Abstract
The O-antigen (OAg) of the Gram-negative bacterium Francisella tularensis (Ft), which is both a capsular polysaccharide and a component of lipopolysaccharide, is comprised of tetrasaccharide repeats and induces antibodies mainly against repeating internal epitopes. We previously reported on several BALB/c mouse monoclonal antibodies (MAbs) that bind to internal Ft OAg epitopes and are protective in mouse models of respiratory tularemia. We now characterize three new internal Ft OAg IgG2a MAbs, N203, N77, and N24, with 10- to 100-fold lower binding potency than previously characterized internal-OAg IgG2a MAbs, despite sharing one or more variable region germline genes with some of them. In a mouse model of respiratory tularemia with the highly virulent Ft type A strain SchuS4, the three new MAbs reduced blood bacterial burden with potencies that mirror their antigen-binding strength; the best binder of the new MAbs, N203, prolonged survival in a dose-dependent manner, but was at least 10-fold less potent than the best previously characterized IgG2a MAb, Ab52. X-ray crystallographic studies of N203 Fab showed a flexible binding site in the form of a partitioned groove, which cannot provide as many contacts to OAg as does the Ab52 binding site. These results reveal structural features of antibodies at the low end of reactivity with multi-repeat microbial carbohydrates and demonstrate that such antibodies still have substantial protective effects against infection.
Introduction
Francisella tularensis (Ft), the Gram-negative intracellular bacterium that causes tularemia, is a category A potential bioterrorism agent, specifically the highly virulent type A subspecies.(1–4) Respiratory tularemia, the most severe form of the disease, causes high morbidity and up to 2% mortality even in antibiotic-treated patients.(1,3–6) Although a live vaccine strain (LVS) is partially protective against Ft in humans, it is not currently licensed due to safety concerns(6,7); hence the need to identify and characterize protective T- and B-cell antigens and epitopes for development of vaccines and antibody therapeutics.
Lipopolysaccharide (LPS), the main component of the Ft outer membrane, which is identical between type A and type B Ft strains,(8–12) is a main protective antigen in mice and circumstantially in humans.(13–22) In addition to lipid A and a core oligosaccharide (C, mainly Hex4HexNAcKdo), the main component of LPS is an O-polysaccharide [O-antigen (OAg)].(8–12,23,24) The OAg consists of variable numbers of the tetrasaccharide repeat [2)-β-D-4,6-dideoxy-4-formamido-D-glucose(1→4)-α-D-2-acetamido-2-deoxy-D-galacturonamide(1→4)-α-D-2-acetamido-2-deoxy-D-galacturonamide(1→3)-β-D-2-acetamido-2,6-dideoxy-D-glucose(1→] (Quip4NFm-GalpNAcAN-GalpNAcAN-QuipNAc), with Quip4NFm at the non-reducing end,(8–12,23) referred to, for simplicity, as ABCD. Because of the variable numbers of the tetrasaccharide repeat in the OAg, the LPS molecules form a ladder pattern in SDS gels and Western blots. Ft capsular polysaccharide also consists of OAg(23,24) and, therefore, antibodies to Ft OAg target both LPS and capsule.
We have previously described mouse MAbs specific for repeating internal epitopes of Ft OAg and have shown that they confer survival to LVS-infected mice and prolong survival of mice infected with the prototypic type A strain SchuS4 in a mouse model of respiratory tularemia.(25–27) These internal-binding OAg MAbs are distinguished from MAbs that bind to the non-reducing terminus of Ft OAg by their reactivity with the LPS ladder on Western blots, showing increased binding with increasing LPS chain length, in contrast to the relatively even binding of MAbs specific for terminal OAg epitopes.(27,28) The ability of each OAg chain to engage many internal-binding OAg antibodies is reflected in the higher prevalence of these antibodies compared with terminal-binding OAg antibodies, presumably due to the engagement of multiple B-cell receptors and efficient B-cell stimulation during natural or artificial immunization.(27)
We have also reported on the Fab X-ray crystal structure of Ab52, the most avid of three IgG2a MAbs that target internal Ft OAg epitopes, and the docking of a calculated model of Ft OAg in its binding site.(29) The site has the shape of an open-ended wide groove that accommodates a V-shaped epitope consisting of six sugar residues spanning two OAg repeats, α-D-GalpNAcAN(1→4)-α-D-GalpNAcAN(1→3)-β-D-QuipNAc(1→2)-β-D-Quip4NFm(1→4)-α-D-GalpNAcAN(1→4)-α-D-GalpNAcAN (BCDA'B'C'). The formyl group of the β-D-Quip4NFm sugar protrudes into a small side-pocket at the center of the site.(29)
In the current study, we obtained three IgG2a MAbs, N24, N77, and N203, which showed reactivity only to high molecular weight Ft LPS chains on our standard Western blots, with no discernible LPS ladder. Further characterization showed these to be much weaker internal-binding anti-OAg MAbs than the previously characterized internal-binding anti-OAg MAbs, with the strongest binding of the new MAbs (found with N203) being two orders of magnitude less potent than Ab52, and less efficacious than Ab52 at prolonging survival of mice infected intranasally (i.n.) with SchuS4. The X-ray crystal structure of N203 Fab was determined for comparison of the N203 and Ab52 binding sites to gain insights into structural features that distinguish high and low affinity Ft OAg binding.
Materials and Methods
Bacterial strains and MAbs
Ft holarctica strain LVS was obtained from Dr. Jeannine Petersen (Centers for Disease Control and Prevention, Fort Collins, CO). Ft tularensis strain SchuS4 was obtained from BEI Resources (Manassas, VA). E. coli strain TG1 was purchased from Stratagene (La Jolla, CA). WbtIG191V (WbtI), an OAg deficient LVS mutant,(30) was obtained from Dr. Thomas Inzana (Virginia Polytechnic Institute and State University, Blacksburg, VA). All strains were propagated and heat-inactivated as previously described.(26) SchuS4 vortexate was prepared by vigorously vortexing a SchuS4 suspension for 15 min. The vortexed suspension was centrifuged at 15,000 g for 60 min at 4°C, the supernatant was collected and subjected to a second centrifugation under the same conditions, and the second supernatant was filtered through a 0.22 μm membrane. Protein concentration in the SchuS4 vortexate was determined by the Lowry assay (DC protein assay kit, Bio-Rad, Hercules, CA).
Protein G-purified mouse IgG2a MAb GTX40330, specific for Escherichia coli J5 LPS, was purchased from GeneTex (Irvine, CA). Generation of internal-binding Ft OAg MAbs Ab2 (IgG3), Ab3 (IgG2a), Ab6 (IgM), Ab7 (IgM), and Ab9 (IgG1)(26) and Ab52 and Ab54,(27) and of the terminal-binding Ft OAg MAbs Ab63 (IgG3), N213 (IgG3), and N62 (IgG2b),(28) was previously reported. All mouse experiments were performed under a protocol approved by the Boston University Medical Center Institutional Animal Care and Use Committee.
The N24, N77, and N203 MAbs were generated in the current study by intradermal (i.d.) immunization of BALB/cJ mice (Jackson Laboratory, Bar Harbor, ME) with the sub-lethal dose of 2×105 – 2×107 CFU of LVS followed 32–56 days later by an intraperitoneal (i.p.) booster immunization with an outer membrane- or capsule-enriched LVS prep, and 3.75 days later by fusion of spleen cells with Sp2/0-Ag14 myeloma cells, as previously described.(26) The LVS membrane prep was prepared from an LVS vortexate by pelleting at 200,000 g for 2 h. The LVS capsule prep was prepared by high salt extraction, as described by Hood.(31) The three new MAbs were derived from three separate mice. All three were determined to be IgG2a(κ) by IsoStrip (Mouse Monoclonal Antibody Isotyping Kit, Roche Diagnostics, Indianapolis, IN).
