The nucleotide excision repair preincision complex assembles on chromatin even in the absence of DNA damage through the sequential and ordered addition of its individual subunits.
Abstract
In nucleotide excision repair (NER), damage recognition by XPC-hHR23b is described as a critical step in the formation of the preincision complex (PInC) further composed of TFIIH, XPA, RPA, XPG, and ERCC1-XPF. To obtain new molecular insights into the assembly of the PInC, we analyzed its formation independently of DNA damage by using the lactose operator/repressor reporter system. We observed a sequential and ordered self-assembly of the PInC operating upon immobilization of individual NER factors on undamaged chromatin and mimicking that functioning on a bona fide NER substrate. We also revealed that the recruitment of the TFIIH subunit TTDA, involved in trichothiodystrophy group A disorder (TTD-A), was key in the completion of the PInC. TTDA recruits XPA through its first 15 amino acids, depleted in some TTD-A patients. More generally, these results show that proteins forming large nuclear complexes can be recruited sequentially on chromatin in the absence of their natural DNA target and with no reciprocity in their recruitment.
Introduction
One of the most versatile mammalian DNA repair pathways is nucleotide excision repair (NER) in which repair factors assemble sequentially on DNA damage. First, XPC-hHR23b locates DNA injuries in a critical step initiating the formation of the preincision complex (PInC). Subsequently, XPC-hHR23b recruits the transcription/repair factor TFIIH (Sugasawa et al., 1998) containing ten subunits (XPB, XPD, p62, p52, p44, p34, cdk7, cyclH, MAT1, and TTDA; Compe and Egly, 2012). Some of them are not always strictly part of the complex (Zhovmer et al., 2010). For instance, TTDA is more strongly associated with TFIIH after induction of NER-specific DNA lesions (Giglia-Mari et al., 2006). Loading of TFIIH promotes the unwinding of the damaged DNA that provides a three-dimensional structure able to recruit XPA and RPA (Oksenych et al., 2009). The edges of the DNA bubble created by the TFIIH helicases are recognized by the two junction-specific endonucleases XPG and ERCC1-XPF. Altogether, XPC, TFIIH, XPA, RPA, XPG, and ERCC1-XPF form the PInC that generates 3′ and 5′ single DNA incisions relative to the damage (O’Donovan et al., 1994; Sijbers et al., 1996). Excision of a 24–32-mer damaged oligonucleotide precedes the gap-filling DNA resynthesis step (Ogi et al., 2010). Although XPC has a high affinity for several NER substrates such as the UV light–induced pyrimidine–pyrimidone (6–4) photoproducts, its binding to some of them can be weak and require the DDB1–DDB2(XPE) complex (Tang et al., 2000). Mutations in genes coding for XPA, XPB, XPC, XPD, DDB2, XPF, XPG, and TTDA give rise to either xeroderma pigmentosum (XP), trichothiodystrophy (TTD), or a combined XP and Cockayne syndrome. These inherited human disorders exhibit a broad spectrum of clinical features including a common photosensitivity of the skin (Lehmann, 2003).
In the current NER model, the recognition of the DNA lesion by XPC-hHR23b nucleates the formation of the PInC (Sugasawa et al., 1998). However, the participation of NER factors in other cellular processes that do not involve DNA lesions such as transcription (Fong et al., 2013) raised the question of the role of DNA injuries in the formation of the PInC.
To obtain molecular insights into the assembly of the NER factors, we analyzed the formation of the PInC independently of the presence of DNA damage using the lactose operator (LacO)/lactose repressor (LacR) reporter system that allows stable targeting of individual NER factors to a defined chromosomal locus in vivo (Tumbar et al., 1999; Soutoglou and Misteli, 2008). To our surprise, we found that the formation of the PInC in the absence of DNA damage takes place in a sequential and ordered manner mimicking that of the NER reaction on a bona fide DNA substrate. Depending on the tethered protein, in some cases the complex formation is abortive because of competition between upstream and downstream partners of the given NER factor. For instance, interaction of XPC with DDB2 or TFIIH is mutually exclusive because DDB2 and TFIIH share the same binding site on XPC. Consequently, a complex with the first three factors of the NER reaction (XPE, XPC, and TFIIH) cannot exist on chromatin. In contrast, tethering the smallest polypeptide of the reaction (TTDA) triggers the completion of the PInC. However, introduction of a mutation found in TTD-A patients, deficient in NER, impairs the recruitment of the immediate downstream factor XPA. Using in vitro NER assay we show that TTDA recruits XPA to damaged DNA through a direct interaction and that TTD-A mutation impairs it, explaining at the molecular level the NER deficiency of the patients. Our observations highlight that the LacO/LacR is a genuine tool to study complex molecular reactions in a pathological situation. More generally, these results show that proteins forming large nuclear complexes can be recruited sequentially on chromatin in the absence of their natural DNA target and with no reciprocity in their recruitment.
