiments. MWA and RT performed data analysis, interpreted the results, discussed the data and drafted the manuscript. All authors have read and approved the final manuscript.
Contributor Information
Ranjan Kumar Sahoo, Email: rksahoo@icgeb.res.in.
Mohammad Wahid Ansari, Email: wahid@icgeb.res.in.
Renu Tuteja, Email: renu@icgeb.res.in.
Narendra Tuteja, Email: narendra@icgeb.res.
Acknowledgments
Work on plant helicases and abiotic stress tolerance in crop plants in NT’s laboratory is partially supported by Department of Biotechnology (DBT), Government of India. The authors are thankful to Dr. Meerambika Mishra (Department of Lifescience, Sambalpur University, Odisha) and Dr. Alok Shukla (G. B. Pant University of Agriculture and Technology, Pant Nagar, Uttarakhand) for critical reading and for improving the English language quality.
References
- Acquaah G. Principles of plant genetics and breeding. Blackwell, Oxford, UK; 2007. [Google Scholar]
- Chen JG, Du XM, Zhao HY, Zhou X. Fluctuation in levels of endogenous plant hormones in ovules of normal and mutant cotton during flowering and their relation to fiber development. J Plant Growth Regul. 1996;7:173–177. doi: 10.1007/BF00190581. [DOI] [Google Scholar]
- Egamberdieva D. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Physiol Plant. 2009;7:861–864. doi: 10.1007/s11738-009-0297-0. [DOI] [Google Scholar]
- Ha S, Vankova R, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS (2012) Cytokinins: metabolism and function in plant adaptation to environmental stresses. Trends Plant Sci doi:10.1016/j.tplants.2011.12.005. [DOI] [PubMed]
- Harrison MA. In: Phytohormones and Abiotic Stress Tolerance in Plants. Khan NA, Nazar R, Iqbal N, Anjum NA, editor. Springer, New York; 2012. Cross-Talk Between Phytohormone Signaling Pathways Under Both Optimal and Stressful Environmental Conditions; pp. 49–73. [DOI] [Google Scholar]
- Kazan K (2013) Auxin and the integration of environmental signals into plant root development. Ann Bot doi:10.1093/aob/mct229. [DOI] [PMC free article] [PubMed]
- Kuppu S, Mishra N, Hu R, Sun L, Zhu X, Shen G, Blumwald E, Payton P, Zhang H. Water-deficit inducible expression of a cytokinin bio-synthetic gene IPT improves drought tolerance in cotton. PLoS One. 2013;7:e64190. doi: 10.1371/journal.pone.0064190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S, Tuteja N. Cold, salinity and drought stresses: an overview. Arch Biochem Biophys. 2005;7:139–158. doi: 10.1016/j.abb.2005.10.018. [DOI] [PubMed] [Google Scholar]
- Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LP. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot. 2013;7:445–458. doi: 10.1093/jxb/ers354. [DOI] [PubMed] [Google Scholar]
- Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 2011;7:1900–1913. doi: 10.1104/pp.111.187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo R, Tuteja N. OsSUV3 functions in cadmium and zinc stress tolerance in rice (Oryza sativa L. cv IR64) Plant Signal Behav. 2014;7:e27389. doi: 10.4161/psb.29377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Sahoo RK, Garg B, Tuteja R (2013) OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery in rice (Oryza sativa L. cv. IR64). The Plant J doi:10.1111/tpj.12277. [DOI] [PubMed]
- Tuteja N. Mechanisms of high salinity tolerance in plants. Meth Enzymol. 2007;7:419–438. doi: 10.1016/S0076-6879(07)28024-3. [DOI] [PubMed] [Google Scholar]
- Zolla G, Heimer YM, Barak S. Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot. 2010;7:211–224. doi: 10.1093/jxb/erp290. [DOI] [PMC free article] [PubMed] [Google Scholar]