Purification of MAbs
Hybridoma cells were cultured in IMDM (Gibco, Grand Island, NY) supplemented with 10% FBS and grown to mass culture in IMDM supplemented with 2% FBS in 10 cm Optilux™ petri dishes (Becton Dickinson Labware, Franklin Lakes, NJ) or in a CELLine classic 1000 two-compartment bioreactor (Wilson Wolf Manufacturing, New Brighton, MN) at 37°C in a humidified environment of 5% CO2/95% air. MAbs were separately purified from culture supernatants on Pierce Protein G Plus (IgG1, IgG2a, IgG2b) or Protein A Plus (IgG3) Agarose (Thermo Scientific, Rockford, IL) according to the manufacturer's instructions (modified to use 0.1 M sodium acetate [pH 5.0] for elution of IgG3) and their purity and specificity were verified by SDS-PAGE and Western blot analysis on SchuS4, respectively.
Immunoassays
Bacterial microagglutination, direct ELISA, and Western blot analysis were performed as previously described.(27) For direct ELISA, 0.04 OD/mL of heat-killed Ft SchuS4 or E. coli TG1, 0.125 μg/mL of Ft LPS, or 5 μg/mL of Ft OAg-core (OAgC, Sussex Research, Ottawa, Canada) were coated onto ELISA plates by overnight drying in a 37°C oven and, after incubation with test MAbs, the reactions were developed with anti-mouse IgG2a (γ2a chain-specific) HRP conjugate (SouthernBiotech, Birmingham, AL).
For isotype-specific competition ELISA, EIA/RIA plates were coated with 100 μL/well of SchuS4 vortexate (prepared as described above) at a protein concentration of 1 μg/mL in 50 mM carbonate buffer (pH 9.6) and allowed to air-dry overnight at 37°C. Anti-mouse IgG2a (γ2a chain-specific) HRP conjugate was used to determine the binding of a fixed concentration (2.5 μg/mL) of N203 reporter in the presence of graded concentrations of competitor MAb (starting at 400 μg/mL). The presence/binding of competitor MAbs to SchuS4 vortexate was verified in a duplicate plate by developing the ELISA with HRP-conjugated anti-mouse isotype-specific secondary antibody. Percent inhibition was calculated (after subtracting the blank from all values using the following formula: [(OD without competitor – OD with competitor)/(OD without competitor)]×100.
For competition ELISA with Ft OAgC, serially diluted Ft OAgC was pre-mixed with 2.5 μg/mL N203, Ab52, or Ab53 (anti-Ft GroEL, IgG2a isotype control), then the mixtures were added to a SchuS4 vortexate-coated, washed, and blocked ELISA plate, followed by HRP-anti-mouse-IgG2a to detect bound MAb. Percent inhibition was calculated as described above.
Nucleotide and deduced amino acid sequence determination
V region nucleotide sequences of MAbs were obtained from RT-PCR products as previously described for the Ab52 MAb.(29) Homology to immunoglobulin (Ig) germline genes was determined by IgBlast (www.ncbi.nlm.nih.gov/igblast) and conversion to amino acid sequences was done by EMBOSS Transeq (www.ebi.ac.uk/Tools/emboss/transeq/index.html).
In vivo efficacy studies
All animal procedures were approved by the Boston University Institutional Animal Care and Use Committee. BALB/cJ female mice were obtained from Jackson Laboratories, at 7–8 weeks of age, and inoculated i.n. with Ft bacteria under ketamine/xylazine anesthesia as previously described.(26) For inoculation of mice, Ft bacteria were serially diluted in PBS to the intended CFU/mL based on OD600 of the starting stock, and administered in 10 μL followed by 10 μL of PBS as described by Klimpel and colleagues.(22) The actual CFU inoculated per mouse was determined retrospectively after each experiment by plating serial dilutions of the bacterial preparation used for inoculation on chocolate agar plates. Two hours post-SchuS4 inoculation, mice were injected i.p. with MAb or vehicle (PBS supplemented with 5% BALB/c mouse serum). Survival was monitored every 12 h. Kaplan-Meier survival curves were plotted using Graph-Pad Prism 5.0 (GraphPad Software, San Diego, CA), and the log rank test was used to compare groups.
For determination of blood bacterial burden, blood was collected from the submandibular vein into a BD Microtainer® tube with Lithium Heparin additive (BD, Franklin Lakes, NJ) 3 days post-bacterial inoculation. Undiluted blood and a 5-fold serial dilution of the blood were streaked on chocolate agar plates, and the plates were incubated at 35°C for 2 days for CFU enumeration. Percent CFU reduction compared with PBS was calculated from the median CFU obtained from the plate with ≥30 CFU or the undiluted-blood plate if both plates had <30 CFU. The p value was determined using the two-tailed Mann Whitney test. p values of <0.05 were considered statistically significant for both the log rank test and the two-tailed Mann Whitney test.
Fab X-ray crystal structure determination
Fab was prepared from purified IgG using the Pierce Fab Preparation Kit (Thermo Scientific). For crystallization trials, the Fab was dialyzed and concentrated/diafiltered in a YM-10 Centricon Centrifugal Filter Device (Millipore, Bedford, MA) to 38 mg/mL into 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 0.02% NaN3, as determined by absorbance at 280 nm. A sparse matrix screen was carried out using the reagents in the MCSG-1 screen (Microlytic North America, Burlington, MA) in a 96-well Corning Crystal EX microplate (Corning, NY).
An initial crystallization condition from the screen was optimized using microseeding techniques. For seeding in hanging drops, 0.5 μL of Fab was mixed with 0.5 μL of reservoir solution (0.1 M sodium citrate [pH 4.0], 1 M lithium chloride, 13–16% PEG 6000) on a siliconized coverslip, which was inverted over a well containing 0.5 mL of reservoir solution and sealed. After overnight equilibration at 17°C, the drop was streak-seeded using a cat whisker that had been dipped into a seed stock mixture made by crushing a crystal cluster in 50 μL of reservoir solution. Prior to freezing in a cold nitrogen stream, the crystal used for diffraction studies was cryoprotected by soaking for 20 s in a mix of 8 μL reservoir solution and 3 μL 10 M lithium chloride. Diffraction data were collected on a RAXIS-IV imaging plate detector mounted on a Rigaku RU-300 rotating anode generator. Data indexing, integration, and merging were performed using the programs Denzo and Scalepack.(32) The structure was solved by molecular replacement using AutoMR in Phenix.(33) The search models used were the light (L) chain of the phosphocholine-binding MAb McPC603 (PDB code 1MCP(34)) and the heavy (H) chain of the HIV neutralizing MAb Yz23 (www.rcsb.org/pdb/home/home; PDB code 3CLE). Each search model was divided into variable and constant domains for a total of four search models. In addition, the CDRH3 loop of the H chain was removed. The molecular replacement solution, comprising two Fab molecules in the asymmetric unit, was then rebuilt with AutoBuild in Phenix, with the starting model not included in the building of the output structure to reduce phase bias. There was clear density to build the missing residues of the CDRH3 loop, which were built in manually. Iterative cycles of manual rebuilding and refinement were performed with Coot(35) and Phenix to obtain the final structure. The elbow angles (θ) of the Fabs in the crystal were determined by superposing the L (residues 1-107) and H (residues 1-114) chain variable domains and the L and H constant domains of each Fab using the program LSQMAN,(36) which calculates the direction cosines of the two pseudo-rotation axes in the Fab (cos α, cos β, cos γ). The elbow angle was then calculated using the formula: cos θ=cos αvar * cos αcon+cos βvar * cos βcon+cos γvar * cos γcon. The crystal structure was used for docking in Glide as previously described, with the same starting structures of the OAg repeats.(29) Images were generated using MAESTRO (v. 9.3.5, Schrödinger, Inc., New York, NY).
