Stabilization of oscillatory UNC-40 clustering by its ligand UNC-6 directs localized F-actin generation and polarization of cells.
Abstract
The receptor deleted in colorectal cancer (DCC) directs dynamic polarizing activities in animals toward its extracellular ligand netrin. How DCC polarizes toward netrin is poorly understood. By performing live-cell imaging of the DCC orthologue UNC-40 during anchor cell invasion in Caenorhabditis elegans, we have found that UNC-40 clusters, recruits F-actin effectors, and generates F-actin in the absence of UNC-6 (netrin). Time-lapse analyses revealed that UNC-40 clusters assemble, disassemble, and reform at periodic intervals in different regions of the cell membrane. This oscillatory behavior indicates that UNC-40 clusters through a mechanism involving interlinked positive (formation) and negative (disassembly) feedback. We show that endogenous UNC-6 and ectopically provided UNC-6 orient and stabilize UNC-40 clustering. Furthermore, the UNC-40–binding protein MADD-2 (a TRIM family protein) promotes ligand-independent clustering and robust UNC-40 polarization toward UNC-6. Together, our data suggest that UNC-6 (netrin) directs polarized responses by stabilizing UNC-40 clustering. We propose that ligand-independent UNC-40 clustering provides a robust and adaptable mechanism to polarize toward netrin.
Introduction
The ability of cells to dynamically polarize toward external cues is critical to many biological processes, including cell migration, neurite outgrowth, synapse formation, and directional growth (Wang, 2009; Arkowitz, 2013). In experimentally accessible, single-cell models of dynamic polarization in yeast, Dictyostelium discoideum, and neutrophils, polarization occurs in random orientations when the directional cue is absent or uniformly present (Sohrmann and Peter, 2003; Arrieumerlou and Meyer, 2005; Ozbudak et al., 2005; Sasaki et al., 2007; Wang, 2009; Arai et al., 2010; Wu and Lew, 2013). External chemical gradients and cues are thought to act in these cases by biasing the orientation of intrinsic polarity systems (Sasaki et al., 2007; Johnson et al., 2011; Huang et al., 2013). Recent studies during yeast mating using high-resolution live-cell imaging have also observed that the polarity-organizing Cdc42 GTPase, which accumulates at the site of polarization, can undergo oscillatory clustering and dispersal at the cell cortex in low pheromone conditions in both Schizosaccharomyces pombe and Saccharomyces cerevisiae (Bendezú and Martin, 2013; Dyer et al., 2013). Only when exposed to high pheromone concentrations or steep gradients does polarity stabilize toward the source of a pheromone. The GTPase Ras also dynamically localizes at random cell surface locations in the absence of the chemoattractant cAMP in D. discoideum (Sasaki et al., 2007). This behavior indicates the existence of a system of positive feedback that promotes cluster formation, which then initiates a delayed negative feedback loop that triggers cluster dispersal (Bendezú and Martin, 2013; Dyer et al., 2013; Wu and Lew, 2013). These systems are thought to have evolved in part to allow robust responses to external cues: positive feedback might allow cells to rapidly amplify responses toward directional external cues, whereas negative feedback may prevent polarization from over-spreading, regulate competition between polarization clusters, and allow reorientation of response toward varying external signals (Houk et al., 2012; Dyer et al., 2013; Wu and Lew, 2013). Whether similar mechanisms underlie pathways that mediate dynamic polarity in multicellular tissue environments is unclear.
The netrin receptor deleted in colorectal cancer (DCC) is a central regulator of diverse polarization processes in multicellular animals (Keino-Masu et al., 1996; Ziel and Sherwood, 2010; Lai Wing Sun et al., 2011). Important insights into the mechanisms of DCC signaling have come from Caenorhabditis elegans, where its simple tissue architecture and the presence of a single netrin ligand, the secreted protein UNC-6, and a single DCC receptor orthologue, UNC-40, have facilitated detailed dissection of this pathway. These studies have revealed roles for UNC-40–mediated polarizing activities in UNC-6–directed guidance of axon outgrowth, synapse formation, axon arborization, and cell migration (Chan et al., 1996; Adler et al., 2006; Asakura et al., 2007; Colón-Ramos et al., 2007; Nelson and Colón-Ramos, 2013). In these events, the receptor UNC-40 polarizes within the plasma membrane toward extracellular sources of UNC-6 (netrin), where it directs F-actin formation at the cell cortex. Studies of UNC-40 receptor localization with static imaging in the hermaphrodite-specific neuron (HSN) and anterior ventral microtubule (AVM) neurons in mutant backgrounds affecting axon outgrowth have suggested that UNC-40 can asymmetrically localize to random domains on the cell membrane independently of UNC-6 (Xu et al., 2009; Kulkarni et al., 2013). These studies have led to the hypothesis that UNC-6 polarizes UNC-40 by biasing the localization of stochastically distributed UNC-40 receptors (Kulkarni et al., 2013). How UNC-6 biases UNC-40 polarity and regulates the dynamics of the UNC-40 receptor during polarization, however, is unknown.
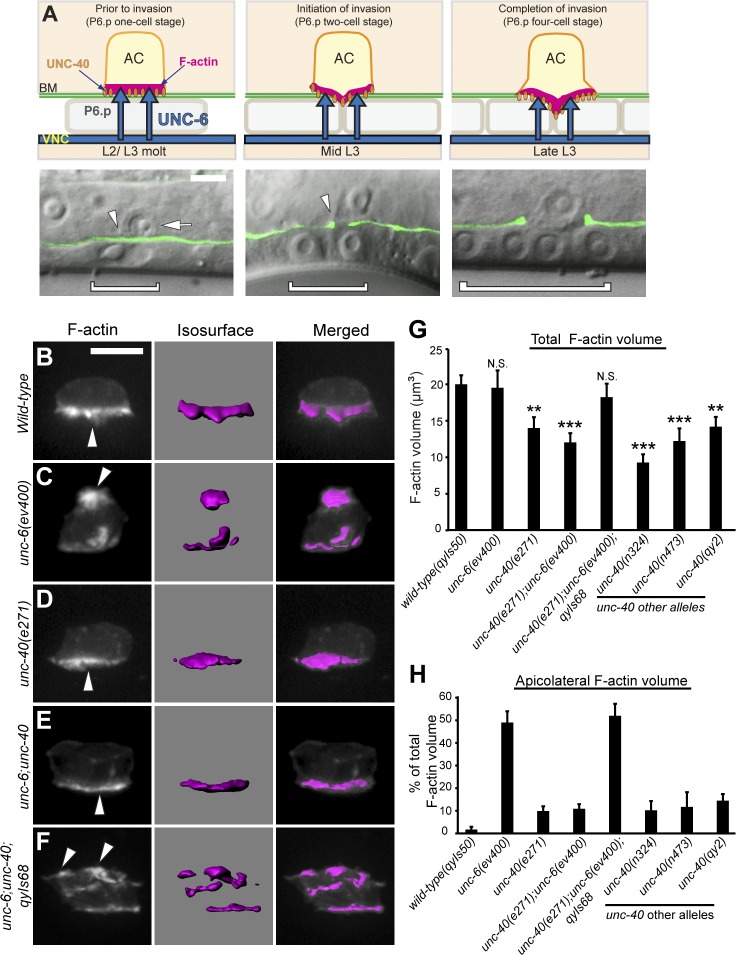
The C. elegans anchor cell (AC) is a specialized gonadal cell that polarizes toward and then invades through the basement membrane separating the uterine and vulval epithelium to initiate uterine–vulval attachment (Fig. 1 A; Sherwood and Sternberg, 2003; Hagedorn and Sherwood, 2011; Ihara et al., 2011; Kelley et al., 2014). UNC-6 (netrin) and UNC-40 (DCC) are crucial mediators of AC polarization. The receptor UNC-40 is enriched at the AC’s invasive cell membrane, where it directs the formation of an invasive protrusion that breaches the basement membrane (Ziel et al., 2009; Hagedorn et al., 2013). UNC-40 polarization relies on UNC-6 (netrin), which is secreted from the ventral nerve cord and accumulates in the basement membrane in contact with the invasive cell membrane of the AC (Ziel et al., 2009). Loss of unc-6 perturbs invasion and results in UNC-40 and F-actin regulators mislocalizing to all regions of the AC’s plasma membrane.
Figure 1.
UNC-40 is mispolarized and active in the absence of UNC-6. Anterior is left; ventral is down. (A) AC invasion in C. elegans (top, schematic; green, differential interference contrast [DIC] microscopy with basement membrane marker laminin::GFP; bottom, overlay). During the L2/L3 molt (left), the AC (bottom, arrow) is attached to the basement membrane (BM; arrowhead) over the P6.p vulval precursor cell (bracket outlines nucleus, bottom). UNC-6 (netrin; top, blue arrows) secreted from the ventral nerve cord (VNC) polarizes UNC-40 (DCC; orange ovals) and F-actin (magenta) to the AC’s basal, invasive cell membrane. After P6.p divides (middle, P6.p two-cell stage), a protrusion breaches (bottom, arrowhead) and then removes basement membrane, and moves between the central P6.p granddaughter cells (right, P6.p four-cell stage). (B–F) Fluorescence (left), corresponding dense F-actin network (isosurface, middle), and overlay (right). (B) In wild-type animals, F-actin (visualized with cdh-3 > mCherry::moeABD) was polarized to the basal membrane (arrowhead). (C) In unc-6 mutants, F-actin was mislocalized to the AC’s apical and lateral membranes (arrowhead). (D) In unc-40 mutants, F-actin volume was reduced but polarized (arrowhead). (E) In unc-6; unc-40 double mutants, F-actin was reduced but polarized (arrowhead), comparable to unc-40. (F) In unc-6; unc-40; qyIs68[cdh-3 > unc-40::GFP] animals, F-actin was mislocalized (arrowheads), resembling unc-6. (G and H) The total volume of F-actin and the percentage that localized apicolaterally, respectively, at the P6.p four-cell stage (n ≥ 15 per genotype). *, P < 0.05; **, P < 0.01; ***, P < 0.001. N.S., no significant difference (P > 0.05, Student’s t test). Error bars indicate ± SEM. Significant differences relative to wild-type are indicated. Bars, 5 µm.
To further understand how UNC-40 (DCC) and UNC-6 (netrin) function during polarization, we have used live-cell imaging and genetic analysis to examine the activity and localization of UNC-40 during AC invasion. Surprisingly, we have discovered that in the absence of UNC-6 (netrin), UNC-40 (DCC) is active and displays oscillatory clustering behavior in the cell membrane: UNC-40 molecules assemble into a large cluster, recruit F-actin effectors, generate F-actin, and then break down and reform in a different location with regular periodicity. This oscillatory behavior suggests that UNC-40 clustering is regulated by a mechanism involving interlinked positive (cluster formation) and delayed negative (disassembly) feedback. We demonstrate that localized presentation of UNC-6 orients and stabilizes UNC-40 clustering. Furthermore, we show that MADD-2, a TRIM protein family member and direct regulator of UNC-40, promotes ligand-independent UNC-40 clustering and is required for robust UNC-40 polarization toward UNC-6. Together, these results indicate that UNC-6 directs polarized responses by stabilizing randomly localized, ligand-independent UNC-40 clustering. We suggest that ligand-independent UNC-40 (DCC) oscillatory clustering provides a rapid and flexible system for polarization toward UNC-6 (netrin).
