Abstract
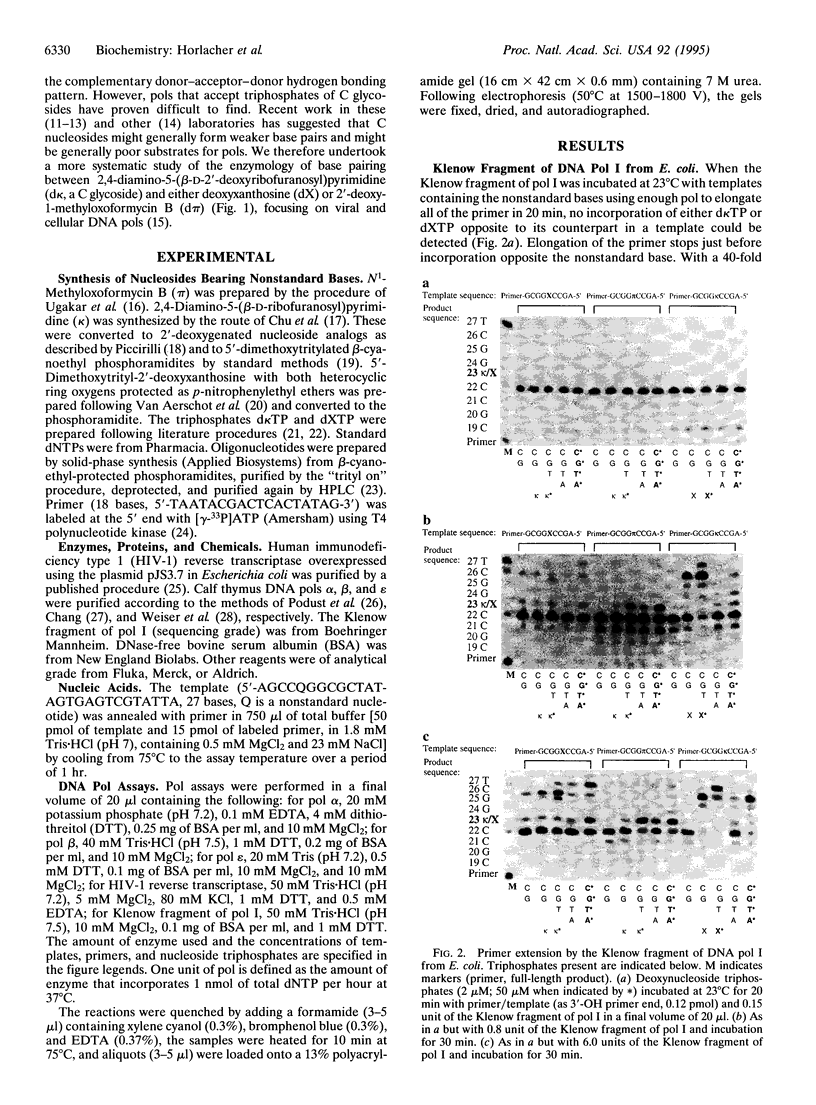
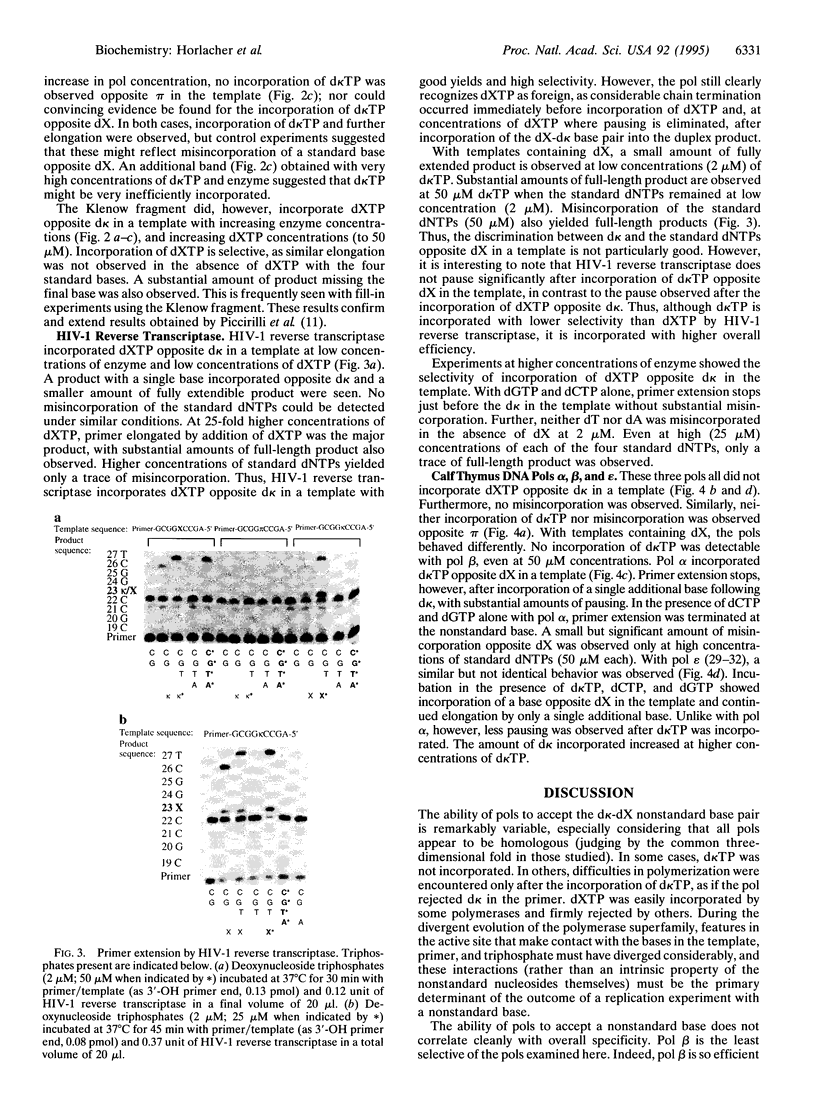
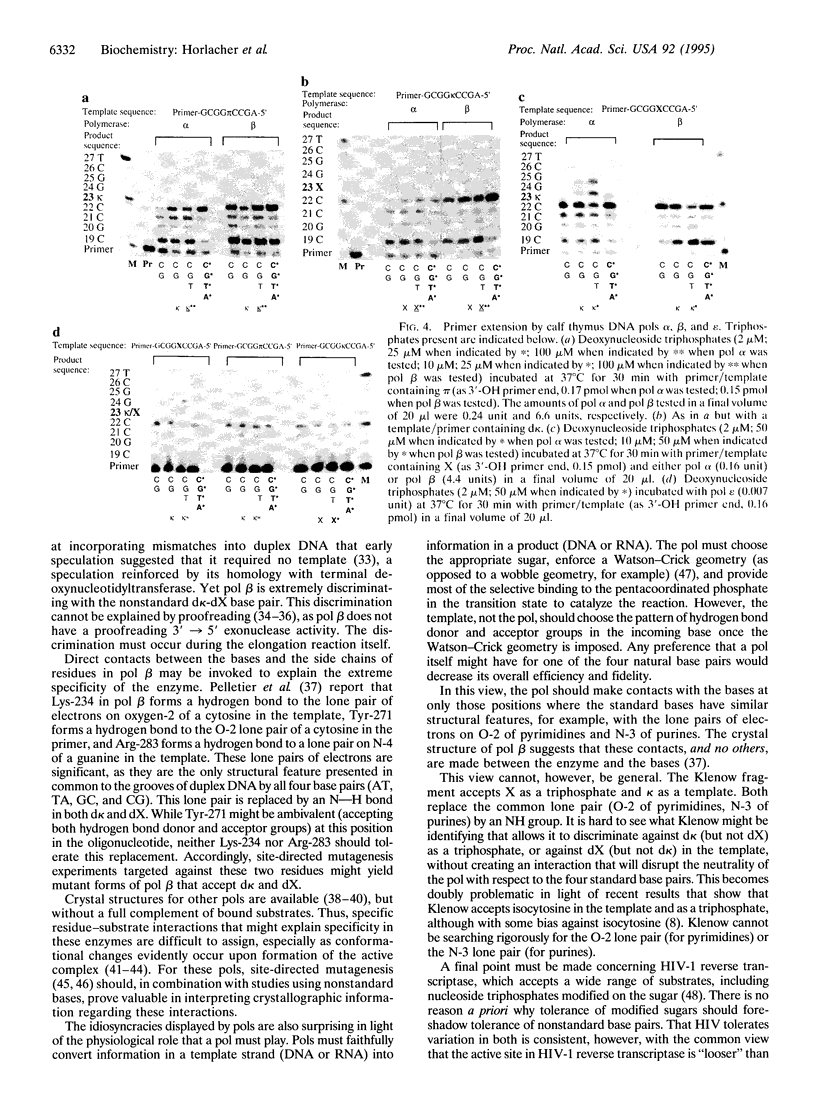
The ability of DNA polymerases (pols) to catalyze the template-directed synthesis of duplex oligonucleotides containing a nonstandard Watson-Crick base pair between a nucleotide bearing a 5-(2,4-diaminopyrimidine) heterocycle (d kappa) and a nucleotide bearing either deoxyxanthosine (dX) or N1-methyloxoformycin B (pi) has been investigated. The kappa-X and kappa-pi base pairs are jointed by a hydrogen bonding pattern different from and exclusive of those joining the AT and GC base pairs. Reverse transcriptase from human immunodeficiency virus type 1 (HIV-1) incorporates dXTP into an oligonucleotide opposite d kappa in a template with good fidelity. With lower efficiency and fidelity, HIV-1 reverse transcriptase also incorporates d kappa TP opposite dX in the template. With d pi in the template, no incorporation of d kappa TP was observed with HIV reverse transcriptase. The Klenow fragment of DNA pol I from Escherichia coli does not incorporate d kappa TP opposite dX in a template but does incorporate dXTP opposite d kappa. Bovine DNA pols alpha, beta, and epsilon accept neither dXTP opposite d kappa nor d kappa TP opposite d pi. DNA pols alpha and epsilon (but not beta) incorporate d kappa TP opposite dX in a template but discontinue elongation after incorporating a single additional base. These results are discussed in light of the crystal structure for pol beta and general considerations of how polymerases must interact with an incoming base pair to faithfully copy genetic information.
Full text
PDF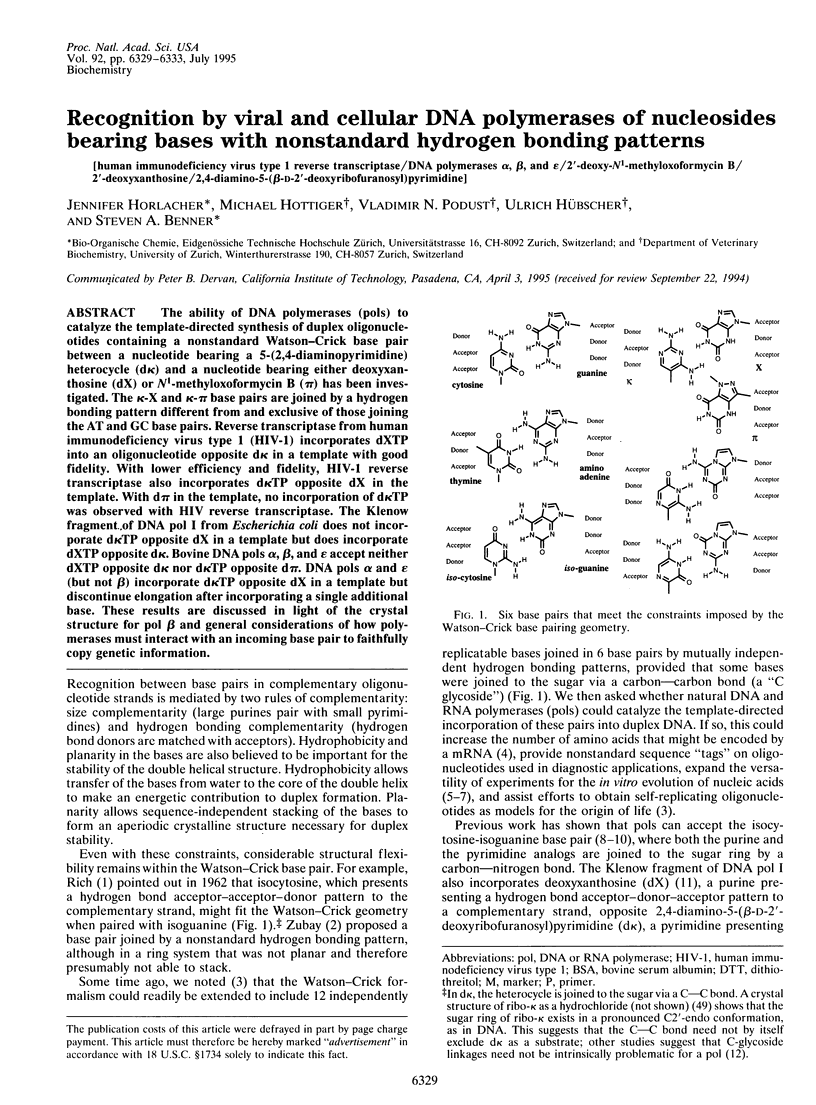