Results
Three new MAbs are specific for internal OAg epitopes but have much lower binding potency than previously characterized internal OAg-binding MAbs
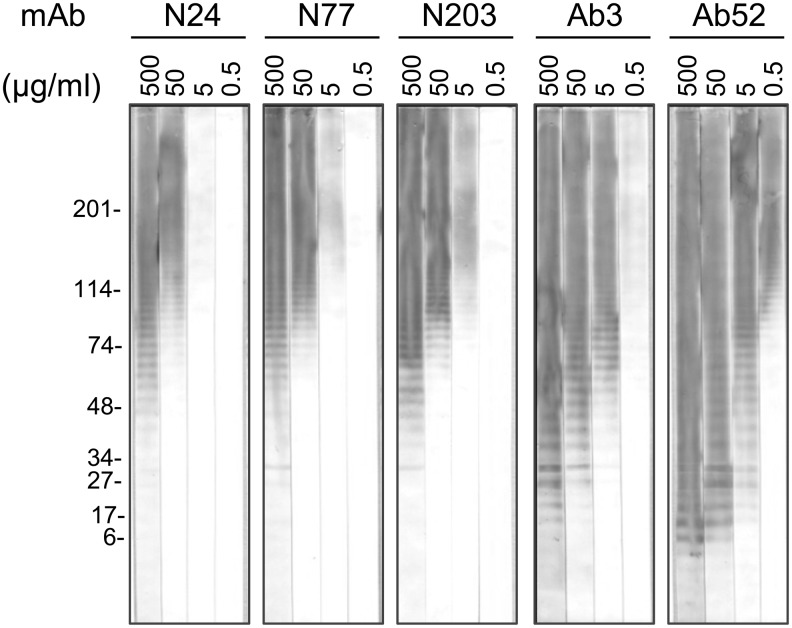
While screening for antigen-specific supernatants of hybridomas derived from BALB/c mice immunized with live LVS and boosted with outer membrane- and/or capsule-enriched Ft preps, we identified three IgG2a MAbs that bound to a diffuse proteinase K-resistant 100–250 kDa band on Western blots of SchuS4 lysates but showed no reactivity on our standard Ft LPS Western blots. Further characterization revealed that the three new MAbs, N24, N77, and N203, were specific for internal OAg epitopes but had much lower antigen-binding potency than that of internal OAg-binding MAbs, which we had previously detected by their binding to the Ft LPS ladder on Western blots. Thus, when the amount of LPS electrophoresed in the preparative polyacrylamide gel was increased from our standard 10–20 μg to 200 μg and the Western blot strips were probed with 10-fold serial dilutions of 0.5–500 μg/mL of purified MAbs, the three new MAbs detected the LPS ladder. However, the MAb concentration required for ladder detection was 100-fold greater than that for the most avid (Ab52) and 10-fold greater than that for the least avid (Ab3) previously characterized MAb, respectively (Fig. 1).
FIG. 1.
N24, N77, and N203 bind poorly to Ft LPS on Western blots compared with Ab52 and Ab3. Ft LPS (200 μg, usually 10–20 μg) was electrophoresed in a preparative 4–15% polyacrylamide gel under denaturing conditions. After transfer to nitrocellulose, strips were probed with the specified MAbs at the indicated concentrations and developed with AP-anti-mouse IgG (H+L) conjugate for 15 min.
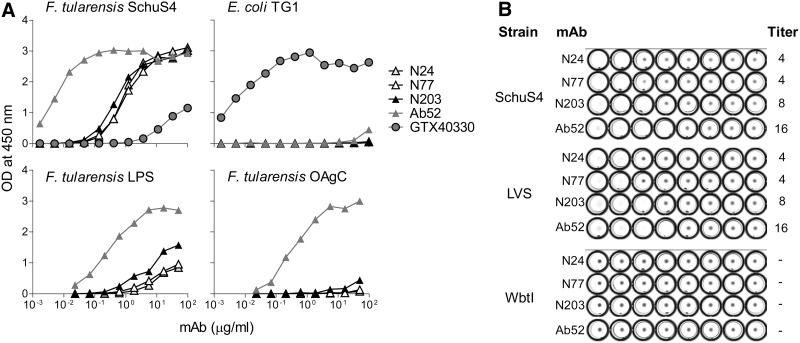
The weaker reactivity of the new MAbs was also evident in ELISA, where the three new MAbs required 100–200-fold greater concentrations for equivalent binding to heat-killed SchuS4 but showed specificity for Ft, as no binding to E. coli was detected (Fig. 2A). Similarly, N203, the best binder of the new MAbs, required a 120-fold greater concentration than Ab52 for equivalent binding to LPS, and showed barely detectable binding to OAgC (LPS-derived OAg connected to core oligosaccharide) even at 50 μg/mL N203 (Fig. 2A), presumably due to the poor adherence of Ft OAgC to ELISA plates. However, the specificity of the new MAbs for OAg was supported by their ability to agglutinate SchuS4 and LVS but not the OAg-deficient LVS mutant WbtI (Fig. 2B) and by the ability of OAgC to inhibit the binding of N203 to SchuS4 vortexate (Fig. 3A).
FIG. 2.
Binding characteristics of the three new MAbs. (A) The three new MAbs bind specifically to virulent Ft and not to E. coli in ELISA, but binding to Ft LPS and OAgC is reduced compared with binding by Ab52. ELISA plates were coated with heat-killed Ft SchuS4, heat-killed E. coli TG1, Ft LPS, or Ft OAgC and were probed with serial dilutions of the indicated MAbs. GTX40330, a commercial MAb specific for E. coli LPS, was used as specificity control. (B) Ft agglutination by N24, N77, and N203, like agglutination by Ab52, requires OAg. Two-fold serial dilutions of different MAbs, starting with a final concentration of 256 μg/mL in well 1, were incubated with equal volumes of fixed bacteria of the indicated strains. Wells showing a carpet-like pattern were recorded as positive and those showing a small, tight button were recorded as negative. Agglutination titers are indicated on the right side of each row.
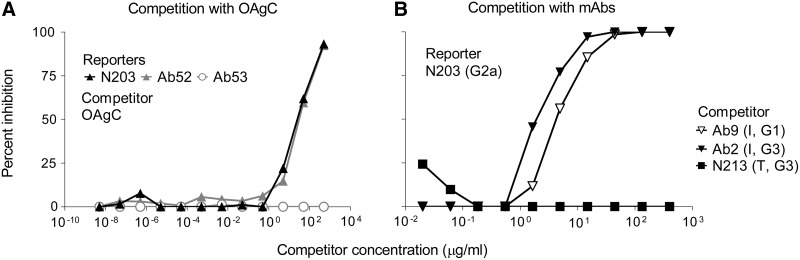
FIG. 3.
Binding of N203 to Ft SchuS4 vortexate is inhibited by OAgC and by internal binding but not terminal binding anti-OAg MAbs. (A) Serially diluted Ft OAgC was pre-mixed with 5 μg/mL N203, Ab52, or Ab53 (anti-Ft GroEL, IgG2a isotype control); then the mixtures were added to a SchuS4 vortexate-coated, washed, and blocked ELISA plate, followed by HRP-a-m-IgG2a to detect bound MAb. (B) Serial dilutions of the indicated internal binding (I) or terminal binding (T) anti-OAg MAbs were used as competitors, and the ELISA was developed with HRP-a-mouse IgG2a antibody.