Results
Loss of UNC-6 (netrin) and UNC-40 (DCC) have different effects on F-actin
UNC-6 and UNC-40 are thought to organize polarized responses in part by modulating F-actin formation through UNC-40 downstream effectors (Gitai et al., 2003; Adler et al., 2006; Lai Wing Sun et al., 2011). To further understand how UNC-6 and UNC-40 modulate polarity, we first characterized F-actin formation and distribution in the absence of UNC-6. We found similar amounts of total F-actin in both wild-type animals and unc-6 mutants, but 50% of the F-actin was mislocalized in the absence of unc-6 and clustered along the apical or lateral membranes of the AC (Fig. 1, B, C, G, and H). To determine whether loss of the UNC-6 receptor UNC-40 (DCC) had a similar phenotype, we examined three putative unc-40–null mutants: n324, n473, and e271. We also generated an additional allele, qy2, a deletion (711 bp) in the 11th intron and 12th exon, which creates a predicted stop codon (Fig. S1). Surprisingly, loss of unc-40 affected F-actin differently than loss of unc-6: F-actin levels were reduced ∼30% and F-actin still remained at the invasive cell membrane in all unc-40 mutant backgrounds (Fig. 1, B, D, G, and H). This residual F-actin was dependent on the integrin heterodimer αINA-1/βPAT-3, which mediates F-actin formation independently of UNC-40 at the invasive membrane (Fig. S2; Hagedorn et al., 2009). Together, these results indicate that UNC-40 (DCC) promotes F-actin generation, whereas UNC-6 (netrin) polarizes this activity toward the AC’s invasive cell membrane.
UNC-40 is mispolarized and active in the absence of UNC-6
To reconcile how loss of UNC-40 and UNC-6 differentially regulate F-actin formation and localization, respectively, we generated an unc-6; unc-40 double mutant to determine their epistatic relationship. Notably, the F-actin phenotype in this double mutant was similar to unc-40 mutants alone (Fig. 1, D, E, G, and H). Thus, both normal levels of F-actin formation at the invasive membrane as well as mislocalization of F-actin patches in unc-6 mutants are dependent on UNC-40 activity. Confirming this notion, transgenic expression of UNC-40 in the AC restored F-actin levels and ectopic F-actin clusters in unc-6; unc-40 double mutants (Fig. 1, F–H). These results indicate that in the absence of UNC-6, UNC-40 is active and directs ectopic F-actin formation.
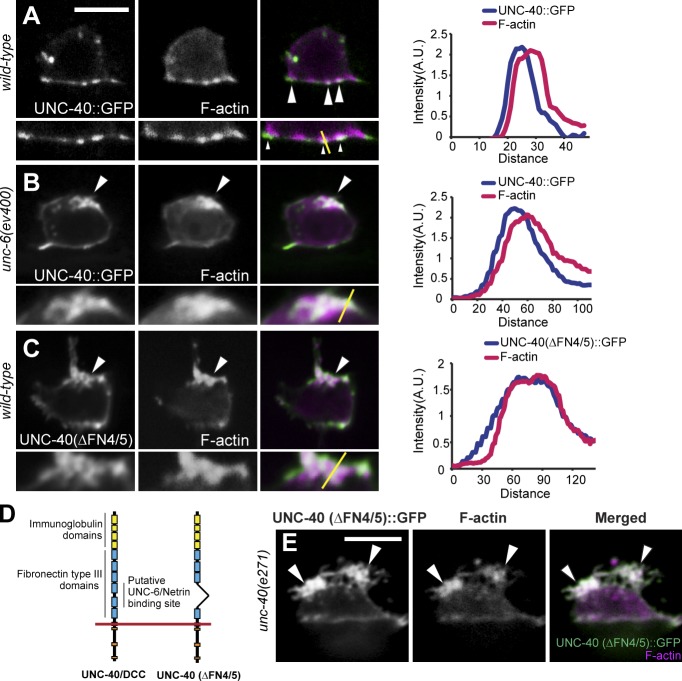
Considering the UNC-6–independent activity of UNC-40, we hypothesized that the ectopic F-actin found in unc-6 mutants might be directly organized by mislocalized, but active UNC-40. Supporting this idea, we found that F-actin patches were strongly colocalized with UNC-40 in both wild-type animals and unc-6 mutants (Fig. 2, A and B; and Fig. 3 D). In addition, a form of UNC-40 lacking the fourth and fifth FNIII repeats, which are necessary for UNC-6 binding (Geisbrecht et al., 2003; Kruger et al., 2004), was also active and mislocalized when expressed in both wild-type and unc-40 mutant ACs (Fig. 2, C–E; and Fig. 3 D). We also found that in the absence of UNC-6, F-actin patches colocalized tightly with membrane-localized clusters of UNC-40 downstream effectors (UNC-34, CED-10, MIG-2, and UNC-115; Fig. 3, A–D; Wang et al., 2014). This genetic, molecular, and colocalization analysis offers strong evidence that the UNC-40 receptor promotes F-actin formation in an UNC-6–independent manner.
Figure 2.
Mispolarized UNC-40 colocalizes with F-actin in the absence of UNC-6. Anterior is left; ventral is down. (A–C) All animals were examined at the P6.p two-cell stage. Shown are UNC-40::GFP and UNC-40(ΔFN4/5)::GFP (left), F-actin (middle), and an overlay (right). Magnification (below) and colocalization quantification graphs (far right) show extensive UNC-40 and F-actin overlapping localization in all cases (arrowheads; colocalization was measured along the yellow line in the insets from the outside to the inside of the cell, fluorescent intensity was plotted in arbitrary units, and distances are given in pixels). (D) A schematic diagram illustrates the structural domains of the wild-type UNC-40 protein and the UNC-40(ΔFN4/5) protein, an engineered form of UNC-40 lacking the putative UNC-6 binding site in the fourth and fifth FNIII domains. (E) In unc-40 mutants, UNC-40(ΔFN4/5)::GFP colocalized with F-actin in the apical and lateral membranes of the AC (arrowheads). Bars, 5 µm.
Figure 3.
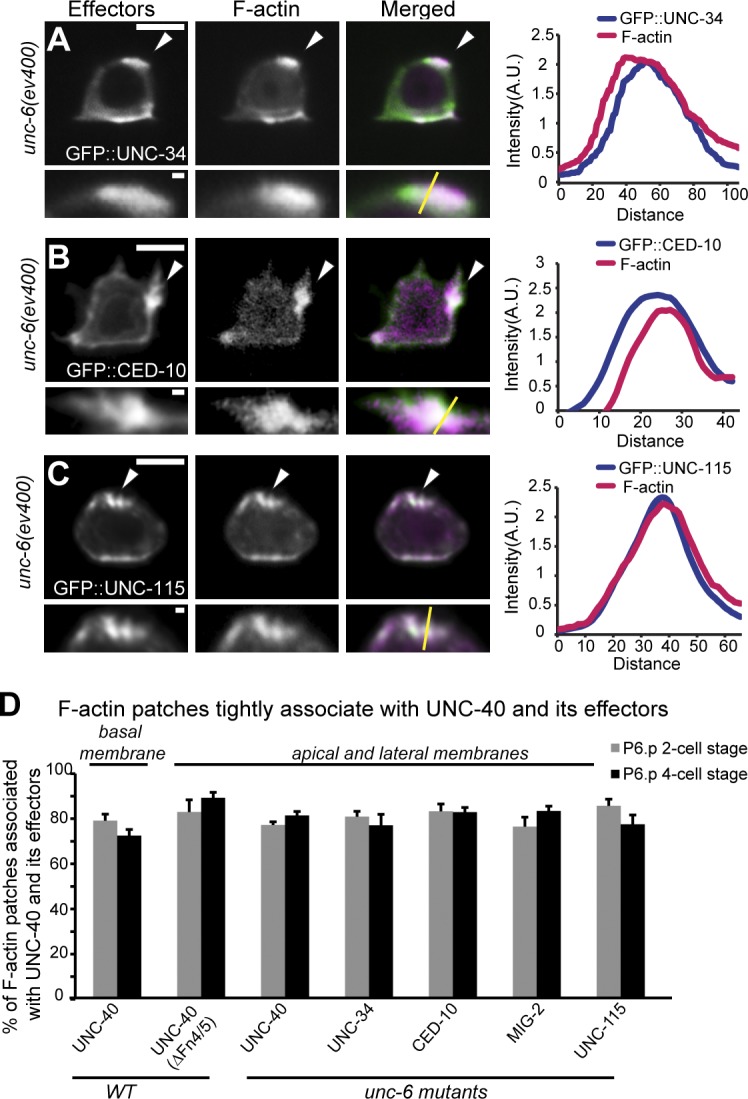
F-actin colocalizes with UNC-40 effectors in the absence of UNC-6. Anterior is left; ventral is down. (A–C) All animals were examined at the P6.p two-cell stage. UNC-40 effectors (left), F-actin (middle), overlay (right), and magnifications (below) are shown. Colocalization graphs (far right) show areas of colocalization (arrowheads; measured along yellow lines in insets; fluorescent intensity is plotted in arbitrary units and distances are given in pixels). In unc-6 mutants, the UNC-40 effectors, GFP::UNC-34 (Ena/VASP), GFP::CED-10 (Rac), and GFP::UNC-115 (abLIM; green), were all colocalized (arrowheads) with F-actin (magenta) at the AC’s apical and lateral membranes. (D) The percentage of the F-actin patches that were associated with the patches of UNC-40::GFP, UNC-40(ΔFN4/5)::GFP, GFP::UNC-34, GFP::CED-10, GFP::MIG-2 (Rac), and GFP::UNC-115 (n ≥ 15 for each stage per genotype). Error bars indicate ± SEM. No significant differences (P > 0.05, Student’s t test) compared with wild-type UNC-40/F-actin were observed. Bars: (main panels) 5 µm; (magnified insets) 0.5 µm.
The activity of the UNC-40 receptor observable in unc-6 (netrin) mutants might result from activation by a second signal. To explore this possibility, we determined whether SLT-1 (slit), MADD-4 (ADAMTSL), and UNC-129 (TGF-β), three secreted proteins showing genetic interactions with UNC-40 (Stein and Tessier-Lavigne, 2001; Yu et al., 2002; MacNeil et al., 2009; Seetharaman et al., 2011), were responsible for activating UNC-40. We found, however, that these signals are not required for activation of UNC-40 in the absence of UNC-6 (Fig. S3). Thus, UNC-40 is activated by an unknown ligand or independently of a ligand.