Results and discussion
Prolonged immobilization of XPC on chromatin leads to abortive PInC formation
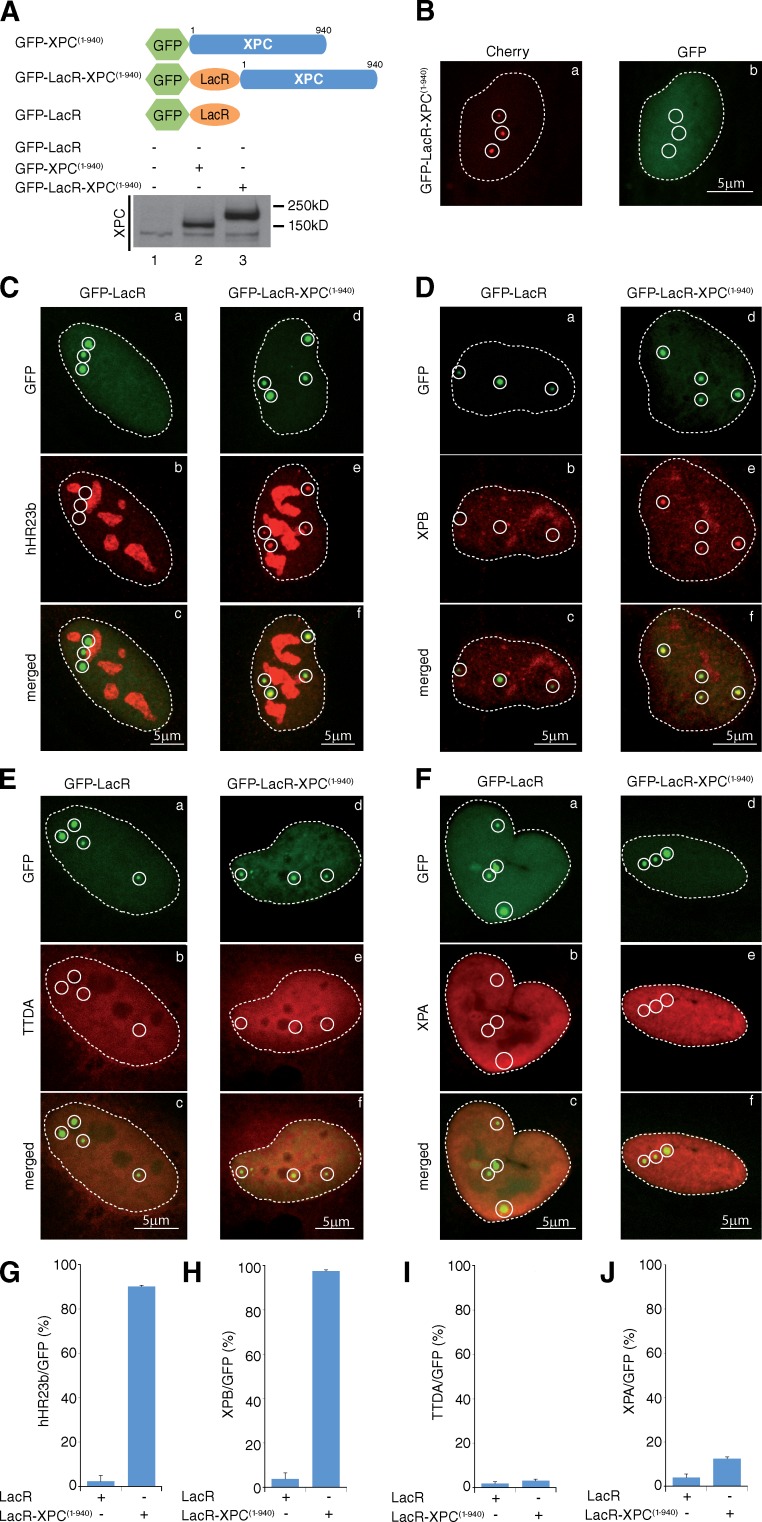
To obtain insights into the formation of the PInC in the absence of DNA damage, we used the LacO/LacR reporter system (Tumbar et al., 1999; Soutoglou and Misteli, 2008). The full-length human GFP-XPC(1–940) construct (Bernardes de Jesus et al., 2008) was fused to the LacR (Fig. 1 A, top). In a local UV irradiation experiment (Coin et al., 2006), GFP-LacR-XPC(1–940) colocalized with XPB on UV-C–induced DNA damage (Fig. S1, A and B), showing its functionality (see also Fig. 2, E and F).
Figure 1.
Tethering XPC to undamaged chromatin triggers the recruitment of TFIIH. (A, top) Schematic representation of the GFP-XPC(1–940), GFP-LacR-XPC(1–940), and GFP-LacR. For clarity, the sizes of the GFP (238 aa) and LacR (367 aa) were omitted. (bottom) Proteins from whole cell extracts (15 µg) of U2OS either expressing GFP-LacR, GFP-XPC(1–940), or GFP-LacR-XPC(1–940) were resolved by SDS-PAGE followed by Western blotting using an anti-XPC antibody. (B) Recruitment of Cherry-LacR (a) and GFP-XPC(1–940) (b) to the LacO repeats in U2OS17 cells. (C–F) Recruitment of hHR23b (C), XPB (D), RFP-TTDA (E; Giglia-Mari et al., 2004), and flag-XPA (F; Unsal-Kaçmaz et al., 2007) to tethered GFP-LacR (a–c) or GFP-LacR-XPC(1–940) (d–f). Note that anti-hHR23b antibody produced an important staining of the nucleoli in U2OS cells. Circles indicate LacO arrays. (G–J) The values on the graphs represent the percentage of colocalization of hHR23b (G), XPB (H), TTDA (I), and XPA (J) with GFP on the array based on three independent experiments with SD.
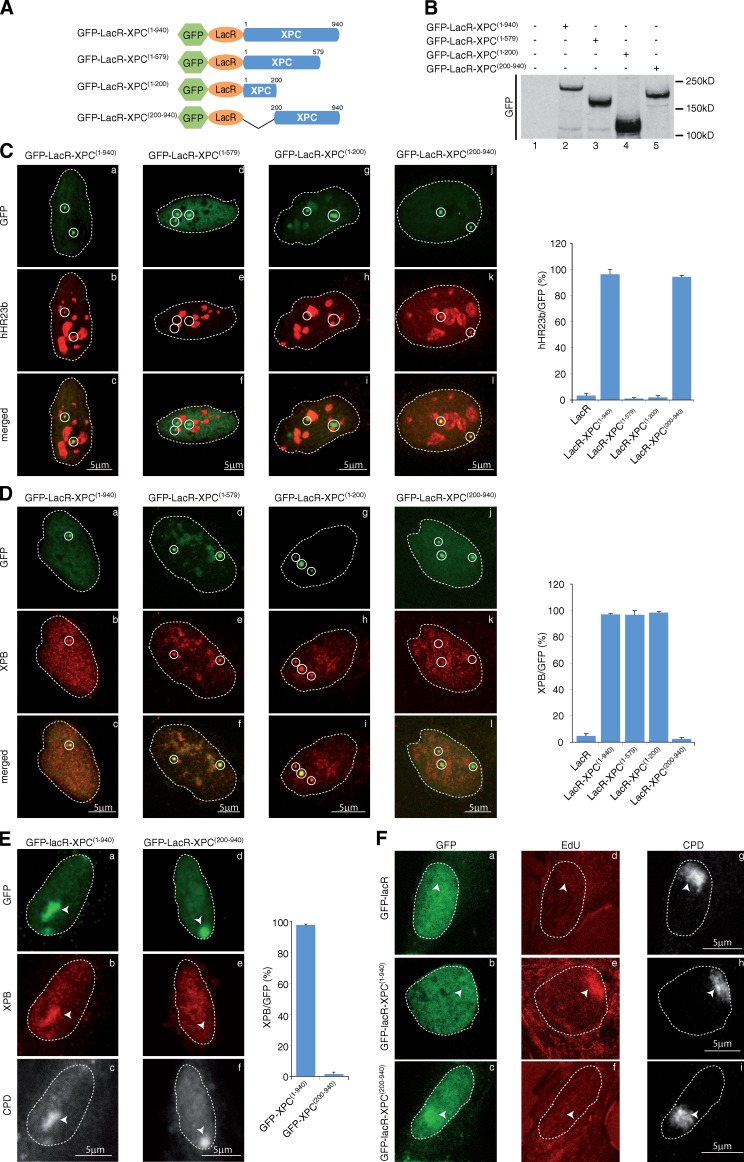
Figure 2.