Furthermore, analysis of the nucleotide sequences of the expressed variable (V) regions of the three new MAbs showed that they are all partially encoded by the same IGKV germline gene, which is also shared by two of seven previously described anti-Ft OAg internal-binding MAbs (Ab3 and Ab9, Table 1). However the IGKVs of all 10 MAbs, except Ab6, belong to the same subgroup (Table 1). Analysis of the nucleotide sequences also showed that one of the new MAbs (N77) is partially encoded by the same IGHV germline gene as two other of the previously described internal OAg-binding MAbs (Ab54 and Ab6), and another of the new MAbs (N203) is partially encoded by the same IGHD gene as the previously described internal OAg-binding MAb Ab3 (Table 1). It is noteworthy that the IGHVs of the new MAbs and all internal-binding OAg MAbs, except Ab3 and Ab2, belong to the same subgroup (IGHV1), and the IGHDs of all three new MAbs and of four of the seven internal-binding OAg MAbs belong to either subgroup IGHD1 or subgroup IGHD4 (Table 1). No shared V or D germline genes were observed with MAbs specific for the terminal non-reducing end of Ft OAg (Ab63, N213, and N62, Table 1). The shared IGKV, IGHV, and IGHD germline genes between the new MAbs and internal OAg-binding MAbs suggested that the three new MAbs also target internal OAg epitopes. This was validated by the ability of internal (I) OAg-binding MAbs Ab9 and Ab2, but not of the terminal (T) OAg-binding MAb N213, to inhibit the binding of the new MAb N203 to SchuS4 vortexate in isotype-specific competition ELISA (Fig. 3B).
Table 1.
Germline Genes Encoding V Regions of the New MAbs Compared with Other MAbs Specific for Ft LPS
| Germline gene (% nucleotide identity or no. of contiguous nucleotide matches for IGHD) | |||||
|---|---|---|---|---|---|
| MAb (non-IgG2a isotype) | IGHV | IGHD | IGHJ | IGKV | IGKJ |
| New | |||||
| N24 | 1-12*01 (95.3) | 1-1*02 (5) | 3*01 (100.0) | 8-19*01 (99.6) | 2*01 (100.0) |
| N77 | 1S29*02 (98.8) | 1-1*02 (5) | 4*01 (100.0) | 8-19*01 (100.0) | 1*01 (100.0) |
| N203 | 1-7*01 (90.9) | 4-1*01 (6) | 2*01 (100.0) | 8-19*01 (100.0) | 5*01 (100.0) |
| Internal | |||||
| Ab3 | 3-2*02 (100.0) | 4-1*01 (6) | 2*01 (100.0) | 8-19*01 (100.0) | 4*01 (100.0) |
| Ab9 (G1) | 1S127*01 (91.2) | 2-3*01 (5) | 4*01 (97.6) | 8-19*01 (98.5) | 2*01 (100.0) |
| Ab54 | 1S29*02 (99.6) | 1-1*01 (15) | 2*01 (100.0) | 8-24*01 (100.0) | 1*01 (100.0) |
| Ab6 (M) | 1S29*02 (99.6) | –a | 4*01 (100.0) | 3-2*01 (100.0) | 2*01 (100.0) |
| Ab2 (G3) | 9-2-1*01 (100.0) | 4-1*02 (5) | 3*01 (100.0) | 8-24*01 (100.0) | 4*01 (100.0) |
| Ab7 (M) | 1S22*01 (98.8) | 1-1*01 (14) | 4*01 (100.0) | 8-24*01 (100.0) | 4*01 (100.0) |
| Ab52 | 1S130*01 (98.8) | –a | 3*01 (100.0) | 8-24*01 (99.6) | 4*01 (100.0) |
| Terminal | |||||
| Ab63 (G3) | 3-8*02 (99.6) | –a | 3*01 (100.0) | 4-80*01 (100.0) | 2*01 (100.0) |
| N213 (G3) | 3-8*02 (95.0) | 2-14*01 (9) | 3*01 (97.8) | 4-80*01 (97.1) | 5*01 (100.0) |
| N62 (G2b) | 3-8*02 (97.7) | 2-14*01 (6) | 3*01 (100.0) | 4-80*01 (99.2) | 2*01 (100.0) |
No D gene was identified by the IgBlast program (www.ncbi.nlm.nih.gov/igblast/) using the minimum requirement of five contiguous nucleotides.
Ab52 is more efficacious than the new MAbs at reducing blood bacterial burden and prolonging survival of BALB/c mice infected intranasally with SchuS4
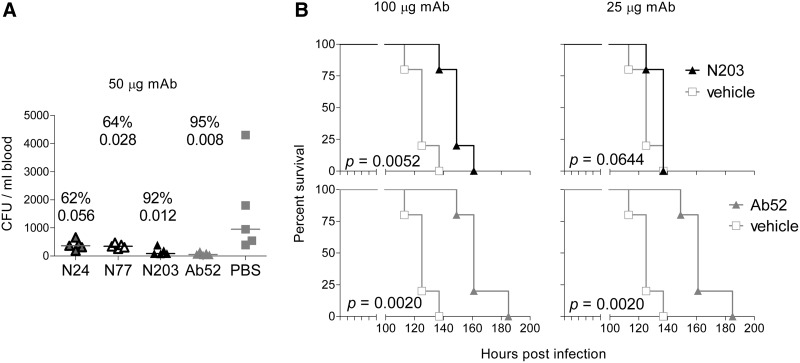
To determine whether the three new MAbs can be efficacious against Ft in vivo, N24, N77, and N203 were tested alongside the previously characterized Ab52 MAb for their ability to reduce blood bacterial burden in BALB/c mice infected i.n. with SchuS4 when administered i.p. 2 h post-infection. As shown in Figure 4A, Ab52, N203, N77, and N24 reduced bacterial burden 95%, 92%, 64%, and 62%, respectively, compared with the PBS control, as assessed 3 days post-infection, with the results for all, except N24, showing statistical significance (p<0.05).
FIG. 4.
Ab52 is more efficacious than N203, N77, and N24 at reducing blood bacterial burden (A) and prolonging survival (B) in BALB/c mice infected intranasally with SchuS4. Mice were inoculated i.n. with 140 CFU of SchuS4 (A) or 25 CFU of SchuS4 (B), then injected i.p. with the indicated doses of the specified antibodies. For determination of blood bacterial burden (A), mice were bled, then euthanized 3 days post-infection, and percent CFU reduction compared with vehicle (PBS) was calculated from the median CFU numbers and the p value was determined using the two-tailed Mann-Whitney test. To test for prolongation of survival (B), mice were monitored for survival every 12 h, Kaplan-Meier survival curves were plotted, and groups treated with MAb were compared with the group treated with vehicle (5% BALB/c mouse serum in PBS), using the log rank test.
When Ab52 and N203 were compared for ability to prolong survival of SchuS4-infected BALB/c mice, if administered i.p. at 100 μg/mouse 2 h post-infection, both MAbs significantly prolonged survival compared with vehicle control, by 36 and 24 h, respectively. However, at 25 μg MAb/mouse, the ability of Ab52 to prolong survival was unchanged, whereas that of N203 was decreased to 12 h, which did not quite reach statistical significance (by the two-tailed analysis used, Fig. 4B). The blood bacterial burden and prolongation of survival results indicate that the in vivo efficacies of Ab52, N203, N77, and N24 correlate directly with their OAg-binding potency.