UNC-40 is randomly polarized in the absence of UNC-6
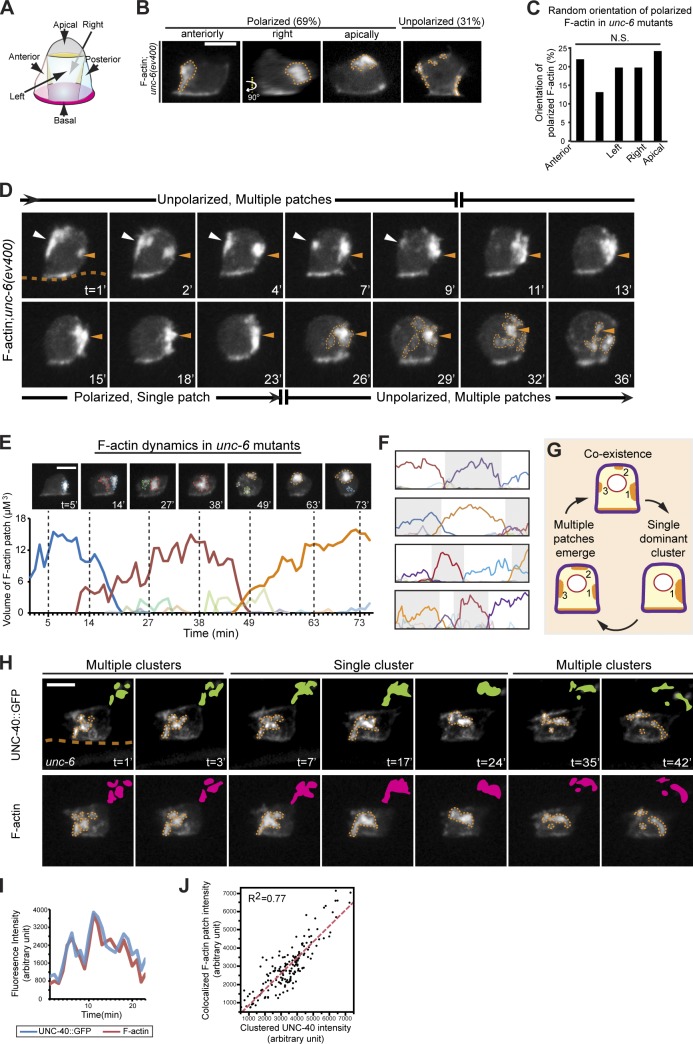
To further explore the UNC-6–independent activity and dynamics of UNC-40 in the AC, we first examined static images of the spatial distribution of ectopic F-actin (the output of UNC-40 activity) during the P6.p two-cell stage (an ∼1-h period) in unc-6 mutants. To facilitate analysis, we partitioned the AC’s apical and lateral cell membranes into five equally sized portions: anterior, posterior, left, right, and apical (Fig. 4 A). The basal cell membrane (in contact with the basement membrane) was not included, as it contained F-actin that was generated by integrin (see Fig. S2). Importantly, we found that 69% of the ACs (45 out of 65 ACs observed) had a single ectopic F-actin patch polarized within one of the five membrane portions. The location of the F-actin cluster was not biased to any of the five membrane domains (Fig. 4, B and C). Thus, in the absence of UNC-6, UNC-40 mediates randomly directed F-actin polarity within the AC, where we define random as unbiased F-actin patch localization in any one of the equally sized membrane regions.
Figure 4.
UNC-40 undergoes periodic clustering and disassembly in the absence of UNC-6. Anterior is left; ventral is down. (A) A schematic diagram shows five membrane portions of the ACs used for scoring F-actin localization. (B and C) Most ACs in unc-6 mutants (69%) had a single dominant F-actin patch (orange broken lines, visualized with the F-actin probe cdh-3 > mCherry::moeABD) polarized randomly toward one of the five divided portions (P > 0.1, χ2 test, n = 65 animals observed). (D) Time series of F-actin localization in the AC of an unc-6 mutant revealed cycling between multiple F-actin patches and a single cluster. Initially, multiple small F-actin patches (white and orange arrowheads) formed randomly. After 9 min, one patch (orange arrowheads) grew and became dominant. By 26 min, this single cluster began disassembling, and multiple new F-actin foci (orange broken lines) formed. The orange broken line in the first frame indicates the position of the basement membrane where integrin maintains a light band of F-actin in the AC. (E) The volume of individual F-actin patches over time in an unc-6 mutant. Each colored line represents an individual patch. Images above show snapshots at times marked by the vertical broken lines (patches are outlined by colors that correspond to the graph). (F) Similar analysis of four other ACs in unc-6 mutants. (G) Schematic of cluster oscillations in unc-6 mutants. (H) A 42-min time series shows that clusters of UNC-40::GFP (top, green) colocalized with F-actin (bottom, magenta). (I) The fluorescence intensity of UNC-40::GFP and colocalized F-actin during the life cycle of a representative F-actin patch in an unc-6 mutant animal. (J) UNC-40::GFP intensity at each time point (black dots) was plotted against colocalized F-actin patch intensity, revealing a tight correlation (measured using coefficient of determination R2) during cluster oscillations. Data were pooled from nine cluster life cycles in seven animals. Bars, 5 µm.
UNC-40 clusters and disassembles with regular periodicity in the absence of UNC-6
To understand the dynamics of UNC-40 polarity in the absence of UNC-6, we performed live-cell imaging of F-actin and UNC-40 in the AC in unc-6 mutants. These observations revealed a pattern of polarized clustering that formed and disassembled in an oscillatory manner in all plasma membrane regions of the cell. The polarity cycle initiated with the formation of multiple small F-actin patches at the cell cortex (mean of 3.1 patches; n = 32 ACs observed). After a short coexistence time (mean of 9.6 min), one patch grew in volume (at a constant rate of ∼0.7 µm3/min; Table 1) and became dominant, while the others disappeared (Fig. 4 D and Video 1). The dominant single patches had a mean lifetime of 25.9 min (Table 1). After this time, there was a breakdown of the dominant patch (at a constant rate of 0.91 µm3/min; Table 1), and its loss correlated with the emergence of multiple F-actin foci in other random locations, reinitiating the polarity cycle (Fig. 4, D–G and Table 1). Importantly, only a few, small, transient F-actin patches occasionally formed in the AC’s apical and lateral membranes in unc-40 mutants (Fig. S4 and Video 1), which indicates that oscillatory F-actin polarity is dependent on UNC-40.
Table 1.
F-actin polarization and formation dynamics
| Genotype | Lifetimea | Frequency of polarization axis switchb | F-actin patch co-existence timec | Dominant F-actin patch formation ratee | Dominant F-actin patch disassembly rate | Peak volume of dominant ectopic F-acting |
| min | Counts per hour | min | µm3/min | µm3/min | µm3 | |
| unc-6(ev400) | 25.9 ± 2.1h (n = 12) | 2.5 ± 0.2h (n = 12) | 9.6 ± 1.1 (n = 32) | 0.697 ± 0.17 (n = 9) R2 = 0.92 | 0.91 ± 0.12 (n = 12) R2 = 0.97 | 15.4 ± 1.1 (n = 20) |
| unc-6(ev400); Ex[dorsal UNC-6] | >69.3 ± 5.9 (n = 8 with one stable UNC-6 source) | 0 (n = 8, with one stable UNC-6 source) | N.A.d | 0.669 ± 0.12 (n = 6) R2 = 0.97 | N.A.f | 18.1 ± 2.7 (n = 7) |
| madd-2(ok2226); unc-6(ev400) | >82.7 ± 12.2 (n = 6) | <0.8 ± 0.1i (n = 6) | ND | 0.200 ± 0.06j (n = 5) R2 = 0.86 | 0.707 ± 0.23 (n = 5) R2 = 0.80 | 16.5 ± 2.3 (n = 6) |
Data are presented as mean ± SEM. N.A., not applicable.
The lifetime of the polarized F-actin patches in most unc-6(ev400); Ex[dorsal UNC-6] and madd-2(ok2226); unc-6(ev400) mutant animals exceeded the corresponding time-lapse length. Thus, we used the recorded times, which are an underestimate.
A switch in polarization axis was defined when the dominant polarity patch broke down and a new dominant patch formed in a different quadrant of the AC (see Fig. 4 A).
The coexistence period was defined as the time when multiple patches were first observed until the moment when only one remained and the others completely disassembled.
Multiple patches were not observed in the presence of stable dorsal UNC-6.
In unc-6 and madd-2; unc-6 animals, formation rate was defined as the time period when a future dominant F-actin patch was first detected until the peak volume was achieved. In unc-6; Ex[dorsal UNC-6] animals, F-actin patch formation rate was defined as the period when the AC membrane first encountered a new source of UNC-6 until peak F-actin volume was achieved. A trend line was fit to the volume of each F-actin patch during assembly to determine the slope (rate of F-actin formation). No significant difference in F-action formation rate was observed between unc-6(ev400) and unc-6(ev400); Ex[dorsal UNC-6] (P > 0.1, Student’s t test).
No disassembly was observed with stable dorsal UNC-6.
No significant differences were observed between unc-6(ev400), unc-6(ev400); Ex[dorsal UNC-6], and madd-2(ok2226); unc-6(ev400) (P > 0.1, Student’s t test).
Relative to unc-6; Ex[dorsal UNC-6] (P < 0.001, Student’s t test).
Relative to unc-6 (P < 0.001, Student’s t test).
Relative to unc-6 (P < 0.05, Student’s t test).
To determine the relationship of the F-actin patches to UNC-40 during these oscillations, we examined UNC-40 and F-actin colocalization in real time. This analysis revealed that UNC-40 and F-actin clusters colocalized both temporally and spatially in unc-6 mutants for the complete oscillatory cycle (Fig. 4, H–J; and Video 2). Collectively, these results indicate that, in the absence of UNC-6, UNC-40 is active, generates F-actin, and has oscillating clustering behavior. As oscillatory behaviors in biological systems are thought to require delayed negative feedback (Park and Bi, 2007; Brandman and Meyer, 2008; Novák and Tyson, 2008; Tsai et al., 2008), this suggests that UNC-6–independent UNC-40 clustering is regulated by a system involving positive feedback (cluster formation), which triggers delayed negative feedback (cluster disassembly), and after dissipation of negative feedback, the restart of the cycle with the initiation of positive feedback–driven clustering.
Localized UNC-6 orients and stabilizes UNC-40 clustering
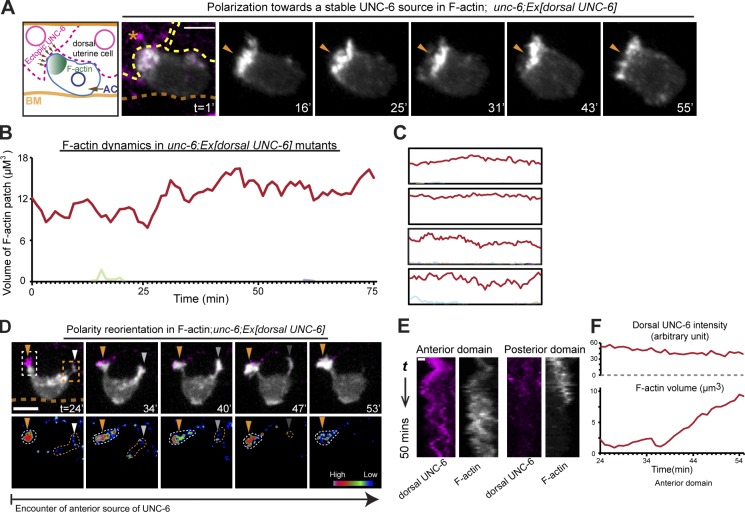
We next sought to determine how UNC-6 might influence the observed ligand-independent UNC-40/F-actin clustering activity. We have previously shown that UNC-40 and F-actin are polarized to the invasive cell membrane of the AC in contact with the basement membrane (where UNC-6 is localized) for ∼5 h before AC invasion (Hagedorn et al., 2009; Ziel et al., 2009). We next wanted to determine whether UNC-6 was sufficient to orient and stabilize UNC-40. To test this, we expressed a membrane-tethered UNC-6::GFP protein in the dorsal uterine cells of an unc-6 mutant, thus presenting the AC with a localized source of UNC-6 opposite to the endogenous ventral presentation of UNC-6 in the basement membrane of wild-type animals (Fig. 5, A and B). This ectopic dorsal presentation of UNC-6 directed UNC-40 and F-actin clustering stably toward the AC’s apical cell membrane in contact with UNC-6 (Fig. 5, A–G). Time-lapse analysis of F-actin indicated that a constant UNC-6 source in dorsal uterine cells stabilized F-actin formation for the entire duration of time-lapse imaging (∼70 min; Fig. 6, A–C; Video 3; and Table 1). The volume of F-actin in these patches was equivalent to the peak volume in UNC-40 clusters in the absence of UNC-6 (Table 1). These results demonstrate that UNC-6 orients and stabilizes UNC-40 clustering and indicate that UNC-6–UNC-40 interactions must counter the negative feedback mechanism that disassembles UNC-40 clusters in the absence of UNC-6.