Defining TFIIH- and hHR23b-interacting domains in XPC. (A) Schematic representation of the wild-type and mutant XPC constructs. For clarity, the sizes of the GFP (238 aa) and LacR (367 aa) were omitted. (B) Proteins from whole cell extracts (15 µg) of U2OS17-expressing wild-type or mutant XPC proteins were resolved by SDS-PAGE followed by Western blotting using anti-GFP antibody. (C and D) Recruitment of hHR23b (C) and XPB (D) to tethered GFP-LacR-XPC(1–940) (a–c), GFP-LacR-XPC(1–579) (d–f), GFP-LacR-XPC(1–200) (g–i), or GFP-LacR-XPC(200−940) (j–l). The values on the graphs represent the percentage of colocalization of each NER factor with GFP on the array based on three independent experiments with SD. Circles indicate LacO arrays. (E) After transfection of GFP-LacR-XPC(1–940) or GFP-LacR-XPC(200–940), XP-C cells were locally UV irradiated (150 J/m2), fixed 15 min later, and stained with antibodies raised against GFP, XPB, and UV-induced CPD. The values on the graphs represent the percentage of colocalization of XPB with GFP on the local UV array based on three independent experiments with SD. Arrowheads indicate locally irradiated areas. (F) After transfection of GFP-LacR, GFP-LacR-XPC(1–940), or GFP-LacR-XPC(200–940) (GFP), XP-C cells were locally UV irradiated (150 J/m2) and repair synthesis was analyzed by EdU incorporation at DNA damage (CPD). Arrowheads indicate locally irradiated areas.
Expression of GFP-LacR-XPC(1–940) in U2OS17 cells (Fig. 1 A, bottom), containing an array of 256 copies of the LacO sequence (Soutoglou and Misteli, 2008), resulted in the localization of the fusion protein to discrete areas of the nucleus, characteristic of the LacO loci (Fig. 1, C–F, d). In contrast, the original GFP-XPC(1–940) construct was not recruited to the array marked by a Cherry-LacR fusion (Fig. 1 B), indicating that the LacO locus did not contain structural alterations recognized by XPC. In GFP-LacR-XPC(1–940)–expressed U2OS17 cells, we observed a colocalization of GFP with hHR23b (Fig. 1, C [e and f] and G) and with the subunits of TFIIH, XPB (Fig. 1, D [e and f] and H), and p44 (Fig. S2) that was not observed after expression of GFP-LacR (Fig. 1, C, D [b and c], G, and H). In marked contrast to XPB or p44, the NER-specific TFIIH subunit TTDA and the TFIIH downstream NER factor XPA were not, or barely, recruited to immobilized GFP-LacR-XPC(1–940) (Fig. 1, E and F [e and f] and I and J). These results show that the sequestration of XPC to undamaged chromatin leads to the recruitment of the immediate downstream factors but not far downstream components of the NER reaction.
Domains of XPC required for hHR23b and TFIIH interactions
The domains of XPC responsible for interactions with TFIIH and hHR23b have been assigned to the C terminus of the protein using in vitro GST pull-down assays (Uchida et al., 2002). To delineate the interacting domains of XPC with hHR23b and TFIIH on chromatin, we generated the mutant protein GFP-LacR-XPC(1–579), deleted of the last 361 aa (Fig. 2, A and B). Surprisingly, tethering of GFP-LacR-XPC(1–579) to chromatin led to a loss of hHR23b recruitment to the array but still promoted the loading of XPB and p44 (Fig. 2, C and D [d–f]; and Fig. S2). Similarly, XPC deleted of the last 740 aa (GFP-LacR-XPC(1–200)) still recruited XPB and p44 (Fig. 2 D [g–i] and Fig. S2). Finally, we deleted in the wild-type XPC the first 200 aa to generate the GFP-LacR-XPC(200–940) mutant (Fig. 2 A) that did not recruit XPB and p44, while it interacted with hHR23b (Fig. 2, C and D [j–l]; and Fig. S2). To reveal the biological significance of these observations, we performed local UV irradiation in XP-C cells transfected either with GFP-LacR-XPC(1–940) or GFP-lacR-XPC(200–940) and showed that the corresponding proteins were recruited to UV-damaged areas labeled by an antibody against cyclobutane pyrimidine dimers (CPD; Fig. 2 E, a, c, d, and f). However, although GFP-LacR-XPC(1–940) recruited XPB to damage, GFP-LacR-XPC(200–940) did not (Fig. 2 E, b and e). Finally, DNA repair synthesis revealed by 5-ethynyl 2-deoxyuridine (EdU) incorporation was restored with GFP-LacR-XPC(1–940) but not with GFP-LacR-XPC(200–940) (Fig. 2 F) in XP-C cells. These data indicate that the first 200 aa of XPC recruit TFIIH to damaged sites to promote DNA repair. Note that LacR-XPC(200–940) remained bound to the UV damage even 4 h after irradiation (Fig. 2 F, c).