Fab X-ray crystal structure of N203 reveals a flexible antigen-binding site that can adopt three different conformations
To determine the structural basis for the much weaker OAg binding of the new MAbs compared with that of Ab52 MAb, the Fab of N203 was analyzed by X-ray crystallography. The initial crystals of N203 Fab obtained from sparse matrix screening grew as clusters. Consecutive cycles of seeding were used to grow large crystals, which formed plates and diffracted X-rays to 2.1 Å resolution. From analysis of the diffraction data (Table 2), the space group was determined to be P212121 with unit cell dimensions a=66.010 Å, b=122.322 Å, c=124.764 Å; α=β=γ=90.00°.
Table 2.
X-ray Crystallographic Data Collection and Refinement Statistics
| Item | Value |
|---|---|
| Data collection | |
| No. of reflections | 58893 (5650)a |
| Data cutoff | I ≤−3σ |
| I/σ(I) | 20.1 (4.4) |
| Completeness (%) | 99.0 (96.3) |
| Redundancy | 4.1 (3.6) |
| Rmerge | 0.037 (0.299) |
| Refinement | |
| Rwork | 0.193 |
| Rfree | 0.228 |
| No. of atoms | 7180 |
| Deviations from ideal values | |
| Bond lengths (Å) | 0.002 |
| Bond angles (°) | 0.674 |
| Ramachandran plot | |
| Favored (%) | 97.7 |
| Allowed (%) | 2.3 |
| Outliers (%) | 0.0 |
The resolution limits overall were 15-2.10 Å. The number in parentheses is the value for the highest resolution shell (2.17–2.10 Å).
The structure was solved using molecular replacement. After several cycles of refinement and manual rebuilding, the final model was constructed, including building in residues removed from the search models. The model contains two Fab molecules comprising residues L1-213 and H1-213. Residues L204-209 (from both Fabs) and residues H129-132 and H129-134 (from the second Fab) were not modeled due to poor electron density. The H1 residues were modeled as pyroglutamic acid as indicated by the electron density. The model also contains 642 water molecules and 18 chloride ions. The final structure has good geometry and agreement with the observed diffraction data in Table 2.
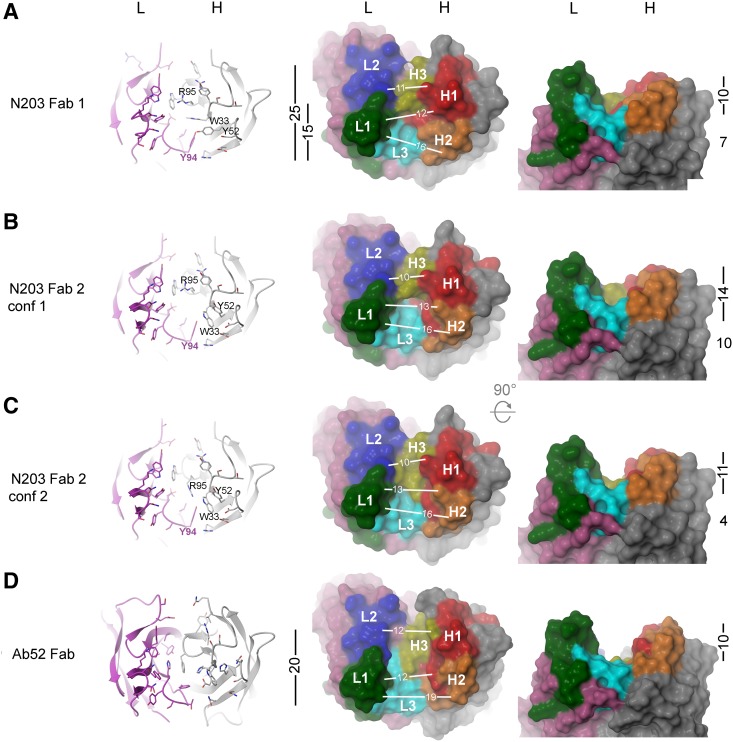
All six complementarity-determining regions (CDRs) are part of the presumed binding site. The CDRL1, CDRL2, CDRL3, CDRH1, and CDRH2 loops fall into canonical folding classes(37) L1-17-1, L2-8-1, L3-9-cis7-1, H1-13-1, and H2-10-1, respectively. The CDRH3 loop is short (6 residues) and does not fall into any canonical class. The general shape of the N203 binding site is a groove with an uneven bottom and a partition of protruding CDRH3 residues. The two Fabs in the asymmetric unit superimpose well with an r.m.s.d. of 0.85 Å calculated using 416 α-carbons as determined by the Iterative Magic Fit function in the program Deepview.(38) However, there are differences between the two copies of the Fab in the asymmetric unit caused by crystal packing contacts that create differences in the binding site topology. The elbow angle of the two Fabs differs by 6° (130° vs 136°), causing a slight shift in the relative orientation of the V domains toward one another. The crystal packing places the two antigen-binding sites face-to-face with one another, making crystal contacts. In particular, the conformations of the side chains of residues H-W33, H-Y52, and H-R95 in both Fabs appear to be affected by the lattice constraints (Fig. 5A). In Fab 1, there is clear density for all three side chains in the maps. The side chain of H-Y52 makes a hydrogen bond to the side chain of L-Y94 and contacts the side chain of H-W33, which makes van der Waals contacts with the CDRH3 loop at the main chain of H-R95 as well as aromatic interactions with L-Y94 (Fig. 5A).
FIG. 5.
The antigen-binding site of N203 is flexible and partitioned. X-ray crystal structures of N203 Fab (A–C) and Ab52 Fab (D). Left, ribbon diagrams with side chains lining the binding sites as stick figures, head-on view; the three N203 H chain residues whose side chains assume different positions in the different conformations are indicated, as is the L chain residue Y94, which is involved in hydrogen bonding to H-Y52. Middle, molecular surface representation, head-on view; the L and H chain CDRs are labeled as L1, L2, L3, and H1, H2, H3, and colored green, blue, cyan, red, orange and yellow, respectively. VL and VH framework regions are colored pink and grey, respectively. Right, molecular surface representation, side view, rotated 90° clockwise about the X-axis. Site dimensions, in Å, as measured in the final, refined coordinates (white lines) or as scaled black bars, are indicated. The number below the bar in the right column is the approximate height of the CDRH3 partition for that conformation.
The second Fab in the asymmetric unit (Fab 2) shows all three of these residues in different conformations (Fig. 5B,C). H-Y52 cannot adopt the same conformation as in Fab 1, because this would clash with the neighboring H-Y52 in the crystal. So, the H-Y52 side chain is rotated clockwise approximately 60° about the Cβ-Cγ bond, breaking the hydrogen bond to L-Y94. In this position, H-Y52 makes aromatic interactions with H-Y52 in the neighboring molecule, which may help to stabilize this conformation. The H-W33 side chain in Fab 2 is rotated approximately 80° counterclockwise about its Cβ-Cγ bond, so that it moves into the space formerly occupied by H-Y52. There, it makes aromatic interactions with L-Y94 and H-Y52 as well as H-Y52 from a neighboring molecule. This reorganization of side chains relative to the Fab 1 conformation opens up a deep cavity in the binding groove, bordering the protruding CDRH3 partition (Fig. 5B). The density for the side chain of H-R95 in Fab 2 is broken and suggests that this residue is somewhat disordered. Refinement of the H-R95 side chain as alternate conformers, one as in Fab 1 (referred to as Fab 2 conf 1, Fig. 5B), and one in which it occupies the space where H-W33 is located in Fab 1 (Fab 2 conf 2, Fig. 5C), shows the occupancy to be about 60% Fab 2 conf 1 and 40% Fab 2 conf 2. The H-R95 in Fab 2 conf 2 fills up the deep cavity, decreasing the height of the CDRH3 partition from 10 Å to 4 Å (Fig. 5C). Despite the different topology of the N203 groove in Fab 1, Fab 2 conf 1, and Fab 2 conf 2, the groove in all three N203 binding site conformations is, on the average, 13 Å wide, its full length is 25 Å, and its length to the CDRH3 partition is 15 Å (Fig. 5A–C). And, in the Fab 1 and Fab 2 conf 2 conformations, the ones assumed to interact with OAg, the depth of the site is 10 Å (Fig. 5A,C). By comparison, the Ab52 binding site(29) is a flat-bottom groove (except for a side pocket), 12 Å wide, 20 Å long, and 10 Å deep (Fig. 5D). Twenty-six amino acid residues line the N203 binding site in Fab 1 and Fab 2 conf 2, 21 of them in the 15 Å compartment (Fig. 5A and 5C left) compared with 33 amino acid residues that line the binding site of Ab52 (Fig. 5D, left).