Figure 5.
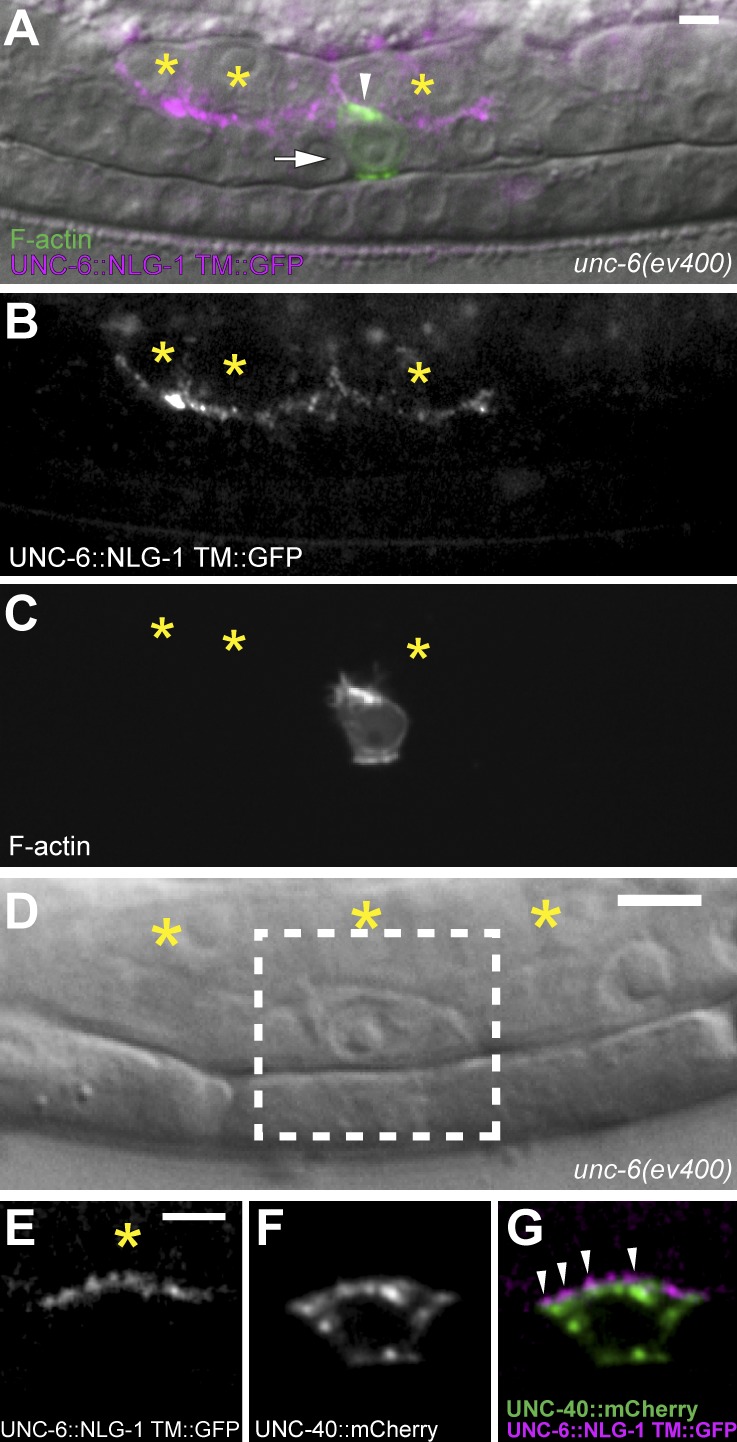
Membrane-tethered dorsal UNC-6 orients and stabilizes UNC-40 clustering. Anterior is left; ventral is down. (A) An unc-6 mutant with membrane-tethered UNC-6 (zmp-5 > unc-6::nlg-1 TM::GFP; magenta) expressed in the dorsal uterine cells (asterisks). F-actin (green, arrowhead) within the AC (arrow) was polarized toward the source of UNC-6 (animal shown at the P6.p four-cell stage). (B) A grayscale image shows that the dorsal uterine cells (asterisks) express and localize a zmp-5–driven, membrane-tethered UNC-6 protein at cell membranes. (C) A grayscale image shows apical polarization of F-actin in the AC toward dorsal UNC-6. (D–G) Images show colocalization of UNC-40::mCherry (arrowheads; green; expressed in the AC) and membrane-tethered UNC-6 (magenta; expressed in the dorsal uterine cells; asterisks). Images in E–G are magnifications of the area outlined by the box in D. Bars, 5 µm.
Figure 6.
UNC-6 stably polarizes UNC-40/F-actin oscillatory clustering. Anterior is left; ventral is down. (A) The time series shows F-actin polarity (orange arrowhead, grayscale) stabilized in the AC of an unc-6 mutant toward UNC-6 (zmp-5 > unc-6::nlg-1 TM::GFP; magenta), which is localized to dorsal uterine cell membranes (outlined with yellow lines; the asterisk marks a dorsal cell expressing UNC-6; the location of basement membrane [BM] is indicated with an orange line). (B) The red line in the graph represents the volume of the dominant F-actin patch in the time series shown above (the light green line is a small, transient F-actin patch localized away from the dominant patch). (C) Similar stabilization in four other cases. (D) The time series shows dynamic reorientation of F-actin polarity (grayscale) toward a changing UNC-6 source (magenta in top panels; outlined by broken lines in the bottom panels with spectral representation of fluorescence intensity). When the AC made contact with a new anterior source of UNC-6 (orange arrowheads), a polarized response was directed toward this new source, and polarity was lost on the posterior dorsal uterine cell as UNC-6 levels diminished (fading white arrowheads). See also Video 4. (E) A kymograph of the two outlined areas in the top of D from time 24 to 55 min of Video 4. (F) Quantification of UNC-6 (top) within the anterior domain and the associated increase in F-actin volume (bottom) during polarity reorientation after contact with this UNC-6 source (white boxed area in D). Bars: (A and D) 5 µm; (E) 0.5 µm.
UNC-40 (DCC) orients toward a dynamic source of UNC-6 (netrin)
DCC guides diverse polarization processes in animals (Lai Wing Sun et al., 2011), some of which involve guidance toward dynamic sources of netrin (Chan et al., 1996; Wadsworth et al., 1996; Asakura et al., 2007). We next wanted to determine if UNC-40 (DCC) in the AC could direct polarized responses toward changing sources of UNC-6 (netrin). The mosaic nature of transgene expression of UNC-6 and the shifting positions of dividing dorsal uterine cells allowed us to visualize UNC-40–mediated F-actin polarization in the AC toward changing sources of UNC-6. In these cases, we observed that UNC-40 directed rapid polarized responses toward changing sources of UNC-6 (Fig. 6, D–F; and Video 4; n = 6/6 animals observed). F-actin formation rates upon contact with a new source of UNC-6 were similar to rates during clustering activity in the absence of UNC-6 (Table 1), which suggests that UNC-6–UNC-40 interactions do not stimulate F-actin formation. Collectively, these results demonstrate that UNC-40 polarity in the AC is adaptable and directs polarization toward stable as well as varying sources of UNC-6 (netrin).
MADD-2 promotes UNC-40 clustering and polarization toward UNC-6
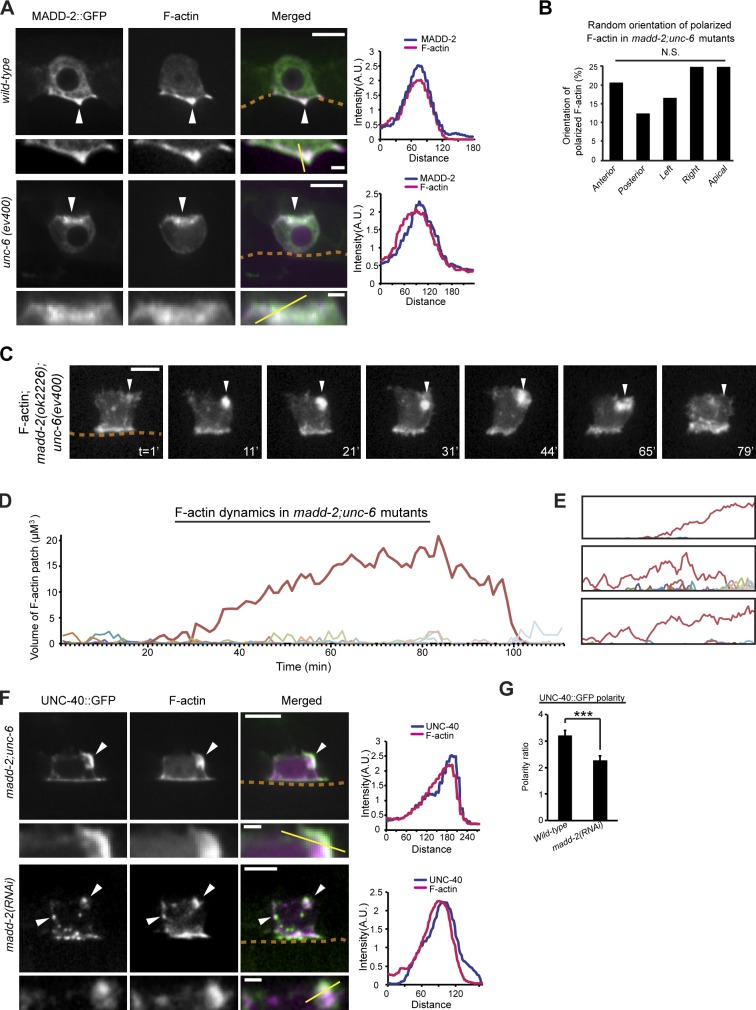
Next, we were interested in understanding how UNC-6–independent UNC-40 clustering is regulated and whether UNC-40 clustering behavior is required for polarization toward UNC-6. One potential candidate is the C. elegans madd-2 gene, which encodes a TRIM protein family member most similar to mammalian TRIM9 and TRIM67 (Hao et al., 2010). The MADD-2 protein binds directly to UNC-40 and promotes UNC-40 function during neuronal outgrowth and muscle arm extension (Alexander et al., 2010; Hao et al., 2010). Unlike other effectors of UNC-40, genetic interaction studies have suggested that MADD-2 does not function downstream of UNC-40, but instead acts upstream or as a cofactor to potentiate UNC-40’s activity in directing guidance responses (Alexander et al., 2010; Hao et al., 2010). Intriguingly, recent studies have shown that madd-2 is expressed in the AC and regulates AC invasion (Morf et al., 2013).