Tethering of XPB and DDB2 reveals ordered assembly of PInC on undamaged chromatin
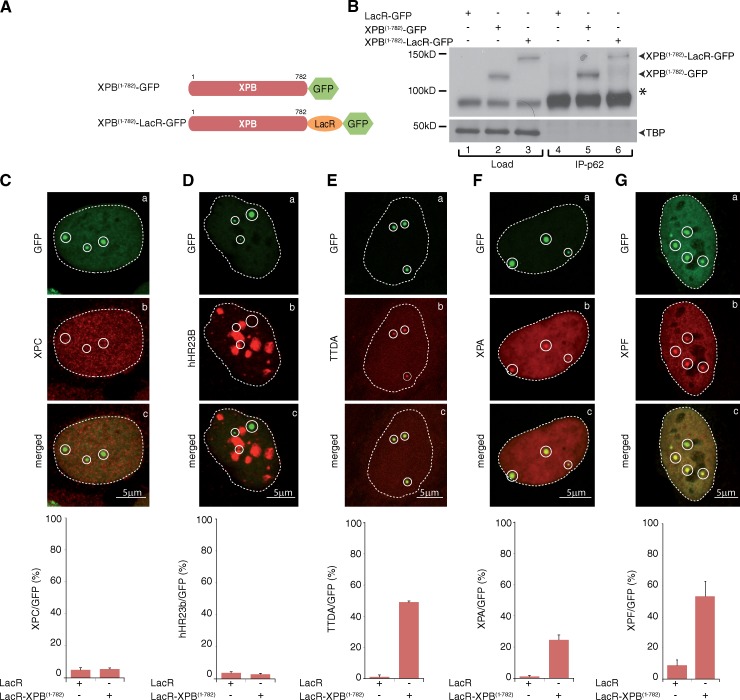
We subsequently tested the reciprocity of the recruitments that we observed by tethering TFIIH (XPB) to the LacO array (Fig. 3 A). To verify the functionality of the chimeric protein, we performed immunoprecipitation experiments using anti-p62, another TFIIH subunit, and observed a similar incorporation of XPB(1–782)-GFP and XPB(1–782)-LacR-GFP into TFIIH (Fig. 3 B). Furthermore, we observed colocalization of XPB(1–782)-LacR-GFP with XPC in a local UV irradiation experiment (Fig. S1 C). Also, TFIIH containing the XPB(1–782)-LacR-GFP was functional in a dual incision assay (unpublished data), showing that the LacR fusion did not inhibit XPB function.
Figure 3.
Tethering XPB to chromatin triggers the recruitment of downstream NER factors. (A) Schematic representation of XPB(1–782)-LacR-GFP and XPB(1–782)-GFP. For clarity, the sizes of the GFP (238 aa) and LacR (367 aa) were omitted. (B, lanes 1–3) Proteins from whole cell extracts (15 µg) of U2OS17-expressing XPB proteins were resolved by SDS-PAGE followed by Western blotting using anti-XPB antibodies. (lanes 4–6) TFIIH from 300 µg of U2OS17 extracts was immunoprecipitated with an anti-p62 antibody and resolved by SDS-PAGE followed by Western blotting with an anti-XPB antibody. The asterisk indicates the endogenous XPB. (C–G) Recruitment of XPC (C), hHR23b (D), RFP-TTDA (E), Flag-XPA (F), and HA-XPF (G; Su et al., 2012) to tethered XPB-LacR-GFP (a–c). The values on the graphs represent the percentage of colocalization of each NER factor with GFP on the array based on three independent experiments with SD. Circles indicate LacO arrays.
Immobilization of XPB(1–782)-LacR-GFP in U2OS17 (Fig. 3, C–G, a) did not result in the recruitment of the upstream NER factors XPC (Fig. 3 C, b) or hHR23b (Fig. 3 D, b). In contrast, two TFIIH subunits, TTDA (Fig. 3 E) and p44 (Fig. S2), and two downstream NER factors, XPA (Fig. 3 F) and XPF (Fig. 3 G), clearly colocalized with GFP on the array in the presence of XPB(1–782)-LacR-GFP. Although XPB(1–782)-LacR-GFP attracted XPF, we were not able to show that incision or DNA resynthesis were taking place at the LacO in these conditions (unpublished data), indicating that the PInC was presumably not functional in this artificial system.
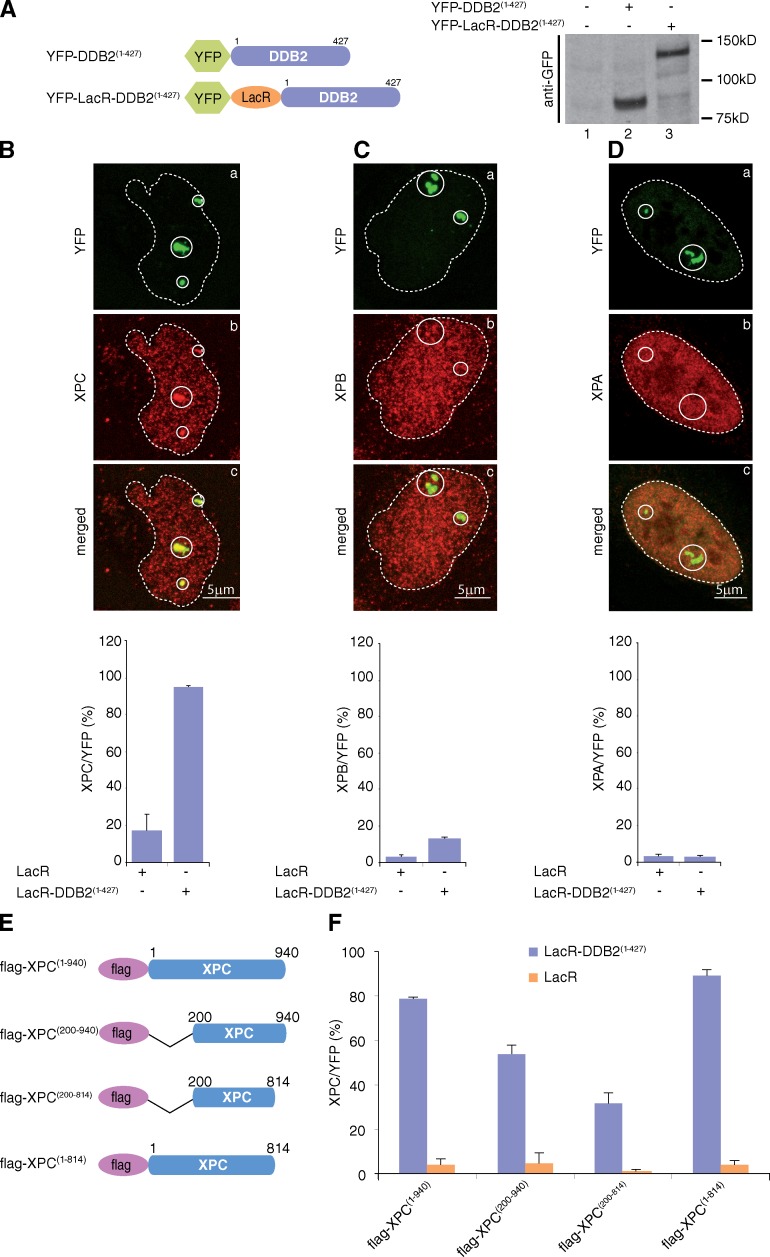
These data suggest that an immobilized NER factor can recruit the immediate downstream factors but not those upstream. To extend this observation, we introduced the LacR cassette into the YFP-DDB2(1–427) construct (Alekseev et al., 2008; Fig. 4 A). The YFP-LacR-DDB2(1–427) functionality was shown in locally irradiated cells (Fig. S1 D). As previously observed, tethering DDB2 to chromatin in U2OS17 cells caused an unfolding of the array visualized by larger spots, characteristic of decondensed chromatin (Luijsterburg et al., 2012). In agreement with our hypothesis, tethered YFP-LacR-DDB2 recruited XPC (Fig. 4 B) but not XPB and XPA (Fig. 4, C and D). These results demonstrate that PInC formation on undamaged chromatin is ordered.