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bain J. D., Switzer C., Chamberlin A. R., Benner S. A. Ribosome-mediated incorporation of a non-standard amino acid into a peptide through expansion of the genetic code. Nature. 1992 Apr 9;356(6369):537–539. doi: 10.1038/356537a0. [DOI] [PubMed] [Google Scholar]
- Benner S. A., Allemann R. K., Ellington A. D., Ge L., Glasfeld A., Leanz G. F., Krauch T., MacPherson L. J., Moroney S., Piccirilli J. A. Natural selection, protein engineering, and the last riboorganism: rational model building in biochemistry. Cold Spring Harb Symp Quant Biol. 1987;52:53–63. doi: 10.1101/sqb.1987.052.01.009. [DOI] [PubMed] [Google Scholar]
- Campbell J. L. Yeast DNA replication. J Biol Chem. 1993 Dec 5;268(34):25261–25264. [PubMed] [Google Scholar]
- Chang L. M. Low molecular weight deoxyribonucleic acid polymerase from calf thymus chromatin. I. Preparation of homogeneous enzyme. J Biol Chem. 1973 Jun 10;248(11):3789–3795. [PubMed] [Google Scholar]
- Chu C. K., Reichman U., Watanabe K. A., Fox J. J. Nucleosides. 104. Synthesis of 4-amino-5-(D-ribofuranosyl)pyrimidine C-nucleosides from 2-(2,3-O-isopropylidene-5-O-trityl-D-ribofuranosyl)acetonitrile. J Org Chem. 1977 Feb 18;42(4):711–714. doi: 10.1021/jo00424a030. [DOI] [PubMed] [Google Scholar]
- Davies J. F., 2nd, Almassy R. J., Hostomska Z., Ferre R. A., Hostomsky Z. 2.3 A crystal structure of the catalytic domain of DNA polymerase beta. Cell. 1994 Mar 25;76(6):1123–1133. doi: 10.1016/0092-8674(94)90388-3. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W. DNA polymerase accuracy and spontaneous mutation rates: frequencies of purine.purine, purine.pyrimidine, and pyrimidine.pyrimidine mismatches during DNA replication. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4251–4255. doi: 10.1073/pnas.78.7.4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giver L., Bartel D. P., Zapp M. L., Green M. R., Ellington A. D. Selection and design of high-affinity RNA ligands for HIV-1 Rev. Gene. 1993 Dec 27;137(1):19–24. doi: 10.1016/0378-1119(93)90246-y. [DOI] [PubMed] [Google Scholar]