The N203 crystals were not amenable to soaking with an Ft OAg oligosaccharide for structural analysis of an antibody-antigen complex due to the observed crystal contacts. In an attempt to determine the epitope accommodated by the N203 binding site, we docked the tetrasaccharide repeat unit models of OAg, previously used to determine the Ab52 epitope,(29) into the putative antigen-binding sites of N203 Fab 1 and Fab 2 conf 2 from the crystal structure. The results of the docking calculations suffer from poor Glide scores—not surprising given the low binding potency of N203 to the Ft OAg, so that a prediction of the binding mode of the OAg to N203 cannot be made from these studies.
Discussion
We generated and characterized three IgG2a MAbs (N24, N77, and N203) specific for a repeating internal epitope of Ft OAg, whose antigen-binding potency, as assessed by Western blot and ELISA, is one to two orders of magnitude lower than that of the previously characterized internal-binding Ft OAg IgG2a MAbs Ab3 and Ab52, respectively.(27) The lower antigen-binding potency of the new MAbs was reflected in their lower efficacy at reducing blood bacterial burden and prolonging survival of BALB/c mice infected intranasally with Ft SchuS4. The specificity of the new MAbs for Ft OAg was demonstrated by their binding to Ft SchuS4 and Ft LPS but not to E. coli and by their agglutination of Ft SchuS4 and Ft LVS but not of WbtI, an OAg-deficient LVS mutant. Their specificity for a repeating internal OAg epitope, rather than for the non-reducing end of OAg, was revealed by the increased binding to increasing-length LPS chains on Western blots and by the ability of internal-binding but not of terminal binding OAg MAbs to inhibit N203 binding to Ft LPS in isotype-specific competition ELISA.
The relative antigen-binding potency of the new MAb set from weakest to strongest was N24, N77, and N203, as evident in Western blot, direct ELISA, and microagglutination assays, and was also reflected in their relative potency at reducing blood bacterial burden in BALB/c mice infected intranasally with SchuS4. However, the intra-set differences were relatively small in all assays when compared with differences between N203 and Ab52, which were over 100-fold in Western blot and direct ELISA. Despite the large affinity difference, N203 and Ab52 are within 2-fold in bacterial agglutination assays and in blood bacterial burden reduction and prolongation of survival (at a dose of 100 μg) in the mouse model of respiratory tularemia. These results underscore the low avidity threshold sufficient for moderate in vivo efficacy of antibodies to microbial antigens with multiple repeating epitopes.
Despite the derivation of the three new MAbs from three different mice, all three are partially encoded by the same IGKV gene (8–19), suggesting that they target the same internal OAg epitope. It is noteworthy that Ab3, the weakest binder of three previously characterized internal-binding Ft OAg IgG2a MAbs(27) but still 10-fold more potent than N203, is also partially encoded by the 8–19 IGKV gene. In contrast, Ab52 and Ab54, the other two of the previously characterized internal-binding Ft OAg IgG2a MAbs,(27) are partially encoded by the 8–24 IGKV gene. Furthermore, the side-pocket in the center of the Ab52 binding-site, which anchors the repeating elbow of Ft OAg, is formed mainly by light chain CDR residues (29); this finding suggests that Ft OAg MAbs with higher affinity, and thereby higher bivalent avidity,(27) use 8–24 IGKV to anchor the OAg elbows, whereas anti-Ft OAg MAbs with lower affinity and different IGKVs, such as 8–19, may be unable to anchor the OAg elbow or may target a different, although necessarily overlapping, OAg epitope. We had previously determined the bivalent KD of the internal-binding Ft OAg IgG2a MAbs Ab52 and Ab54 for Ft LPS to be 3.6×10−9 M and 1.0×10−8 M, respectively, but could not determine the bivalent KD of the lower-affinity internal-binding anti-Ft OAg IgG2a MAb Ab3, using the same plasmon resonance assay.(27) Because the antigen-binding potency of the new internal-binding Ft OAg IgG2a MAbs is at least 10-fold lower than that of Ab3, as evident on Western blots, determination of the bivalent KDs of the new MAbs was not attempted.
The difference in the N203 and Ab52 epitopes is supported by X-ray crystallographic studies. The X-ray crystal structure of N203 Fab, as described here, is the second reported structure of an Ft OAg antibody that binds to a repeating internal epitope, the first being that of Ab52.(29) Although the structure of an N203 Fab–antigen complex could not be obtained, the free N203 Fab structure offers some insights into the poorer antigen-binding of N203 compared with Ab52. The Ab52 binding site is an open-ended groove (12 Å wide, 20Å long, and 10 Å deep) that accommodates a 6-sugar V-shaped epitope BCDA'B'C', with the formyl group of the A' sugar at the point of the V buried in a side pocket, formed mainly by L chain residues, at the center of the groove.(29) In contrast, the N203 binding site exists in the crystal in three different conformations, all of which have a width of about 13 Å and a protruding CDRH3 partition that divides the site into two unequal compartments—15 Å and 10 Å long. The three conformations of the N203 binding site differ in the positions of the side chains of three residues (H-W33, H-Y52, and H-R95), resulting in a deep cavity in the 15 Å compartment of one conformation but remaining as a 10 Å deep groove in the other two conformations. Although one or more of these conformations could be over-represented due to stabilization by crystal contacts, their existence in the crystal indicates that N203 has a flexible site, suggestive of an induced-fit antigen-binding mechanism. This could explain the relatively poor binding of N203 to Ft OAg, as studies have shown that mutations that lead to a lock and key binding mechanism over an induced-fit mechanism result in higher affinity due to decreased entropic losses during binding.(39–42)
Antibodies to internal epitopes of OAgs were predicted to be groove-type binding sites that could accommodate a long carbohydrate chain, whereas terminal binders were predicted to have cavity-type antigen-binding sites.(43,44) Several structures of complexes of antibodies to internal as well as terminal epitopes have so far confirmed this prediction.(45–49) The immunochemical evidence shows a pattern of binding consistent with an internal epitope for N203. Therefore, N203 was expected to have a groove-type antigen-binding site. But the CDRH3 partition in the N203 site was not expected and may interfere with antigen binding.