Figure 4.
Tethering DDB2 to chromatin recruits XPC through its N- and C-terminal extremities. (A, left) Schematic representation of YFP-DDB2(1–427) and YFP-LacR-DDB2(1–427). For clarity, the sizes of the YFP (238 aa) and LacR (367 aa) were omitted. (right) Proteins from whole cell extracts (15 µg) of U2OS17 expressing either YFP-DDB2(1–427) or YFP-LacR-DDB2(1–427) were resolved by SDS-PAGE followed by Western blotting using anti-GFP antibodies. (B–D) Recruitment of XPC (B), XPB (C), or flag-XPA (D) to tethered YFP-LacR-DDB2(1–427). The values on the graphs represent the percentage of colocalization of each NER factor with YFP on the array based on three independent experiments with SD. Circles indicate LacO arrays. (E) Schematic representation of wild-type and deleted flag-XPC constructs. (F) Recruitment of flag-XPC constructs to tethered YFP-LacR-DDB2(1–427). The values represent the percentage of colocalization of each Flag-XPC constructs with YFP on the array based on three independent experiments with SD.
Although a DDB2–XPC interaction has been reported (Sugasawa et al., 2005), the domains of interaction were not determined. We used flag-tagged XPC to introduce deletions in the N- and/or C-terminal regions (Fig. 4 E). Deletion of the first 200 aa of XPC significantly impaired its recruitment to tethered DDB2 (Fig. 4 F). Codeletion of the C-terminal domain of XPC (flag-XPC(200–814)) increased this effect, whereas C-terminal deletion alone did not affect the recruitment of XPC to tethered DDB2 (Fig. 4 F). These results show that XPC interacts with DDB2 through its N- and C-terminal domains and suggest a direct competition between DDB2 and TFIIH for binding to XPC.
Recruitment of TTDA to chromatin is key in PInC assembly
The aforementioned data show that the presence of TTDA in TFIIH on the LacO array correlates with the recruitment of XPA and XPF (Figs. 1 and 3), pinpointing to a crucial role of this small polypeptide in the completion of the PInC. To study this hypothesis, we constructed the TTDA(1–71)-LacR-GFP fusion protein (Fig. 5, A and B) that colocalized with XPC on local UV-induced DNA damage (Fig. S1 E). Prolonged immobilization of TTDA(1–71)-LacR-GFP in U2OS17 cells induced the recruitment of XPB, XPA, and XPF but not that of XPC or hHR23b (Fig. 5, C–E; and Fig. S2 B).
Figure 5.
The N-terminal domain of TTDA recruits XPA to damaged DNA. (A) Schematic representation of the TTDA(1–71)-GFP, TTDA(1–71)-LacR-GFP, and TTDA(15–71)-LacR-GFP. For clarity, the sizes of the GFP (238 aa) and LacR (367 aa) were omitted. (B) Proteins from whole cell extracts (15 µg) of U2OS17-expressing TTDA proteins were resolved by SDS-PAGE followed by Western blotting using anti-GFP antibodies. (C–E) Recruitment of XPC (C), XPB (D), and Flag-XPA (E) to either tethered TTDA(1–71)-LacR-GFP or TTDA(15–71)-LacR-GFP as indicated. The values on the graphs represent the percentage of colocalization of each NER factor with GFP on the array based on three independent experiments with SD. Circles indicate LacO arrays. (F) Flag-TTDA(1–71) or Flag-TTDA(15–71) were expressed in E. coli and immunoprecipitated with anti-flag antibody–covered beads. After washes, the beads were incubated with bacterial extracts expressing XPD, p52, or XPA. Pull-down fractions were washed, resolved by SDS-PAGE, and immunoblotted with anti-XPD, p52, and XPA antibodies (top) or anti-Flag antibody (bottom). (G) 10 and 30 ng of either flag-TTDA(1–71) or flag-TTDA(15–71) were tested in a dual incision assay (NER) containing the recombinant XPC-HR23b, recombinant TFIIH lacking TTDA (IIH9; Coin et al., 2006), XPA, RPA, XPG, ERCC1-XPF factors, and a closed circular plasmid containing a single 1,3-intra strand d(GpTpG) cisplatin-DNA cross-link (Pt-DNA) as a template. Sizes of the incision products are indicated. (H) Immobilized biotinylated damaged DNA was incubated with XPC/hHR23B, IIH9, XPA, and either flag-TTDA(1–71) or flag-TTDA(15–71) as indicated (PreInc) for 15 min at 30°C with ATP. DNA was washed, supplemented with XPC/hHR23b, purified TFIIH from HeLa, RPA, ERCC1-XPF, and XPG without XPA, and subjected to dual incision assay. In lane 3, all the NER factors were added in a single step with immobilized damaged DNA and subjected to dual incision.