- Greene R., Korn D. Partial purification and characterization of deoxyribonucleic acid polymerase from KB cells. J Biol Chem. 1970 Jan 25;245(2):254–261. [PubMed] [Google Scholar]
- Hafkemeyer P., Ferrari E., Brecher J., Hübscher U. The p15 carboxyl-terminal proteolysis product of the human immunodeficiency virus type 1 reverse transcriptase p66 has DNA polymerase activity. Proc Natl Acad Sci U S A. 1991 Jun 15;88(12):5262–5266. doi: 10.1073/pnas.88.12.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübscher U., Spadari S. DNA replication and chemotherapy. Physiol Rev. 1994 Apr;74(2):259–304. doi: 10.1152/physrev.1994.74.2.259. [DOI] [PubMed] [Google Scholar]
- Hübscher U., Thömmes P. DNA polymerase epsilon: in search of a function. Trends Biochem Sci. 1992 Feb;17(2):55–58. doi: 10.1016/0968-0004(92)90499-y. [DOI] [PubMed] [Google Scholar]
- Irvine D., Tuerk C., Gold L. SELEXION. Systematic evolution of ligands by exponential enrichment with integrated optimization by non-linear analysis. J Mol Biol. 1991 Dec 5;222(3):739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- Jacobo-Molina A., Ding J., Nanni R. G., Clark A. D., Jr, Lu X., Tantillo C., Williams R. L., Kamer G., Ferris A. L., Clark P. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993 Jul 1;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger R., Podust V., Hübscher U., Berg P. A mammalian protein complex that repairs double-strand breaks and deletions by recombination. J Biol Chem. 1993 Jul 15;268(20):15070–15079. [PubMed] [Google Scholar]
