The cofilin CFL-1 recruits actin to the P-type calcium ATPase SPCA1 at the trans-Golgi network, thereby activating the ATPase, promoting Ca2+ influx, and driving secretory cargo sorting.
Abstract
The actin filament severing protein cofilin-1 (CFL-1) is required for actin and P-type ATPase secretory pathway calcium ATPase (SPCA)-dependent sorting of secretory proteins at the trans-Golgi network (TGN). How these proteins interact and activate the pump to facilitate cargo sorting, however, is not known. We used purified proteins to assess interaction of the cytoplasmic domains of SPCA1 with actin and CFL-1. A 132–amino acid portion of the SPCA1 phosphorylation domain (P-domain) interacted with actin in a CFL-1–dependent manner. This domain, coupled to nickel nitrilotriacetic acid (Ni-NTA) agarose beads, specifically recruited F-actin in the presence of CFL-1 and, when expressed in HeLa cells, inhibited Ca2+ entry into the TGN and secretory cargo sorting. Mutagenesis of four amino acids in SPCA1 that represent the CFL-1 binding site also affected Ca2+ import into the TGN and secretory cargo sorting. Altogether, our findings reveal the mechanism of CFL-1–dependent recruitment of actin to SPCA1 and the significance of this interaction for Ca2+ influx and secretory cargo sorting.
Introduction
Newly synthesized secretory cargoes are transported from the ER to the Golgi apparatus. Upon reaching the most distal compartment of the Golgi complex, also called the TGN, secretory cargoes are sorted and packaged into different carriers for transport to the cell surface (De Matteis and Luini, 2008; Pfeffer, 2011; Campelo and Malhotra, 2012; Wakana et al., 2012). Many types of carriers originate from the TGN, including clathrin-coated carriers (Doray et al., 2002), CARTS (CARriers of the TGN to the cell Surface; Wakana et al., 2012), and immature secretory granules of professional secretory cells (Dikeakos and Reudelhuber, 2007). The process of packaging of different cargo molecules in distinct transport carriers at the TGN is highly sophisticated and, for most proteins, poorly understood (Kienzle and von Blume, 2014).
The sorting of transmembrane proteins has been well studied. Many of these proteins contain cytosolic domains with tyrosine- or dileucine-based sorting motifs that are recognized by adaptor proteins, which facilitate the formation of clathrin-coated vesicles (Fölsch et al., 1999; Ang et al., 2003, 2004; Mellman and Nelson, 2008; Burgos et al., 2010). In yeast, it has been shown that exomer, a coat protein complex, regulates the transport of Chs3p and Fus1p from the late Golgi membrane to the plasma membrane (Wang et al., 2006; Barfield et al., 2009; Zanolari et al., 2011; Rockenbauch et al., 2012). However, there are no known orthologues of exomer in higher eukaryotes.
The sorting of soluble proteins at the TGN is more complex, and the simplest concept invokes a ligand receptor interaction where a sorting signal on the cargo binds to a receptor in the TGN. This sorting principle has been well established for lysosomal hydrolases that carry a Mannose-6-Phosphate (M6P) by the Mannose 6-phosphate receptor (M6PR) in the TGN membrane into clathrin-coated vesicles (Reitman and Kornfeld, 1981; Kornfeld and Mellman, 1989; Le Borgne and Hoflack, 1997). In contrast to this well-studied system, the sorting and packaging of secretory cargoes remains less understood. Notably, no sorting receptor such as the M6PR has been identified for the sorting of secretory cargoes.
It has been shown that in Drosophila melanogaster (Bard et al., 2006), in yeast (Curwin et al., 2012), and in mammalian cells (von Blume et al., 2009), the actin-severing protein cofilin orthologues (twinstar, cof1, and ADF/cofilin-1 [CFL-1], respectively) are required for secretory protein sorting at the TGN. CFL-1 and F-actin interact with the TGN-localized secretory pathway calcium ATPase 1 (SPCA1), a P-Type Ca2+-ATPase. Therefore, we hypothesized that F-actin and CFL-1 must be required for pumping of Ca2+ into the lumen of the TGN (von Blume et al., 2011). The transient increase of the luminal Ca2+ concentration induces the binding of secretory proteins to Cab45, a Golgi-resident protein, and subsequent sorting into a transport carrier (von Blume et al., 2012).