Some TTD-A patients carry a mutation in the start codon of TTDA (Giglia-Mari et al., 2004). However, a downstream start codon at position 16 can be used and produces a truncated polypeptide lacking the first 15 aa (Giglia-Mari et al., 2004). Tethering TTDA(15–71)-LacR-GFP to chromatin decreased the recruitment efficiency of the TFIIH subunit XPB as well as the downstream NER factors XPA and XPF (Fig. 5, D and E; and Fig. S2 B), indicating that the N-terminal region of TTDA is directly involved in protein–protein interactions required for PInC completion. TTDA interacts with XPD and p52 in TFIIH (Coin et al., 2006). Each of these core TFIIH proteins was expressed in Escherichia coli and tested for their interaction with recombinant and purified flag-TTDA(1–71) or flag-TTDA(15–71) proteins. XPD and p52 were pulled down with both wild-type and mutant TTDA (Fig. 5 F, lanes 1–4). In contrast, the immediate TFIIH downstream NER factor XPA interacted only with flag-TTDA(1–71) but not with flag-TTDA(15–71). In an in vitro dual incision assay (Coin et al., 2006), deletion of the first 15 aa of TTDA strongly decreased the repair function of TTDA (Fig. 5 G). We also analyzed the recruitment of XPA by preincubating an immobilized biotinylated mono-damaged DNA (Riedl et al., 2003) with XPC/hHR23B, IIH9 (recombinant TFIIH without TTDA), and XPA, with either flag-TTDA(1–71) or flag-TTDA(15–71). The immobilized mono-damaged DNA was subsequently washed to remove the nonspecifically bound proteins and reincubated with XPC-hHR23b, purified TFIIH from HeLa, RPA, XPG, and ERCC1-XPF without XPA, in a dual incision assay. Dual incision activity was observed when flag-TTDA(1–71) was present in the preincubation mix (Fig. 5 H, lane 2), indicating that XPA was retained on the damaged DNA. In contrast, deletion of the first 15 aa of TTDA impaired the recruitment of XPA to DNA damage and subsequently dual incision (Fig. 5 H, lane 1). These results suggest that XPA is recruited to DNA damage through the N terminus of TTDA, which is deleted in some TTD-A patients.
We show here that PInC formation can occur on an undamaged DNA by a sequential and ordered assembly of NER factors mimicking that existing on a damaged DNA (Volker et al., 2001; Riedl et al., 2003). The rules regulating the formation of the PInC on the LacO locus are then clearly different from what has been observed so far with the LacO/LacR system. It was shown that tethering components of large complexes such as those of the Cajal body led to the self-organization of a full complex independently of any order of assembly through a phenomenon called molecular crowding (Kaiser et al., 2008). In another work, it was shown that tethering MDC1, a component of the DNA damage response pathway, led to the recruitment of the upstream factors NBS1 and MRE11 (Soutoglou and Misteli, 2008).
In our artificial system, XPC bound to tethered DDB2 but not to tethered XPB. In contrast, tethered XPC recruited XPB to the array, indicating an absence of reciprocity in the recruitments. If XPB did not recruit XPC, it efficiently attracted the downstream factors XPA or XPF to the chromatin (Fig. S3 A). Presumably, the absence of recruitment of XPC to XPB is caused by steric constraints as a result of the presence of downstream NER factors that may display higher affinity for TFIIH than XPC. Indeed, in a classical NER reaction, the recruitment of XPA and/or XPG leads to the exclusion of XPC from the damaged DNA (Riedl et al., 2003; You et al., 2003). Another characteristic of our system is that an upstream factor like DDB2 can recruit immediate downstream factor like XPC, but not beyond. Interestingly, we found that the N-terminal domain of XPC is important for its interaction both with DDB2 and TFIIH, arguing that interaction of XPC with DDB2 and TFIIH are mutually exclusive and that a complex composed of the first three proteins of the NER reaction cannot exist on DNA damage.
Similarly, we observed that tethered XPC recruited TFIIH (XPB and p44) but not beyond, including TTDA and the downstream factors XPA-XPF (Fig. S3 B). In agreement with a crucial role for TTDA in the completion of PInC, tethering of TTDA to the array led to the recruitment of XPA and XPF. The fact that PInC formation was aborted when TTDA was absent may suggest that bona fide NER substrates elicit the recruitment of TTDA to the XPC–TFIIH complex to complete the formation of the PInC in a classical NER reaction. Indeed, Giglia-Mari et al. (2006) demonstrated that substrates that were recognized by XPC but not removed by NER were not able to induce a more stable incorporation of TTDA into TFIIH. The LacO/LacR reporter system may mimic the situation that takes place with such abortive substrates, showing the role of the DNA damage that may indirectly induce the recruitment of TTDA to the XPC–TFIIH complex and the subsequent completion of the PInC. Furthermore, we showed that TTDA directly interacted with XPA through its N-terminal domain that is absent in some TTD-A patients. These data explain partially the DNA repair defect harbored by TTD-A patients and show that data obtained with the LacO/LacR reporter system could be used to understand the molecular defects of a genuine system in a pathological situation.
Materials and methods
Cell lines and transfection
The U2OS17 clone (containing sequences of 256× repetitions of LacO) was generated as described previously (Lematîre et al., 2012). U2OS and U2OS17 cell lines were cultured in DMEM (4.5 g/l glucose) supplemented with 10% FCS. GM14867 (XP-C cells; Bernardes de Jesus et al., 2008) were cultured in MEM supplemented with 15% FCS, AANE, and vitamins. All the cells were cultured at 37°C in a 5% CO2 incubator.
Plasmids
Full-length cDNA of XPC was amplified by PCR and ligated into the BamHI–EcoRI restriction sites of pCMV vector giving the pCMV-flag-XPC construct. pYFP-DDB2, pEGFP-XPC, pEGFP-XPB, and pEGFP-TTDA were described previously (Coin et al., 2006; Alekseev et al., 2008; Bernardes de Jesus et al., 2008; Oksenych et al., 2009). In brief, full-length XPC coding sequence was amplified and ligated into the blunted BglII site of pEGFP-C1 giving the pEGFP-XPC. Full-length XPB coding sequence was amplified and ligated into the EcoRI–BamHI restriction sites of the pEGFP-N1 vector giving the pEGFP-XPB construct. Full-length TTDA coding sequence was amplified and ligated into the BamHI restriction site of the pEGFP-N1 vector giving the pEGFP-TTDA construct. To generate fusions between LacR, repair factors, and GFP/YFP tag, cDNA of LacR was amplified by PCR and cloned in pYFP-DDB2, pEGFP-XPC, pEGFP-XPB, or pEGPF-TTDA vectors. For truncated forms of XPC and TTDA, site-directed mutagenesis was performed on flag-XPC, GFP-LacR-XPC, or TTDA-LacR-GFP plasmids using QuickChange II XL Site-Directed Mutagenesis kit (Agilent Technologies). For transfection, cells were cultured on coverslips and transfected using the FuGENE6 transfection reagent (Promega) with the appropriate DNA plasmids, and proteins were visualized 16–24 h after transfection by immunofluorescence.