To explore whether MADD-2 might mediate UNC-6–independent UNC-40 clustering, we first examined MADD-2 localization. Consistent with direct regulation of UNC-40, MADD-2::GFP was concentrated at regions of the cell where UNC-40 localizes: at the invasive cell membrane in wild-type animals and with ectopic patches of F-actin in unc-6 mutants (Fig. 7 A). To determine whether MADD-2 regulates UNC-6–independent UNC-40 clustering, we examined F-actin dynamics in unc-6 mutants harboring the putative madd-2–null allele madd-2(ok2226) (Alexander et al., 2010). In madd-2(ok2226); unc-6(ev400) double mutant animals, dominant F-actin clusters still formed and were localized in random regions in the cell (Fig. 7, B and C). Furthermore, the F-actin patches reached a peak volume comparable to that observed in unc-6 mutants and disassembled at a similar rate (Table 1; Fig. 7, C–E). In madd-2(ok2226); unc-6(ev400) mutants, UNC-40 still colocalized with ectopic F-actin clusters (Fig. 7 F). Strikingly, however, madd-2(ok2226); unc-6(ev400) animals had a threefold decrease in the rate of cluster assembly (Fig. 7, C–E; and Table 1). In addition, the F-actin patches had a significantly longer lifetime before disassembly (>80 min versus ∼26 min in unc-6 mutants alone; Table 1). These results indicate that MADD-2 promotes UNC-6–independent UNC-40 cluster assembly. In addition, these observations suggest that the negative feedback mechanism that triggers cluster disassembly is induced at peak UNC-40 clustering.
Figure 7.
MADD-2 promotes UNC-40 clustering and polarization toward UNC-6. Anterior is left; ventral is down. (A) MADD-2::GFP was colocalized with F-actin (visualized with mCherry::moeABD) at the AC’s invasive cell membrane in wild-type animals and in ectopic F-actin patches in unc-6 mutants, locations where UNC-40 resides (arrowheads; the location of basement membrane [BM] is indicated with broken lines; colocalization graphs on the right were measured along the yellow lines in the magnified insets below). (B) The dominant UNC-40–mediated F-actin patch polarized in random sections of the plasma membrane in the ACs of madd-2; unc-6 mutants (P > 0.1, χ2 test, n = 24 animals observed; see Fig. 4 C). (C) F-actin cluster formation (arrowheads) was slower before disassembly in a madd-2; unc-6 mutant (shown over a 79-min time lapse). (D) The volume of F-actin patch formation (red line, dominant patch) in the madd-2; unc-6 animal shown in C. (E) Similar analysis of three additional madd-2; unc-6 mutants. (F) UNC-40::GFP and F-actin colocalized (arrowheads) in madd-2; unc-6 mutants (top) and in animals with RNAi-induced loss of madd-2 (bottom). UNC-40::GFP and F-actin were mispolarized after loss of madd-2 (arrowheads, bottom). (G) Quantification of UNC-40 polarity in wild-type and madd-2 (RNAi)-treated animals (n > 20 for each stage per genotype; ***, P < 0.001, Student’s t test). Bars: (main panels) 5 µm; (magnified insets) 1 µm.
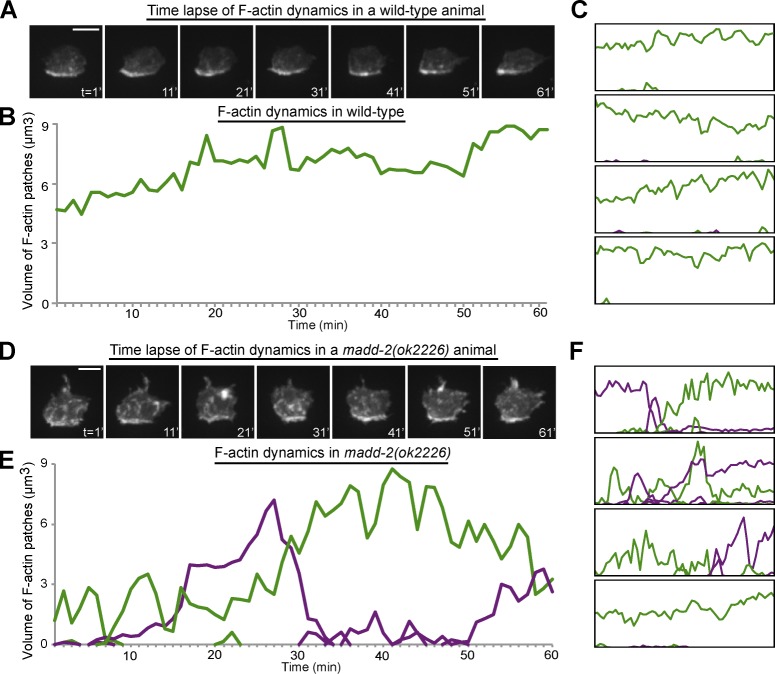
The interplay of positive and negative feedback is important in robust polarization responses in yeast (Howell et al., 2012; Bendezú and Martin, 2013; Dyer et al., 2013; Wu and Lew, 2013). Given that MADD-2 regulates this balance with UNC-40 clustering, we hypothesized that madd-2 mutants may show defects in UNC-40 polarization toward UNC-6. To test this idea, we examined F-actin and UNC-40 polarization toward endogenous UNC-6 in madd-2 mutants. Notably, UNC-40 and F-actin were no longer tightly localized to the invasive cell membrane of the AC in madd-2 mutants (Fig. 7, F and G). Furthermore, in contrast to the robust localization of F-actin along the invasive cell membrane in wild-type animals (n = 10/10 ACs; Fig. 8, A–C; and Video 5), time-lapse analyses revealed that madd-2 mutants often had transient, mislocalized patches of F-actin along the AC’s apical and lateral domains (n = 6/12 animals; Fig. 8, D–F; and Video 5). The specific function of MADD-2 in regulating UNC-6–independent UNC-40 clustering and the perturbation in UNC-40 polarization toward UNC-6 in madd-2 mutants offer compelling evidence that ligand-independent UNC-40 (DCC) clustering is required for robust polarization toward UNC-6 (netrin).
Figure 8.
MADD-2 promotes stable UNC-40–mediated F-actin polarization. Anterior is left; ventral is down. (A) Time series shows F-actin stably polarized at the basal surface of the AC of a wild-type animal. (B) The volume of the stable basal F-actin patch (green line). (C) Similar localization in four other cases. (D) The time series shows multiple mislocalized F-actin patches in the AC of a madd-2 mutant. (E) The volume of individual F-actin patches in a madd-2 mutant. Green lines represent basal patches, and purple lines represent mislocalized patches. (F) Similar analysis of four additional madd-2 mutants. Large mislocalized F-actin patches were observed in 50% of madd-2 mutants (n = 6/12) but were not observed in any wild-type animals (n = 10/10). Isosurface thresholds were set to a stringent level that delineated ectopic and basal patches in madd-2 animals to evaluate the AC’s ability to polarize F-actin at the invasive membrane toward endogenous UNC-6 (see Materials and methods). This thresholding resulted in a reduction of reported F-actin volume compared with previous figures. All animals were examined at the P6.p two-cell stage. Bars, 5 µm.
Discussion
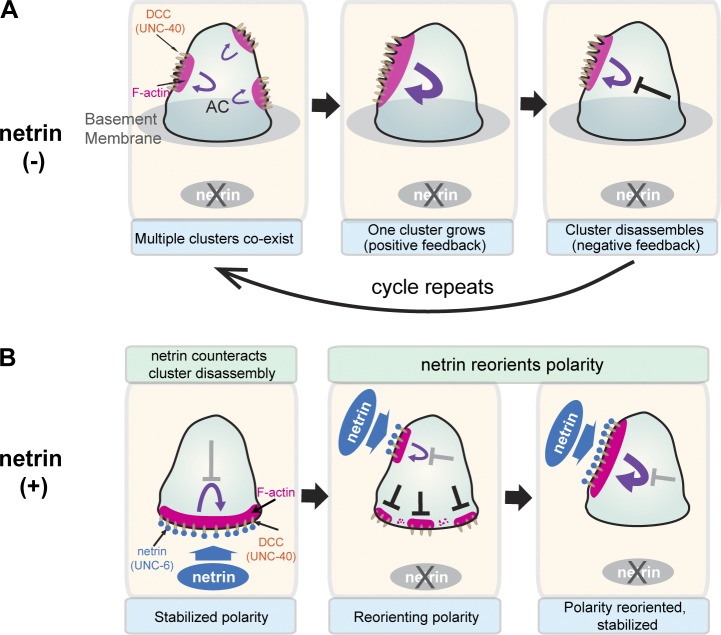
The mechanism by which the receptor UNC-40 (DCC) polarizes toward its ligand UNC-6 (netrin) to mediate diverse guidance functions is poorly understood. Using C. elegans AC invasion as a model for UNC-40/UNC-6 polarization, we have found that in the absence of UNC-6, UNC-40 is active, generates F-actin, and undergoes oscillatory clustering characterized by repeated cycles of cluster assembly and disassembly at different regions of the cell surface. We show that UNC-6 orients and stabilizes UNC-40 clustering, and that the protein MADD-2, a direct regulator of UNC-40, promotes UNC-40 clustering and the ability of UNC-40 to polarize toward UNC-6. Together, our data suggest that UNC-6 (netrin) directs polarized responses by stabilizing the localization of oscillatory clustering of the DCC receptor (Fig. 9).
Figure 9.
A model for UNC-40 (DCC)–mediated polarization toward UNC-6 (netrin). (A) In the absence of UNC-6, UNC-40 (DCC) undergoes oscillations in clustering (polarization). Positive feedback and competition mediates growth and dominance of a single UNC-40/F-actin cluster (middle). Cluster growth then leads to a delayed form of negative feedback (right, black cross) that breaks apart the cluster. After dissipation of negative feedback, the cycle repeats. (B, left) UNC-6 (netrin) in contact with UNC-40 (DCC) at the cell surface counteracts negative feedback, orienting and stabilizing polarity. (middle) Loss of the initial source of netrin leads to breakdown of polarity via negative feedback. (middle, right) A new source of netrin rapidly reorients and stabilizes polarity in a different location by countering negative feedback.
By performing real-time imaging of UNC-40 and its activity (F-actin generation) during AC invasion in C. elegans, we have unexpectedly found that UNC-40 is a required component of a randomly directed polarity system independent of UNC-6. In the absence of UNC-6, UNC-40 clustered in the cell membrane, recruited F-actin effectors, and generated F-actin. UNC-40/F-actin clusters assembled into a single patch, disassembled, and reformed at periodic intervals in different regions of the cell membrane. Animals lacking UNC-40 did not display oscillations in F-actin polarization, demonstrating a direct requirement for UNC-40 in mediating this oscillatory polarity. This dynamic oscillatory behavior indicates that UNC-40 clusters through a positive feedback mechanism that leads to growth and dominance of a single cluster (Howell et al., 2009), followed by delayed negative feedback that disassembles the cluster (Park and Bi, 2007; Brandman and Meyer, 2008; Novák and Tyson, 2008; Tsai et al., 2008). Our data also demonstrated that endogenous and ectopically localized UNC-6 (netrin) polarizes UNC-40 (DCC) by stabilizing UNC-40 clustering to regions of the cell membrane in contact with UNC-6. This stabilization was dependent on the UNC-6–binding domain of UNC-40, which suggests that a direct UNC-6–UNC-40 interaction orients polarization.