- Joyce C. M., Sun X. C., Grindley N. D. Reactions at the polymerase active site that contribute to the fidelity of Escherichia coli DNA polymerase I (Klenow fragment). J Biol Chem. 1992 Dec 5;267(34):24485–24500. [PubMed] [Google Scholar]
- Kati W. M., Johnson K. A., Jerva L. F., Anderson K. S. Mechanism and fidelity of HIV reverse transcriptase. J Biol Chem. 1992 Dec 25;267(36):25988–25997. [PubMed] [Google Scholar]
- Kohlstaedt L. A., Wang J., Friedman J. M., Rice P. A., Steitz T. A. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992 Jun 26;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Schaaper R. M., Beckman R. A., Loeb L. A. On the fidelity of DNA replication. Effect of the next nucleotide on proofreading. J Biol Chem. 1981 Oct 10;256(19):9883–9889. [PubMed] [Google Scholar]
- Ludwig W., Follmann H. The specificity of ribonucleoside triphosphate reductase. Multiple induced activity changes and implications for deoxyribonucleotide formation. Eur J Biochem. 1978 Jan 16;82(2):393–403. doi: 10.1111/j.1432-1033.1978.tb12034.x. [DOI] [PubMed] [Google Scholar]
- Mizrahi V., Henrie R. N., Marlier J. F., Johnson K. A., Benkovic S. J. Rate-limiting steps in the DNA polymerase I reaction pathway. Biochemistry. 1985 Jul 16;24(15):4010–4018. doi: 10.1021/bi00336a031. [DOI] [PubMed] [Google Scholar]
- Muzyczka N., Poland R. L., Bessman M. J. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972 Nov 25;247(22):7116–7122. [PubMed] [Google Scholar]
- Pelletier H., Sawaya M. R., Kumar A., Wilson S. H., Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994 Jun 24;264(5167):1891–1903. [PubMed] [Google Scholar]
- Piccirilli J. A., Krauch T., Moroney S. E., Benner S. A. Enzymatic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature. 1990 Jan 4;343(6253):33–37. doi: 10.1038/343033a0. [DOI] [PubMed] [Google Scholar]