Immunofluorescence in the LacO/LacR reporter system
16–24 h after transfection with the appropriate plasmid DNA, cells were fixed with 2% PFA in PBS (10 min at RT), and then permeabilized with 0.5% Triton X-100 (5 min at RT). For XPC, XPB, and p44 staining, cells were incubated for 40 s with ice-cold nuclear preextraction buffer (50 mM Hepes, pH 7, 150 mM NaCl, 10 mM EGTA, 2 mM MgCl2, and 0.5% Triton X-100) before fixation. After incubation in blocking buffer (PBS, 10% FCS, and 0.2% Triton X-100), cells were stained with the appropriate primary antibody diluted in blocking buffer (2 h at RT), washed three times, and then stained with Alexa Fluor 546 secondary antibody (Invitrogen; 45 min at RT). After four washes with PBS + 0.2% Triton X-100 and staining with DAPI, cells were mounted with ProLong Gold antifade reagent (Molecular Probes) and observed with a confocal system (TCS SP2; Leica) based on an inverted microscope (DMIRBE; Leica; 63× Plan Apochromat, NA 1.4; LCS software [Leica]). Z-stack width was set to 0.5 µM. For the U2OS17 cells, two, three, or four LacO array spots can be observed depending on the cell cycle stage.
Cell extract and Western blot
24 h after transfection, cells were scraped in PBS, lysed in RIPA buffer (10 mM Tris-Cl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and 140 mM NaCl), and centrifuged at 10 Krpm for 20 min at 4°C. For loading control, 15 µg of each extract was loaded on an SDS-PAGE gel. For immunoprecipitation, 300 µg of each extract was incubated overnight with rabbit anti-p62 antibody and Dynabeads protein G (Life Technologies). After three washes the beads were heated to 95°C in loading buffer and loaded on an SDS-PAGE gel, and the membrane was blotted with anti-XPB antibody.
Local UV irradiation
24 h after transfection, cells were irradiated with UV-C at 150 J/m2 dose through an isopore polycarbonate filter with pores of 5-µm diameter (EMD Millipore). The cells were fixed with PFA 15 min later and immunofluorescence staining was performed with primary antibodies as indicated in the figure legends. After incubation with the corresponding secondary antibodies (Alexa Fluor 546 or 488 secondary antibody [Invitrogen]), cells were fixed and DNA was denatured with 2 M HCl. UV lesions were marked with an anti-CPD antibody followed by Alexa Fluor 647 secondary antibody (Invitrogen).
Unscheduled DNA repair synthesis
24 h after transfection, cells were locally UV irradiated and incubated with EdU for 4 h. EdU was revealed using the Click-iT EdU Alexa Fluor 594 Imaging kit (Invitrogen) according to the manufacturer’s instructions.
Pull-down assay
Flag-TTDA polypeptides were expressed in E. coli and incubated with anti-flag–covered beads in RIPA buffer. XPD, p52, and XPA were expressed in E. coli and cell lysates were incubated at 4°C for 1 h in RIPA buffer, with 500 ng of washed flag-TTDA proteins bound to beads. Pull-downs were washed with RIPA and analyzed by Western blotting.
NER assay
Dual incision assay was performed in 25 µl of incision buffer (50 mM Hepes-KOH, pH 7.6, 20 mM Tris-HCl, pH 7.6, 50 mM KCl, 2.5 mM MgCl2, 0.5 mM DTT, 0.5 mM EDTA, and 10% glycerol supplemented with 2 mM ATP). Each reaction contained 100 ng of recombinant TFIIH factor containing nine subunits and lacking TTDA (IIH9; Coin et al., 2006), 5 ng of XPG, 15 ng of XPF/ERCC1, 10 ng of XPC-hHR23B, 50 ng of RPA, and 25 ng of XPA. After 10-min preincubation at 30°C, 30 ng of Pt-DNA was added and the reaction was continued for 90 min at 30°C. The excised fragment was detected on 14% urea-acrylamide after annealing with 9 ng of the complementary oligonucleotide and addition of four radiolabeled dCMPα-P32 (3,000 µCi/mmol) residues by Sequenase v.2.1 (USB).
XPA binding study on immobilized DNA
Immobilized damaged DNA was generated by digesting the Pt-DNA plasmid with HincII and BanI, resulting in a 288-bp fragment. The nondamaged strand was biotinylated at the BanI site by Klenow fill-in reaction with Bio-dUTP and purified after separation on agarose gel with a QIAEX gel extraction kit (QIAGEN; Riedl et al., 2003). M-280 Streptavidin dynabeads (Invitrogen) were coupled to damaged DNA according to the manufacturer’s instructions and were subsequently incubated in 10 mM Hepes, 100 mM glutamate, 10 mM MgOAc, 5 mM EGTA, 3.5% glycerol, 60 mg/ml casein, 5 mg/ml PVP, and 2.5 mM DTT for 15 min at RT to limit unspecific binding of proteins. Immobilized DNA was then incubated with 100 ng of recombinant XPC-hR23B, 50 ng of XPA, 100 ng of IIH9, and 5 ng of either Flag-TTDA(1–71) or Flag-TTDA(15–71) at 30°C for 15 min with 2 mM ATP. After incubation, magnetic beads were collected on a Magnetic Particle Concentrator (Invitrogen) and supernatants were removed. Beads were then washed five times in 4 vol of cold incision buffer and resuspended in the same buffer for functional analysis of bound XPA in a subsequent dual incision assay containing all the NER factors except XPA (Riedl et al., 2003).
Online supplemental material
Fig. S1 shows the recruitment of DDB2, XPC, XPB, or TTDA to local UV-irradiated area. Fig. S2 A shows the recruitment of the subunit of TFIIH p44 to various lacR-XPC constructs tethered to the LacO. Fig. S2 B shows the recruitment of XPF to tethered wild-type or mutant TTDA. Fig. S3 shows a model of the PInC assembly after the tethering of various NER factors to chromatin. Table S1 shows the various antibodies used in the paper and the dilutions used in Western blot or immunofluorescence. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201403096/DC1.
Supplementary Material
Acknowledgments
We thank Leon Mullenders, Ambra Giglia-Mari, and Orlando Scharer for material and Annabel Larnicol for the technical help.
This study was supported by the Agence Nationale de Recherche and the Ligue Contre le Cancer, Equipe Labélisée.