UNC-40 polarization toward UNC-6 has similarities to well-characterized cell polarity systems, including Cdc42-mediated polarization in yeast and Ras/Rac/PI3 kinase–mediated neutrophil and D. discoideum chemotaxis, where external cues bias intrinsic, randomly directed internal polarity systems (Sohrmann and Peter, 2003; Maeda et al., 2008; Johnson et al., 2011; Huang et al., 2013). Furthermore, analogous to UNC-40, positive and negative feedback–driven oscillations in clustering of the polarity-regulating GTPase Cdc42 during yeast mating have been observed that are stabilized and oriented toward high levels of pheromone (Bendezú and Martin, 2013; Dyer et al., 2013). In these systems, positive and negative feedback loops are thought to tune the polarized response toward the signal: positive feedback may rapidly amplify signals toward extracellular cues, and negative feedback might limit polarization, regulate competition between independent polarity sites, and allow cells to quickly break down polarity to redirect polarity toward a changing cue source (Brandman and Meyer, 2008; Houk et al., 2012; Dyer et al., 2013; Wu and Lew, 2013). Our observations add weight to the idea that interlinked positive and negative feedback loops are used in many distinct pathways and contexts to facilitate polarization responses toward external cues.
The stability of UNC-40 clusters in contact with UNC-6 indicates that UNC-6–UNC-40 interactions must counteract negative feedback–driven cluster disassembly. UNC-6–UNC-40 interactions may directly counter negative feedback. Alternatively, UNC-6–UNC-40 could overcome cluster disassembly by increasing the positive feedback mechanisms that drive clustering. Importantly, interactions with UNC-6 did not increase the rate of F-actin generation, which suggests that UNC-6–UNC-40 interactions do not stabilize polarity through increasing F-actin formation. UNC-6 opposition to cluster disassembly would likely provide regions of the plasma membrane in contact with UNC-6 with a competitive advantage for rapid positive feedback–driven clustering. Negative feedback might also contribute to robust polarization by preventing UNC-40 clustering from overexpanding, as well as dismantling UNC-40 after loss of contact with UNC-6 (Wang, 2009; Howell et al., 2012; Wu and Lew, 2013). Supporting the idea that the positive and negative feedback dynamics observed are important for polarizing UNC-40, loss of the C. elegans protein MADD-2, which is similar to the mammalian TRIM9 and TRIM67 proteins (Alexander et al., 2010; Hao et al., 2010), altered feedback dynamics and perturbed the ability of UNC-40 to polarize toward UNC-6. Together, these observations suggest a model of how UNC-6 (netrin) orients UNC-40 (DCC): by counteracting the disassembly of UNC-40 clustering, UNC-6 stabilizes randomly localized UNC-40 clustering to regions of the cell membrane in contact with UNC-6, which are rapidly amplified through positive feedback. Negative feedback might also facilitate reorientation of polarity toward changing sources of UNC-6 (netrin; summarized in Fig. 9).
Because of the challenges of experimental examination, the underlying mechanisms of positive and negative feedback loops in many dynamic polarity systems are unclear (Brandman and Meyer, 2008; Wang, 2009; Wu and Lew, 2013). Our data suggest that the UNC-40–interacting protein MADD-2 promotes positive feedback–mediated UNC-40 clustering: in a madd-2 mutant, there was a specific threefold decrease in the rate of cluster formation. In addition to UNC-40, MADD-2 also directly interacts with the Rac/Rho GEF UNC-73/Trio, which has previously been implicated in UNC-40 trafficking to the cell surface (Levy-Strumpf and Culotti, 2007; Watari-Goshima et al., 2007). This suggests that MADD-2 might promote positive feedback by directing UNC-40 trafficking to the cell surface. Alternatively, MADD-2 might dismantle competing UNC-40 clusters, which would promote assembly of a single dominant cluster by freeing UNC-40. Notably, MADD-2 has a RING finger domain that may play a role in ubiquitination (Song et al., 2011). Thus, it is also possible that MADD-2 could promote cluster assembly by targeting an inhibitor of UNC-40 clustering for degradation. Our results also suggest that the mechanism of negative feedback is triggered at high levels of UNC-40/F-actin clustering activity: in madd-2 mutants, which show significantly slower clustering, initiation of negative feedback was delayed until normal peak levels of clustering occurred. Although the mechanism of negative feedback in cluster disassembly is unknown, induction of negative feedback at high levels of signal is a common design principle that interlinks positive and negative feedback loops (Brandman and Meyer, 2008).
The dynamic localization and activity of UNC-40 (DCC) in the absence of UNC-6 (netrin) in other in vivo contexts has not been determined. Importantly, in the HSN neuron, static imaging has revealed that the UNC-40 receptor also clusters in the absence of UNC-6. This clustering, however, has only been observed in the UNC-40 variant UNC-40 (A1056V) and in animals lacking UNC-53 (NAV2), a conserved cytoskeletal binding protein (Kulkarni et al., 2013). It is thus possible that positive and negative feedback is operating in the HSN neuron on UNC-40 and that UNC-40 (A1056V) and UNC-53 alter these feedback mechanisms such that more stable UNC-40 clusters can be detected. Positive and negative feedback might be differently tuned in the HSN neuron to provide a cell type–specific polarity response (Novák and Tyson, 2008; Howell et al., 2012), and thus oscillations in UNC-40 cluster formation might be absent or more difficult to detect. Given that real-time imaging techniques have not yet been developed in the HSN neuron (Xu et al., 2009; Kulkarni et al., 2013), it is also possible that oscillations in UNC-40 clustering are present but have yet to be resolved. Nevertheless, the striking parallels observed in UNC-40 clustering in the HSN neuron and the AC, the shared role of UNC-6 in localizing UNC-40, and a function for MADD-2 in promoting UNC-40 polarization in both cell types (Kulkarni et al., 2013; Xu et al., 2009; Ziel et al., 2009) support the idea that similar mechanisms underlie the polarization of UNC-40 toward UNC-6 in diverse cellular contexts.
Materials and methods
Worm handling and strains
Worms were reared under standard conditions at 15°C, 20°C, or 25°C (Brenner, 1974). N2 Bristol strain was used as the wild type. Strains were reared and viewed at 20°C or 25°C using standard techniques. In the text and figures, we use a “>” symbol for linkages to a promoter and a “::” symbol for linkages that fuse open reading frames. The following alleles and transgenes were used: qyEx257 [zmp-5 > unc-6::nlg-1 TM::GFP], qyIs57 [cdh-3 > mCherry::moeABD], qyIs61 [cdh-3 > GFP::unc-34], qyIs67 [cdh-3 > unc-40::GFP], qyIs68 [cdh-3 > unc-40::GFP], qyIs155 [cdh-3 > unc-40(ΔFN4/5)::GFP], qyIs182 [cdh-3 > GFP::unc-115], qyIs220 [cdh-3 > GFP::mig-2], qyIs221 [cdh-3 > GFP::ced-10], qyIs262 [cdh-3 > unc-40::mCherry], trIs31 [madd-2 > madd-2::GFP]. LGI, unc-40(e271), unc-40(n324), unc-40(n473), madd-4(ok2854). LGIV, qyIs10 [laminin::GFP], unc-129(ev554). LGV, qyIs50 [cdh-3 > mCherry::moeABD], madd-2(ok2226). LGX, unc-6(ev400), slt-1(eh15).
Microscopy, image acquisition, and image processing
Confocal images were acquired using a microscope (Axio Imager; Carl Zeiss) with a 100× Plan-Apochromat objective lens (1.4 NA) equipped with an EM charge-coupled device (CCD) or ORCA-R2 CCD camera (Hamamatsu Photonics) and a spinning disc confocal scan head (CSU-10; Yokogawa Electric Corporation) controlled by iVision software (Biovision Technologies) or Micro-Manager software (Edelstein et al., 2010). The point-spread function (PSF) of imaging GFP fluorescence on the confocal microscopes used was calculated and well within the acceptable range. Compound light and fluorescent microscopy images were acquired using a microscope (AxioImager; Carl Zeiss) with a 100× Plan-Apochromat objective lens (1.4 NA) and with a CCD camera (AxioCam MRm; Carl Zeiss) controlled by Axiovision software (Carl Zeiss). Acquired images were processed using ImageJ 1.40 and Photoshop CS6 Extended software (Adobe Systems Inc.). 3D reconstructions were built from confocal z stacks, analyzed, and exported using Imaris 7 (Bitplane). Movies were annotated using Photoshop software (Adobe Systems Inc.). Graphs and figures were built using Illustrator software (CS6 Extended; Adobe Systems Inc.).
Quantification of AC polarity
Quantitative measurements of AC polarity were determined as the ratio of the mean fluorescence intensity from a five-pixel-wide line drawn along the invasive (basal) versus the noninvasive (apical and lateral) membranes of the AC using ImageJ 1.40.
RNAi
Double-stranded RNA (dsRNA)-mediated gene interference (RNAi) was performed by feeding larvae with bacteria expressing dsRNA using standard procedures (Kamath et al., 2001). To bypass the embryonic lethality of pat-3 RNAi, synchronized L1-arrested larvae of unc-40(e271);qyIs50 were grown for 7 h on regular OP50 bacteria at 20°C before being transferred to pat-3 RNAi plates.
Quantitative analysis of F-actin volume and time-lapse imaging
F-actin volume was measured by collecting confocal z stacks of F-actin networks in ACs expressing the F-actin binding probe, mCherry::moeABD. Imaris 7.4 was used to build 3D reconstructions of F-actin in the AC using the “isosurface rendering” function of Imaris. Isosurface renderings of mCherry::moeABD were created, setting a threshold that outlined the dense F-actin network at the invasive membrane in wild-type ACs. This same threshold was used in unc-40, unc-6, and madd-2; unc-6 mutants. Quantitative measurements were then made for the volume of fluorescent intensity with these isosurface renderings (Hagedorn et al., 2009).
Isosurface renderings for mCherry::MoeABD in madd-2 mutants and wild-type animals shown in Fig. 8 were created with more stringent thresholds set to delineate ectopic and basal F-actin patches in madd-2 animals. This was done to evaluate F-actin polarization at the invasive membrane in the presence of endogenous UNC-6. Stringent thresholding resulted in a reduction in reported F-actin volume (compared with the the preceding paragraph). The madd-2 and wild-type dataset was smoothed with a 0.11 µm Gaussian filter and processed in Imaris 7.6.5.
To quantify F-actin localization in unc-6 and unc-6; madd-2 mutants, five portions of the AC’s apical and lateral membranes were used for scoring localization. The plasma membrane covering the top third of the AC was considered “apical”; the membrane covering the anterior or posterior third of the AC was the “anterior” or “posterior”; the lateral membrane between the anterior and posterior portions was designated “left” or “right.” An F-actin patch was considered “dominant” only when its volume accounted for >80% of the total F-actin at the apical and lateral membranes.