- Piccirilli J. A., Moroney S. E., Benner S. A. A C-nucleotide base pair: methylpseudouridine-directed incorporation of formycin triphosphate into RNA catalyzed by T7 RNA polymerase. Biochemistry. 1991 Oct 22;30(42):10350–10356. doi: 10.1021/bi00106a037. [DOI] [PubMed] [Google Scholar]
- Podust V. N., Hübscher U. Lagging strand DNA synthesis by calf thymus DNA polymerases alpha, beta, delta and epsilon in the presence of auxiliary proteins. Nucleic Acids Res. 1993 Feb 25;21(4):841–846. doi: 10.1093/nar/21.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesky A. H., Dahlberg M. E., Benkovic S. J., Grindley N. D., Joyce C. M. Side chains involved in catalysis of the polymerase reaction of DNA polymerase I from Escherichia coli. J Biol Chem. 1992 Apr 25;267(12):8417–8428. [PubMed] [Google Scholar]
- Switzer C. Y., Moroney S. E., Benner S. A. Enzymatic recognition of the base pair between isocytidine and isoguanosine. Biochemistry. 1993 Oct 5;32(39):10489–10496. doi: 10.1021/bi00090a027. [DOI] [PubMed] [Google Scholar]
- Szostak J. W. In vitro genetics. Trends Biochem Sci. 1992 Mar;17(3):89–93. doi: 10.1016/0968-0004(92)90242-2. [DOI] [PubMed] [Google Scholar]
- Tanabe K., Bohn E. W., Wilson S. H. Steady-state kinetics of mouse DNA polymerase beta. Biochemistry. 1979 Jul 24;18(15):3401–3406. doi: 10.1021/bi00582a029. [DOI] [PubMed] [Google Scholar]
- Weiser T., Gassmann M., Thömmes P., Ferrari E., Hafkemeyer P., Hübscher U. Biochemical and functional comparison of DNA polymerases alpha, delta, and epsilon from calf thymus. J Biol Chem. 1991 Jun 5;266(16):10420–10428. [PubMed] [Google Scholar]
- Wong I., Patel S. S., Johnson K. A. An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry. 1991 Jan 15;30(2):526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]