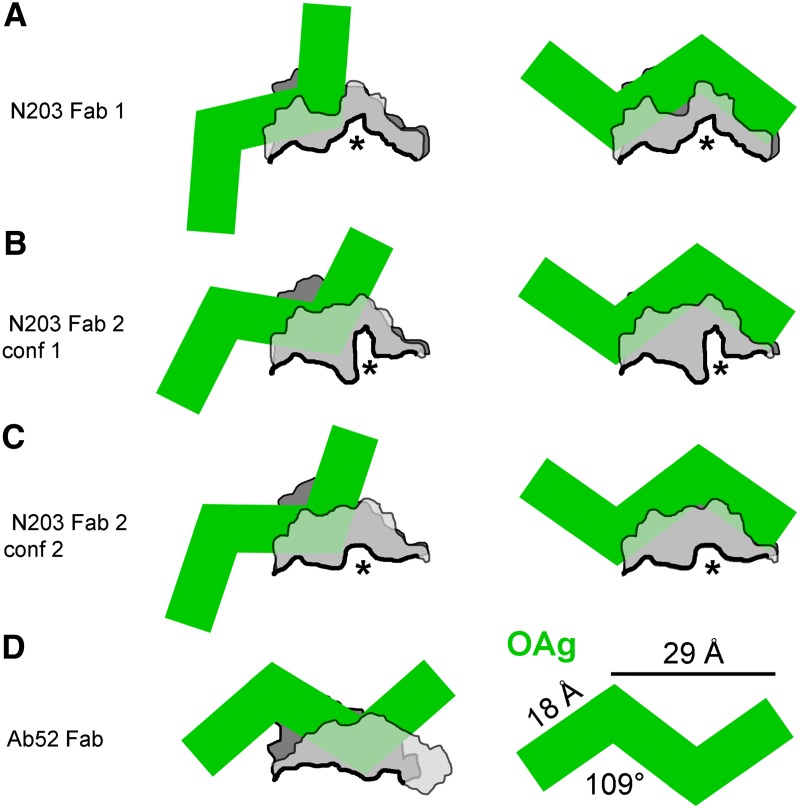
Based on the topology of the N203 binding site and the length of one OAg tetrasaccharide repeat, determined to be 18 Å long with a span of 29 Å for two repeats and an angle of 109° between repeats,(29) it is likely that the N203 epitope binds to the 15 Å compartment as shown for the three N203 conformations found in the crystal (Fig. 6, left). This compartment has only 21 surface-exposed residues compared to 33 surface-exposed residues in the Ab52 site (shown for comparison), suggestive of a smaller epitope size and consistent with the lower affinity of N203. Although it is conceivable that a 2-repeat OAg epitope could bind to the edges of the 25 Å N203 groove over the CDRH3 partition, as an inverted V (Fig. 6, right), such binding would be predicted to have very limited contacts. Thus, the relatively poor binding of N203 to OAg chains may be due to the constraints of the CDRH3 partition, requiring a flexible binding site and an induced-fit binding mode.
FIG. 6.
Ft OAg likely binds to the 15 Å compartment of the N203 site. Schematic representations, side view, rotated about 90° clockwise about the Y-axis from Fig. 5, right, so that the H chain (light gray) is close to the viewer and the L chain (dark gray) is in back. The outlines represent the bottom (thick black) and side surfaces of the antibody binding-site traced by hand in Photoshop from cutaway views of the surfaces generated using the MAESTRO program. The CDRH3 partition in the N203 site is indicated by an asterisk. The Ft OAg is represented by green line segments drawn to scale and positioned manually. Left, Three-repeat OAg chain binding to the 15 Å compartment of the N203 site in the three crystal conformations and to the Ab52 site. (A–C, right) Three-repeat OAg chain binding to edges of the 25 Å N203 site over the CDRH3 partition, as an inverted V, in the three crystal conformations. (D, right) Annotated three-repeat OAg.
Based on the crystal structure of an antibody which can adopt different binding-site conformations that interact with different antigens,(50) antibodies with flexible binding-sites have been suggested to increase the effective size of the antibody repertoire. Antibodies like N203, N77, and N24 may fall under this category of multi-specific antibodies, which bind to multi-repeat microbial carbohydrates with low affinity but, if present in sufficient dose, can still confer a protective effect in vivo due to the high density of the target epitope on the microbe.
Acknowledgment
This work was supported in its entirety with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract no. HHSN272200900054C).
Author Disclosure Statement
The authors have no financial interests to disclose.
Accession Code
Crystallographic data for N203 Fab have been deposited with the Protein Data Bank. The entry has been assigned PDB ID code 4OTX.
References
- 1.Thomas LD, and Schaffner W: Tularemia pneumonia. Infect Dis Clin North Am 2010;24:43–55 [DOI] [PubMed] [Google Scholar]
- 2.McLendon MK, Apicella MA, and Allen LA: Francisella tularensis: taxonomy, genetics, and Immunopathogenesis of a potential agent of biowarfare. Annu Rev Microbiol 2006;60:167–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sjostedt A: Tularemia: history, epidemiology, pathogen physiology, and clinical manifestations. Ann NY Acad Sci 2007;1105:1–29 [DOI] [PubMed] [Google Scholar]
- 4.Tarnvik A, and Chu MC: New approaches to diagnosis and therapy of tularemia. Ann NY Acad Sci 2007;1105:378–404 [DOI] [PubMed] [Google Scholar]
- 5.Dennis DT, Inglesby TV, Henderson DA, Bartlett JG, Ascher MS, Eitzen E, et al. : Tularemia as a biological weapon: medical and public health management. JAMA 2001;285:2763–2773 [DOI] [PubMed] [Google Scholar]
- 6.Conlan JW: Tularemia vaccines: recent developments and remaining hurdles. Future Microbiol 2011;6:391–405 [DOI] [PubMed] [Google Scholar]
- 7.Oyston PC: Francisella tularensis vaccines. Vaccine 2009;27(Suppl 4):D48–51 [DOI] [PubMed] [Google Scholar]
- 8.Conlan JW, Shen H, Webb A, and Perry MB: Mice vaccinated with the O-antigen of Francisella tularensis LVS lipopolysaccharide conjugated to bovine serum albumin develop varying degrees of protective immunity against systemic or aerosol challenge with virulent type A and type B strains of the pathogen. Vaccine 2002;20:3465–3471 [DOI] [PubMed] [Google Scholar]
- 9.Gunn JS, and Ernst RK: The structure and function of Francisella lipopolysaccharide. Ann NY Acad Sci 2007;1105:202–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prior JL, Prior RG, Hitchen PG, Diaper H, Griffin KF, Morris HR, et al. : Characterization of the O antigen gene cluster and structural analysis of the O antigen of Francisella tularensis subsp. tularensis. J Med Microbiol 2003;52:845–851 [DOI] [PubMed] [Google Scholar]
- 11.Thirumalapura NR, Goad DW, Mort A, Morton RJ, Clarke J, and Malayer J: Structural analysis of the O-antigen of Francisella tularensis subspecies tularensis strain OSU 10. J Med Microbiol 2005;54:693–695 [DOI] [PubMed] [Google Scholar]
- 12.Vinogradov EV, Shashkov AS, Knirel YA, Kochetkov NK, Tochtamysheva NV, Averin SF, et al. : Structure of the O-antigen of Francisella tularensis strain 15. Carbohydr Res 1991;214:289–297 [DOI] [PubMed] [Google Scholar]
- 13.Drabick JJ, Narayanan RB, Williams JC, Leduc JW, and Nacy CA: Passive protection of mice against lethal Francisella tularensis (live tularemia vaccine strain) infection by the sera of human recipients of the live tularemia vaccine. Am J Med Sci 1994;308:83–87 [DOI] [PubMed] [Google Scholar]
- 14.Fortier AH, Slayter MV, Ziemba R, Meltzer MS, and Nacy CA: Live vaccine strain of Francisella tularensis: infection and immunity in mice. Infect Immun 1991;59:2922–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foshay L: Tularemia: A summary of certain aspects of disease including methods for early diagnosis and the results of serum treatment in 600 patients. Medicine 1940;19:1–83 [Google Scholar]
- 16.Fulop M, Mastroeni P, Green M, and Titball RW: Role of antibody to lipopolysaccharide in protection against low- and high-virulence strains of Francisella tularensis. Vaccine 2001;19:4465–4472 [DOI] [PubMed] [Google Scholar]
- 17.Kirimanjeswara GS, Golden JM, Bakshi CS, and Metzger DW: Prophylactic and therapeutic use of antibodies for protection against respiratory infection with Francisella tularensis. J Immunol 2007;179:532–539 [DOI] [PubMed] [Google Scholar]
- 18.Rhinehart-Jones TR, Fortier AH, and Elkins KL: Transfer of immunity against lethal murine Francisella infection by specific antibody depends on host gamma interferon and T cells. Infect Immun 1994;62:3129–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenmark S, Lindgren H, Tarnvik A, and Sjostedt A: Specific antibodies contribute to the host protection against strains of Francisella tularensis subspecies holarctica. Microb Pathog 2003;35:73–80 [DOI] [PubMed] [Google Scholar]