For time-lapse imaging, worms were anesthetized using 0.2% tricaine (E10521; Sigma-Aldrich) and 0.02% levamisole (196142; Sigma-Aldrich) in M9 buffer and gently transferred onto a 5% noble agar pad solidified on a glass slide. The slide was then covered with a 22 × 22-mm glass coverslip. Valap was applied to the four sides of the coverslip to minimize liquid evaporation. Worms at the P6.p two-cell stage were used for time-lapse imaging. The duration of time-lapse imaging ranged from 60 to 120 min. Confocal z stacks were taken at 1-min intervals. The following parameters were set for imaging: binning 2 by 2, image size 1,344 pixels by 1,024 pixels, 1 µm z slice interval (7–8 slices per stack), an exposure time of 200 ms for F-actin and UNC-40::GFP, and an exposure time of 150 ms for UNC-6::NLG-1 TM::GFP. Time-lapse imaging of madd-2 mutants and wild-type animals in Fig. 8 was acquired with the following parameters: binning 1 by 1, image size 512 pixels by 512 pixels, 1 µm z slice interval (9–12 slices each stack), and exposure time 300 ms for F-actin. Images from time-lapse analysis were reconstructed in Imaris 7.4 or 7.6 for 4D analysis. Drifts during image acquisition were corrected using the “correct drift” function of Imaris. Isosurface renderings of F-actin were performed similarly to the F-actin volume measurement procedure. Each isosurface was individually tracked for F-actin dynamics analysis. The fluorescence intensity of UNC-40 clusters and F-actin patches was measured using ImageJ.
Quantitative analysis of F-actin patch association
Association analysis of F-actin patches with those of UNC-40 and UNC-40 effectors in unc-6 mutants was performed using the “spot tracking” function of Imaris. In brief, a confocal z stack of an AC expressing both an F-actin mCherry probe and a GFP fusion protein (UNC-40::GFP, UNC-40(ΔFN4/5)::GFP, GFP::UNC-34, GFP::CED-10, GFP::MIG-2, or GFP::UNC-115) was acquired and then reconstructed into a 3D image. A threshold was determined for each AC such that fluorescent patches were assigned with 3D spots (radius of 350 nm). Two adjacent spots were considered to be “associated” if they were in contact or overlapped.
Molecular biology and transgenic strains
Standard techniques were used to generate PCR fusion products (Hobert, 2002), plasmids, and transgenic animals (Mello and Fire, 1995). Templates and specific PCR primers for promoters and genes, and transgenic extrachromosomal (Ex) lines and integrated strains (Is) generated in this study are listed in Tables S1 and S2. For cdh-3 > unc-40(ΔFN4/5)::GFP, the amplicon containing unc-40 cDNA with a deletion of the fourth and fifth FNIII domains (amino acids 738–939 of UNC-40 protein, the putative UNC-6 binding site) in frame to GFP sequence was amplified from the unc-40::GFP expression vector described previously (Ziel et al., 2009). This amplicon unc-40(ΔFN4/5)::GFP was then fused using PCR with the cdh-3 promoter. To express membrane-tethered UNC-6 in dorsal uterine cells (zmp-5 > unc-6::nlg-1 TM::GFP), we first generated the expression vector that produced UNC-6 protein fused with the transmembrane (TM) domain of NLG-1 in frame to GFP. The promoter of the zmp-5 gene (∼8.7kb) was cut from the fosmid WRM066bb10 via restriction enzyme digestion at BamHI (8,641 bp upstream of H18L16.1 ATG start codon) and KpnI (22 bp downstream of H18L16.1 ATG start codon) sites. This promoter was subcloned into pPD95.75 (GFP vector) at BamHI and KpnI sites to generate the zmp-5 > GFP expression vector. The sequence encoding UNC-6::NLG-1 TM amplified from the plasmid DR99 (unc-6 > unc-6::nlg-1 TM::mCherry, obtained from K. Shen, Stanford University, Palo Alto, CA) was inserted after the zmp-5 promoter and in frame to the GFP sequence at SnaBI (65 bp upstream of KpnI) and KpnI sites of the zmp-5 > GFP expression vector. For cdh-3 > unc-40::mCherry, unc-40 cDNA was subcloned into the plasmid pBS-mCherry (pBluescript II vector with mCherry inserted) at HindIII and XmaI sites in frame to mCherry. The sequence of unc-40::mCherry was amplified and fused using PCR with the cdh-3 promoter.
Transgenic worms were created by coinjection of expression constructs with the transformation marker pPD#MM016B (unc-119+) alone, or with the coinjection marker (myo-2 > GFP) into the germline of unc-119(ed4) mutants. These markers were injected with EcoRI-digested salmon sperm DNA and pBluescript II at 50 ng/µl as carrier DNA along with the expression constructs, which were injected at 10–50 ng/µl. Integrated strains were generated as described previously (Inoue et al., 2002).
Generation and isolation of the deletion mutant unc-40(qy2)
The unc-40 deletion mutant was generated using a standard 4,5′,8-trimethylpsoralen (TMP)/UV light mutagenesis protocol as described previously (Jansen et al., 1997, 1999; Liu et al., 1999). In brief, we screened 30,000 wild-type (N2) genomes using TMP/UV mutagenesis. A “poison primer”–based PCR strategy (Edgley et al., 2002) was used to detect and isolate deletion mutants. Four regions of the unc-40 genomic locus (11,075 bp ranging from ATG start codon to TAA stop codon: region I, 4,129–5,345 bp; region II, 5,301–6,694 bp; region III, 6,654–8,226 bp; region IV, 8,184–9,554 bp) were selected for PCR-based deletion detection. For each region, three sets of primers, termed “outer primers,” “poison primers,” and “inner primers,” were designed (Table S3). The outer and poison primers were used in the first PCR round. The inner primers were used in the second round. A deletion in the unc-40 genomic sequence was identified and assigned with the allele name qy2. This deletion removing 711 bp in the unc-40 genomic sequence (7,202–7,912 bp; see Fig. S1) is predicted to cause a frame shift, forming a premature stop codon in the exon immediately after the deletion region. The protein encoded by this allele is predicted to have amino acids 709–1,031 deleted and replaced with 50 amino acids encoded by the shifted frame. The TM and intracellular domains are truncated.
Statistics
Statistical analysis was performed using Student’s t tests or χ2 tests as indicated in the text. The coefficients of determinations (R2) were calculated using JMP version 9.0 (SAS Institute) or Excel software (Microsoft).
Online supplemental material
Fig. S1 contains schematic diagrams of unc-40 mutant alleles. Fig. S2 shows that integrin mediates unc-40–independent polarization and F-actin formation at the invasive cell membrane. Fig. S3 shows that SLT-1, MADD-4, and UNC-129 do not activate UNC-40. Fig. S4 shows that F-actin is stably polarized in unc-40 mutants. Video 1 shows time-lapse analysis of F-actin dynamics in wild-type, unc-40, and unc-6 mutant ACs. Video 2 shows that F-actin colocalizes with UNC-40 in unc-6 mutants. Video 3 shows that dorsally localized UNC-6 orients and stabilizes UNC-40–mediated F-actin clustering in unc-6 mutants. Video 4 shows that UNC-40 reorients F-actin toward a changing source of UNC-6. Video 5 shows that MADD-2 promotes UNC-40–mediated F-actin polarization toward UNC-6. Table S1 contains primer sequences for PCR fusions and plasmid constructs. Table S2 describes extrachromosomal arrays and integrated strains. Table S3 contains primers used to screen for the unc-40(qy2) deletion mutant. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201405026/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201405026.dv.
Supplementary Material
Acknowledgments
We are grateful to K. Shen for the unc-6 > unc-6::nlg-1 TM::mCherry vector; W. Wadsworth, R. Li and D. Lew for discussions; and L. Kelley, L. Lilley, M. Morrissey, A. Schindler, and W. Zou for comments on the manuscript.
Some strains were provided by the Caenorhabditis Genetics Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by a Pew Scholars Award, and National Institutes of Health grant GM100083 to D.R. Sherwood.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AC
- anchor cell
- DCC
- deleted in colorectal cancer
- HSN
- hermaphrodite-specific neuron
- TM
- transmembrane
References
- Adler, C.E., Fetter R.D., and Bargmann C.I.. 2006. UNC-6/Netrin induces neuronal asymmetry and defines the site of axon formation. Nat. Neurosci. 9:511–518 10.1038/nn1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, M., Selman G., Seetharaman A., Chan K.K., D’Souza S.A., Byrne A.B., and Roy P.J.. 2010. MADD-2, a homolog of the Opitz syndrome protein MID1, regulates guidance to the midline through UNC-40 in Caenorhabditis elegans. Dev. Cell. 18:961–972 10.1016/j.devcel.2010.05.016 [DOI] [PubMed] [Google Scholar]
- Arai, Y., Shibata T., Matsuoka S., Sato M.J., Yanagida T., and Ueda M.. 2010. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc. Natl. Acad. Sci. USA. 107:12399–12404 10.1073/pnas.0908278107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkowitz, R.A.2013. Cell polarity: wanderful exploration in yeast sex. Curr. Biol. 23:R10–R12 10.1016/j.cub.2012.11.037 [DOI] [PubMed] [Google Scholar]
- Arrieumerlou, C., and Meyer T.. 2005. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev. Cell. 8:215–227 10.1016/j.devcel.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Asakura, T., Ogura K., and Goshima Y.. 2007. UNC-6 expression by the vulval precursor cells of Caenorhabditis elegans is required for the complex axon guidance of the HSN neurons. Dev. Biol. 304:800–810 10.1016/j.ydbio.2007.01.028 [DOI] [PubMed] [Google Scholar]
- Bendezú, F.O., and Martin S.G.. 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 23:42–47 10.1016/j.cub.2012.10.042 [DOI] [PubMed] [Google Scholar]
- Brandman, O., and Meyer T.. 2008. Feedback loops shape cellular signals in space and time. Science. 322:390–395 10.1126/science.1160617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S.1974. The genetics of Caenorhabditis elegans. Genetics. 77:71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S.S., Zheng H., Su M.W., Wilk R., Killeen M.T., Hedgecock E.M., and Culotti J.G.. 1996. UNC-40, a C. elegans homolog of DCC (Deleted in Colorectal Cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 87:187–195 10.1016/S0092-8674(00)81337-9 [DOI] [PubMed] [Google Scholar]
- Colón-Ramos, D.A., Margeta M.A., and Shen K.. 2007. Glia promote local synaptogenesis through UNC-6 (netrin) signaling in C. elegans. Science. 318:103–106 10.1126/science.1143762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer, J.M., Savage N.S., Jin M., Zyla T.R., Elston T.C., and Lew D.J.. 2013. Tracking shallow chemical gradients by actin-driven wandering of the polarization site. Curr. Biol. 23:32–41 10.1016/j.cub.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein, A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. Chapter 14:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgley, M., D’Souza A., Moulder G., McKay S., Shen B., Gilchrist E., Moerman D., and Barstead R.. 2002. Improved detection of small deletions in complex pools of DNA. Nucleic Acids Res. 30:e52 10.1093/nar/gnf051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht, B.V., Dowd K.A., Barfield R.W., Longo P.A., and Leahy D.J.. 2003. Netrin binds discrete subdomains of DCC and UNC5 and mediates interactions between DCC and heparin. J. Biol. Chem. 278:32561–32568 10.1074/jbc.M302943200 [DOI] [PubMed] [Google Scholar]