S. Ziani, Z. Nagy, S. Alekseev, and F. Coin designed and performed the experiments and analyzed the data. E. Soutoglou and J.-M. Egly provided cell line and antibodies. F. Coin wrote the paper.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- CPD
- cyclobutane pyrimidine dimers
- EdU
- 5-ethynyl 2-deoxyuridine
- LacO
- lactose operator
- LacR
- lactose repressor
- NER
- nucleotide excision repair
- PInC
- preincision complex
- TTD
- trichothiodystrophy
- XP
- xeroderma pigmentosum
References
- Alekseev, S., Luijsterburg M.S., Pines A., Geverts B., Mari P.O., Giglia-Mari G., Lans H., Houtsmuller A.B., Mullenders L.H., Hoeijmakers J.H., and Vermeulen W.. 2008. Cellular concentrations of DDB2 regulate dynamic binding of DDB1 at UV-induced DNA damage. Mol. Cell. Biol. 28:7402–7413 10.1128/MCB.01108-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes de Jesus, B.M., Bjørås M., Coin F., and Egly J.M.. 2008. Dissection of the molecular defects caused by pathogenic mutations in the DNA repair factor XPC. Mol. Cell. Biol. 28:7225–7235 10.1128/MCB.00781-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coin, F., Proietti De Santis L., Nardo T., Zlobinskaya O., Stefanini M., and Egly J.M.. 2006. p8/TTD-A as a repair-specific TFIIH subunit. Mol. Cell. 21:215–226 10.1016/j.molcel.2005.10.024 [DOI] [PubMed] [Google Scholar]
- Compe, E., and Egly J.M.. 2012. TFIIH: when transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 13:343–354 10.1038/nrm3350 [DOI] [PubMed] [Google Scholar]
- Fong, Y.W., Cattoglio C., and Tjian R.. 2013. The intertwined roles of transcription and repair proteins. Mol. Cell. 52:291–302 10.1016/j.molcel.2013.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari, G., Coin F., Ranish J.A., Hoogstraten D., Theil A., Wijgers N., Jaspers N.G., Raams A., Argentini M., van der Spek P.J., et al. 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36:714–719 10.1038/ng1387 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari, G., Miquel C., Theil A.F., Mari P.O., Hoogstraten D., Ng J.M., Dinant C., Hoeijmakers J.H., and Vermeulen W.. 2006. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol. 4:e156 10.1371/journal.pbio.0040156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser, T.E., Intine R.V., and Dundr M.. 2008. De novo formation of a subnuclear body. Science. 322:1713–1717 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- Lehmann, A.R.2003. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 85:1101–1111 10.1016/j.biochi.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Lemaître, C., Fischer B., Kalousi A., Hoffbeck A.S., Guirouilh-Barbat J., Shahar O.D., Genet D., Goldberg M., Betrand P., Lopez B., et al. 2012. The nucleoporin 153, a novel factor in double-strand break repair and DNA damage response. Oncogene. 31:4803–4809 10.1038/onc.2011.638 [DOI] [PubMed] [Google Scholar]
- Luijsterburg, M.S., Lindh M., Acs K., Vrouwe M.G., Pines A., van Attikum H., Mullenders L.H., and Dantuma N.P.. 2012. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J. Cell Biol. 197:267–281 10.1083/jcb.201106074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan, A., Davies A.A., Moggs J.G., West S.C., and Wood R.D.. 1994. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 371:432–435 10.1038/371432a0 [DOI] [PubMed] [Google Scholar]
- Ogi, T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G., et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 37:714–727 10.1016/j.molcel.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Oksenych, V., Bernardes de Jesus B., Zhovmer A., Egly J.M., and Coin F.. 2009. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J. 28:2971–2980 10.1038/emboj.2009.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl, T., Hanaoka F., and Egly J.M.. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22:5293–5303 10.1093/emboj/cdg489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijbers, A.M., de Laat W.L., Ariza R.R., Biggerstaff M., Wei Y.F., Moggs J.G., Carter K.C., Shell B.K., Evans E., de Jong M.C., et al. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 86:811–822 10.1016/S0092-8674(00)80155-5 [DOI] [PubMed] [Google Scholar]
- Soutoglou, E., and Misteli T.. 2008. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 320:1507–1510 10.1126/science.1159051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, Y., Orelli B., Madireddy A., Niedernhofer L.J., and Schärer O.D.. 2012. Multiple DNA binding domains mediate the function of the ERCC1-XPF protein in nucleotide excision repair. J. Biol. Chem. 287:21846–21855 10.1074/jbc.M111.337899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa, K., Ng J.M., Masutani C., Iwai S., van der Spek P.J., Eker A.P., Hanaoka F., Bootsma D., and Hoeijmakers J.H.. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 2:223–232 10.1016/S1097-2765(00)80132-X [DOI] [PubMed] [Google Scholar]
- Sugasawa, K., Okuda Y., Saijo M., Nishi R., Matsuda N., Chu G., Mori T., Iwai S., Tanaka K., Tanaka K., and Hanaoka F.. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 121:387–400 10.1016/j.cell.2005.02.035 [DOI] [PubMed] [Google Scholar]
- Tang, J.Y., Hwang B.J., Ford J.M., Hanawalt P.C., and Chu G.. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell. 5:737–744 10.1016/S1097-2765(00)80252-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumbar, T., Sudlow G., and Belmont A.S.. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341–1354 10.1083/jcb.145.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida, A., Sugasawa K., Masutani C., Dohmae N., Araki M., Yokoi M., Ohkuma Y., and Hanaoka F.. 2002. The carboxy-terminal domain of the XPC protein plays a crucial role in nucleotide excision repair through interactions with transcription factor IIH. DNA Repair (Amst.). 1:449–461 10.1016/S1568-7864(02)00031-9 [DOI] [PubMed] [Google Scholar]
- Unsal-Kaçmaz, K., Chastain P.D., Qu P.P., Minoo P., Cordeiro-Stone M., Sancar A., and Kaufmann W.K.. 2007. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell. Biol. 27:3131–3142 10.1128/MCB.02190-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volker, M., Moné M.J., Karmakar P., van Hoffen A., Schul W., Vermeulen W., Hoeijmakers J.H., van Driel R., van Zeeland A.A., and Mullenders L.H.. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 8:213–224 10.1016/S1097-2765(01)00281-7 [DOI] [PubMed] [Google Scholar]
- You, J.S., Wang M., and Lee S.H.. 2003. Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem. 278:7476–7485 10.1074/jbc.M210603200 [DOI] [PubMed] [Google Scholar]
- Zhovmer, A., Oksenych V., and Coin F.. 2010. Two sides of the same coin: TFIIH complexes in transcription and DNA repair. ScientificWorldJournal. 10:633–643 10.1100/tsw.2010.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.