- 20.Sebastian S, Dillon ST, Lynch JG, Blalock LT, Balon E, Lee KT, et al. : A defined O-antigen polysaccharide mutant of Francisella tularensis live vaccine strain has attenuated virulence while retaining its protective capacity. Infect Immun 2007;75:2591–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirimanjeswara GS, Olmos S, Bakshi CS, and Metzger DW: Humoral and cell-mediated immunity to the intracellular pathogen Francisella tularensis. Immunol Rev 2008;225:244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klimpel GR, Eaves-Pyles T, Moen ST, Taormina J, Peterson JW, Chopra AK, et al. : Levofloxacin rescues mice from lethal intra-nasal infections with virulent Francisella tularensis and induces immunity and production of protective antibody. Vaccine 2008;26:6874–6882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Shi X, Leymarie N, Madico G, Sharon J, Costello CE, et al. : A typical preparation of Francisella tularensis O-antigen yields a mixture of three types of saccharides. Biochemistry 2011;50:10941–10950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Apicella MA, Post DM, Fowler AC, Jones BD, Rasmussen JA, Hunt JR, et al. : Identification, characterization and immunogenicity of an O-antigen capsular polysaccharide of Francisella tularensis. PLoS One 2010;5:e11060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu Z, Madico G, Roche MI, Wang Q, Hui JH, Perkins HM, et al. : Protective B-cell epitopes of Francisella tularensis O-polysaccharide in a mouse model of respiratory tularaemia. Immunology 2012;136:352–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu Z, Roche MI, Hui JH, Unal B, Felgner PL, Gulati S, et al. : Generation and characterization of hybridoma antibodies for immunotherapy of tularemia. Immunol Lett 2007;112:92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche MI, Lu Z, Hui JH, and Sharon J: Characterization of monoclonal antibodies to terminal and internal O-antigen epitopes of Francisella tularensis lipopolysaccharide. Hybridoma 2011;30:19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Z, Rynkiewicz MJ, Yang CY, Madico G, Perkins HM, Wang Q, et al. : The binding sites of monoclonal antibodies to the nonreducing end of Francisella tularensis O-antigen accommodate mainly the terminal saccharide. Immunology 2013;140:374–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rynkiewicz MJ, Lu Z, Hui JH, Sharon J, and Seaton BA: Structural analysis of a protective epitope of the Francisella tularensis O-Polysaccharide. Biochemistry 2012;51:5684–5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Ryder C, Mandal M, Ahmed F, Azadi P, Snyder DS, et al. : Attenuation and protective efficacy of an O-antigen-deficient mutant of Francisella tularensis LVS. Microbiology 2007;153:3141–3153 [DOI] [PubMed] [Google Scholar]
- 31.Hood AM: Virulence factors of Francisella tularensis. J Hyg 1977;79:47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z, and Minor W: Processing of X-ray diffraction data collected in the oscillation mode. Methods Enzymol 1997;276:307–326 [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, et al. : PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010;66:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Satow Y, Cohen GH, Padlan EA, and Davies DR: Phosphocholine binding immunoglobulin Fab McPC603. An X-ray diffraction study at 2.7 A. J Mol Biol 1986;190:593–604 [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, and Cowtan K: Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004;60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 36.Kleywegt G, and Jones T: A super position. CCP4/ESF-EACBM Newsletter Protein Crystallogr 1994;31:9–14 [Google Scholar]
- 37.North B, Lehmann A, and Dunbrack RL, Jr: A new clustering of antibody CDR loop conformations. J Mol Biol 2011;406:228–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guex N, and Peitsch MC: SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 1997;18:2714–23 [DOI] [PubMed] [Google Scholar]
- 39.Manivel V, Sahoo NC, Salunke DM, and Rao KV: Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immunity 2000;13:611–620 [DOI] [PubMed] [Google Scholar]
- 40.Sagawa T, Oda M, Ishimura M, Furukawa K, and Azuma T: Thermodynamic and kinetic aspects of antibody evolution during the immune response to hapten. Mol Immunol 2003;39:801–808 [DOI] [PubMed] [Google Scholar]
- 41.Yin J, Beuscher AEt, Andryski SE, Stevens RC, and Schultz PG: Structural plasticity and the evolution of antibody affinity and specificity. J Mol Biol 2003;330:651–656 [DOI] [PubMed] [Google Scholar]
- 42.Yin J, Mundorff EC, Yang PL, Wendt KU, Hanway D, Stevens RC, et al. : A comparative analysis of the immunological evolution of antibody 28B4. Biochemistry 2001;40:10764–10773 [DOI] [PubMed] [Google Scholar]
- 43.Cisar J, Kabat EA, Dorner MM, and Liao J: Binding properties of immunoglobulin combining sites specific for terminal or nonterminal antigenic determinants in dextran. J Exp Med 1975;142:435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Padlan EA, and Kabat EA: Model-building study of the combining sites of two antibodies to alpha (1→6) dextran. Proc Natl Acad Sci USA 1988;85:6885–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cygler M, Wu S, Zdanov A, Bundle DR, and Rose DR: Recognition of a carbohydrate antigenic determinant of Salmonella by an antibody. Biochem Soc Trans 1993;21:437–441 [DOI] [PubMed] [Google Scholar]
- 46.Villeneuve S, Souchon H, Riottot MM, Mazie JC, Lei P, Glaudemans CP, et al. : Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: molecular basis for serotype specificity. Proc Natl Acad Sci USA 2000;97:8433–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vulliez-Le Normand B, Saul FA, Phalipon A, Belot F, Guerreiro C, Mulard LA, et al. : Structures of synthetic O-antigen fragments from serotype 2a Shigella flexneri in complex with a protective monoclonal antibody. Proc Natl Acad Sci USA 2008;105:9976–9981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vyas NK, Vyas MN, Chervenak MC, Johnson MA, Pinto BM, Bundle DR, et al. : Molecular recognition of oligosaccharide epitopes by a monoclonal Fab specific for Shigella flexneri Y lipopolysaccharide: X-ray structures and thermodynamics. Biochemistry 2002;41:13575–13586 [DOI] [PubMed] [Google Scholar]
- 49.Zdanov A, Li Y, Bundle DR, Deng SJ, MacKenzie CR, Narang SA, et al. : Structure of a single-chain antibody variable domain (Fv) fragment complexed with a carbohydrate antigen at 1.7-A resolution. Proc Natl Acad Sci USA 1994;91:6423–6427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.James LC, Roversi P, and Tawfik DS: Antibody multispecificity mediated by conformational diversity. Science 2003;299:1362–1367 [DOI] [PubMed] [Google Scholar]