- Gitai, Z., Yu T.W., Lundquist E.A., Tessier-Lavigne M., and Bargmann C.I.. 2003. The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron. 37:53–65 10.1016/S0896-6273(02)01149-2 [DOI] [PubMed] [Google Scholar]
- Hagedorn, E.J., and Sherwood D.R.. 2011. Cell invasion through basement membrane: the anchor cell breaches the barrier. Curr. Opin. Cell Biol. 23:589–596 10.1016/j.ceb.2011.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn, E.J., Yashiro H., Ziel J.W., Ihara S., Wang Z., and Sherwood D.R.. 2009. Integrin acts upstream of netrin signaling to regulate formation of the anchor cell’s invasive membrane in C. elegans. Dev. Cell. 17:187–198 10.1016/j.devcel.2009.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagedorn, E.J., Ziel J.W., Morrissey M.A., Linden L.M., Wang Z., Chi Q., Johnson S.A., and Sherwood D.R.. 2013. The netrin receptor DCC focuses invadopodia-driven basement membrane transmigration in vivo. J. Cell Biol. 201:903–913 10.1083/jcb.201301091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao, J.C., Adler C.E., Mebane L., Gertler F.B., Bargmann C.I., and Tessier-Lavigne M.. 2010. The tripartite motif protein MADD-2 functions with the receptor UNC-40 (DCC) in Netrin-mediated axon attraction and branching. Dev. Cell. 18:950–960 10.1016/j.devcel.2010.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert, O.2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 32:728–730 [DOI] [PubMed] [Google Scholar]
- Houk, A.R., Jilkine A., Mejean C.O., Boltyanskiy R., Dufresne E.R., Angenent S.B., Altschuler S.J., Wu L.F., and Weiner O.D.. 2012. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 148:175–188 10.1016/j.cell.2011.10.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, A.S., Savage N.S., Johnson S.A., Bose I., Wagner A.W., Zyla T.R., Nijhout H.F., Reed M.C., Goryachev A.B., and Lew D.J.. 2009. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 139:731–743 10.1016/j.cell.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, A.S., Jin M., Wu C.F., Zyla T.R., Elston T.C., and Lew D.J.. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 149:322–333 10.1016/j.cell.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C.H., Tang M., Shi C., Iglesias P.A., and Devreotes P.N.. 2013. An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration. Nat. Cell Biol. 15:1307–1316 10.1038/ncb2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara, S., Hagedorn E.J., Morrissey M.A., Chi Q., Motegi F., Kramer J.M., and Sherwood D.R.. 2011. Basement membrane sliding and targeted adhesion remodels tissue boundaries during uterine-vulval attachment in Caenorhabditis elegans. Nat. Cell Biol. 13:641–651 10.1038/ncb2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, T., Sherwood D.R., Aspöck G., Butler J.A., Gupta B.P., Kirouac M., Wang M., Lee P.-Y., Kramer J.M., Hope I., et al. 2002. Gene expression markers for Caenorhabditis elegans vulval cells. Mech. Dev. 119(Suppl 1):S203–S209 10.1016/S0925-4773(03)00117-5 [DOI] [PubMed] [Google Scholar]
- Jansen, G., Hazendonk E., Thijssen K.L., and Plasterk R.H.A.. 1997. Reverse genetics by chemical mutagenesis in Caenorhabditis elegans. Nat. Genet. 17:119–121 10.1038/ng0997-119 [DOI] [PubMed] [Google Scholar]
- Jansen, G., Thijssen K.L., Werner P., van der Horst M., Hazendonk E., and Plasterk R.H.A.. 1999. The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat. Genet. 21:414–419 10.1038/7753 [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Jin M., and Lew D.J.. 2011. Symmetry breaking and the establishment of cell polarity in budding yeast. Curr. Opin. Genet. Dev. 21:740–746 10.1016/j.gde.2011.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., Martinez-Campos M., Zipperlen P., Fraser A.G., and Ahringer J.. 2001. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2: research0002–research0002.10 10.1186/gb-2000-2-1-research0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keino-Masu, K., Masu M., Hinck L., Leonardo E.D., Chan S.S., Culotti J.G., and Tessier-Lavigne M.. 1996. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 87:175–185 10.1016/S0092-8674(00)81336-7 [DOI] [PubMed] [Google Scholar]
- Kelley, L.C., Lohmer L.L., Hagedorn E.J., and Sherwood D.R.. 2014. Traversing the basement membrane in vivo: a diversity of strategies. J. Cell Biol. 204:291–302 10.1083/jcb.201311112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger, R.P., Lee J., Li W., and Guan K.L.. 2004. Mapping netrin receptor binding reveals domains of Unc5 regulating its tyrosine phosphorylation. J. Neurosci. 24:10826–10834 10.1523/JNEUROSCI.3715-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, G., Xu Z., Mohamed A.M., Li H., Tang X., Limerick G., and Wadsworth W.G.. 2013. Experimental evidence for UNC-6 (netrin) axon guidance by stochastic fluctuations of intracellular UNC-40 (DCC) outgrowth activity. Biol. Open. 2:1300–1312 10.1242/bio.20136346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Wing Sun, K., Correia J.P., and Kennedy T.E.. 2011. Netrins: versatile extracellular cues with diverse functions. Development. 138:2153–2169 10.1242/dev.044529 [DOI] [PubMed] [Google Scholar]
- Levy-Strumpf, N., and Culotti J.G.. 2007. VAB-8, UNC-73 and MIG-2 regulate axon polarity and cell migration functions of UNC-40 in C. elegans. Nat. Neurosci. 10:161–168 10.1038/nn1835 [DOI] [PubMed] [Google Scholar]
- Liu, L.X., Spoerke J.M., Mulligan E.L., Chen J., Reardon B., Westlund B., Sun L., Abel K., Armstrong B., Hardiman G., et al. 1999. High-throughput isolation of Caenorhabditis elegans deletion mutants. Genome Res. 9:859–867 10.1101/gr.9.9.859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeil, L.T., Hardy W.R., Pawson T., Wrana J.L., and Culotti J.G.. 2009. UNC-129 regulates the balance between UNC-40 dependent and independent UNC-5 signaling pathways. Nat. Neurosci. 12:150–155 10.1038/nn.2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, Y.T., Inose J., Matsuo M.Y., Iwaya S., and Sano M.. 2008. Ordered patterns of cell shape and orientational correlation during spontaneous cell migration. PLoS ONE. 3:e3734 10.1371/journal.pone.0003734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello, C., and Fire A.. 1995. DNA transformation. Methods Cell Biol. 48:451–482 10.1016/S0091-679X(08)61399-0 [DOI] [PubMed] [Google Scholar]
- Morf, M.K., Rimann I., Alexander M., Roy P., and Hajnal A.. 2013. The Caenorhabditis elegans homolog of the Opitz syndrome gene, madd-2/Mid1, regulates anchor cell invasion during vulval development. Dev. Biol. 374:108–114 10.1016/j.ydbio.2012.11.019 [DOI] [PubMed] [Google Scholar]
- Nelson, J.C., and Colón-Ramos D.A.. 2013. Serotonergic neurosecretory synapse targeting is controlled by netrin-releasing guidepost neurons in Caenorhabditis elegans. J. Neurosci. 33:1366–1376 10.1523/JNEUROSCI.3471-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák, B., and Tyson J.J.. 2008. Design principles of biochemical oscillators. Nat. Rev. Mol. Cell Biol. 9:981–991 10.1038/nrm2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak, E.M., Becskei A., and van Oudenaarden A.. 2005. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell. 9:565–571 10.1016/j.devcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Park, H.O., and Bi E.. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 10.1128/MMBR.00028-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, A.T., Janetopoulos C., Lee S., Charest P.G., Takeda K., Sundheimer L.W., Meili R., Devreotes P.N., and Firtel R.A.. 2007. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J. Cell Biol. 178:185–191 10.1083/jcb.200611138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman, A., Selman G., Puckrin R., Barbier L., Wong E., D’Souza S.A., and Roy P.J.. 2011. MADD-4 is a secreted cue required for midline-oriented guidance in Caenorhabditis elegans. Dev. Cell. 21:669–680 10.1016/j.devcel.2011.07.020 [DOI] [PubMed] [Google Scholar]
- Sherwood, D.R., and Sternberg P.W.. 2003. Anchor cell invasion into the vulval epithelium in C. elegans. Dev. Cell. 5:21–31 10.1016/S1534-5807(03)00168-0 [DOI] [PubMed] [Google Scholar]
- Sohrmann, M., and Peter M.. 2003. Polarizing without a c(l)ue. Trends Cell Biol. 13:526–533 10.1016/j.tcb.2003.08.006 [DOI] [PubMed] [Google Scholar]
- Song, S., Ge Q., Wang J., Chen H., Tang S., Bi J., Li X., Xie Q., and Huang X.. 2011. TRIM-9 functions in the UNC-6/UNC-40 pathway to regulate ventral guidance. J. Genet. Genomics. 38:1–11 10.1016/j.jcg.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Stein, E., and Tessier-Lavigne M.. 2001. Hierarchical organization of guidance receptors: silencing of netrin attraction by slit through a Robo/DCC receptor complex. Science. 291:1928–1938 10.1126/science.1058445 [DOI] [PubMed] [Google Scholar]
- Tsai, T.Y., Choi Y.S., Ma W., Pomerening J.R., Tang C., and Ferrell J.E. Jr. 2008. Robust, tunable biological oscillations from interlinked positive and negative feedback loops. Science. 321:126–129 10.1126/science.1156951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth, W.G., Bhatt H., and Hedgecock E.M.. 1996. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 16:35–46 10.1016/S0896-6273(00)80021-5 [DOI] [PubMed] [Google Scholar]
- Wang, F.2009. The signaling mechanisms underlying cell polarity and chemotaxis. Cold Spring Harb. Perspect. Biol. 1:a002980 10.1101/cshperspect.a002980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Chi Q., and Sherwood D.R.. 2014. MIG-10 (lamellipodin) has netrin-independent functions and is a FOS-1A transcriptional target during anchor cell invasion in C. elegans. Development. 141:1342–1353 10.1242/dev.102434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watari-Goshima, N., Ogura K., Wolf F.W., Goshima Y., and Garriga G.. 2007. C. elegans VAB-8 and UNC-73 regulate the SAX-3 receptor to direct cell and growth-cone migrations. Nat. Neurosci. 10:169–176 10.1038/nn1834 [DOI] [PubMed] [Google Scholar]
- Wu, C.F., and Lew D.J.. 2013. Beyond symmetry-breaking: competition and negative feedback in GTPase regulation. Trends Cell Biol. 23:476–483 10.1016/j.tcb.2013.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Z., Li H., and Wadsworth W.G.. 2009. The roles of multiple UNC-40 (DCC) receptor-mediated signals in determining neuronal asymmetry induced by the UNC-6 (netrin) ligand. Genetics. 183:941–949 10.1534/genetics.109.108654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, T.W., Hao J.C., Lim W., Tessier-Lavigne M., and Bargmann C.I.. 2002. Shared receptors in axon guidance: SAX-3/Robo signals via UNC-34/Enabled and a Netrin-independent UNC-40/DCC function. Nat. Neurosci. 5:1147–1154 10.1038/nn956 [DOI] [PubMed] [Google Scholar]
- Ziel, J.W., and Sherwood D.R.. 2010. Roles for netrin signaling outside of axon guidance: a view from the worm. Dev. Dyn. 239:1296–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziel, J.W., Hagedorn E.J., Audhya A., and Sherwood D.R.. 2009. UNC-6 (netrin) orients the invasive membrane of the anchor cell in C. elegans. Nat. Cell Biol. 11:183–189 10.1038/ncb1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.