In the current study, we explored the mechanism by which F-actin and CFL-1 interact with SPCA1. We identified the key domain in SPCA1 that is required for direct binding to CFL-1. In addition, we reconstituted the interaction of the involved components in vitro, which showed that CFL-1 connects SPCA1 with F-actin. Furthermore, the experiments showed that this interaction is crucial for Ca2+ influx into the TGN and, consequently, secretory cargo sorting in living cells. Finally, we identified the crucial amino acids in SPCA1 that are required for the binding to CFL-1 and subsequent Ca2+ pumping of SPCA1.
Results
Expression and purification of the cytosolic domains of SPCA1: putative actin–CFL-1 interaction domains
SPCA1 is a type II phosphorylation P-type Ca2+ transporter ATPase that is encoded by the ATP2C1 gene (Van Baelen et al., 2004). SPCA1 is a TGN-resident protein, and contains 10 hydrophobic transmembrane helices. SPCA1 N and C termini (N-term and C-term) are exposed to the cytosol as well as four inter-transmembrane domains, labeled Loop1 through Loop4, respectively (L1–L4; Fig. S1 A). The N-term and L1 associate with each other to form an actuator domain (A-domain). The largest cytosolic domain (L2) contains a nucleotide binding domain (N-domain) and a phosphorylation domain (P-domain; Kühlbrandt, 2004; Missiaen et al., 2007; Fig. S1 A).
We have recently reported that immunoprecipitated SPCA1 from HeLa cell lysates coprecipitated with CFL-1 (von Blume et al., 2011). To further prove the physical interaction between these proteins, we immunoprecipitated CFL-1 from HeLa cell lysates. As a control, we incubated the cell lysates with an anti-rabbit IgG antibody. Endogenous CFL-1 interacts with SPCA1 and actin, whereas in the IgG immunoprecipitate no protein was detectable (Fig. S1 B). This interaction may be required for the activation of SPCA1 (von Blume et al., 2011).
CFL-1 directly interacts with a small region in the P-domain of SPCA1
The hypothesis is that actin and CFL-1 are required for the activation of SPCA1. However, it remained unclear how the cytosolic proteins gather at the TGN and what are the essential components required to facilitate this Ca2+ uptake and sorting. Therefore, we intended to gain a deeper understanding of the binding mechanism, the involvement of the components, and their role in facilitating TGN Ca2+ influx and cargo sorting.
First, we attempted to determine the domain of SPCA1 that interacts with CFL-1 and actin. To this end, we expressed and purified various cytosolic domains (SPCA1-L1, SPCA1-L2, SPCA1-L3, and SPCA1-L4) as well as the N and C termini (SPCA1-N-term and SPCA1-C-term) of SPCA1 as His-Sumo fusion proteins from Escherichia coli (Fig. S1 C). Analysis of the recombinant proteins by SDS-PAGE and Coomassie staining revealed that each domain is expressed and can be purified (Fig. S1 D).
Thereafter, we incubated the major three cytosolic domains known to be required to pump Ca2+ (SPCA1-N-term, A-domain; SPCA1-L1-, A-domain; and SPCA-L2, N- and P-domains; Fig. 1 A) coupled to nickel nitrilotriacetic acid (Ni-NTA) agarose beads with equal amounts of HeLa cell lysates. The beads were then washed, and bound proteins were eluted. Analysis of the samples by Western blotting with antibodies against CFL-1 and β-actin showed that CFL-1 and actin specifically interacted only with SPCA1-L2 (Fig. 1 A).
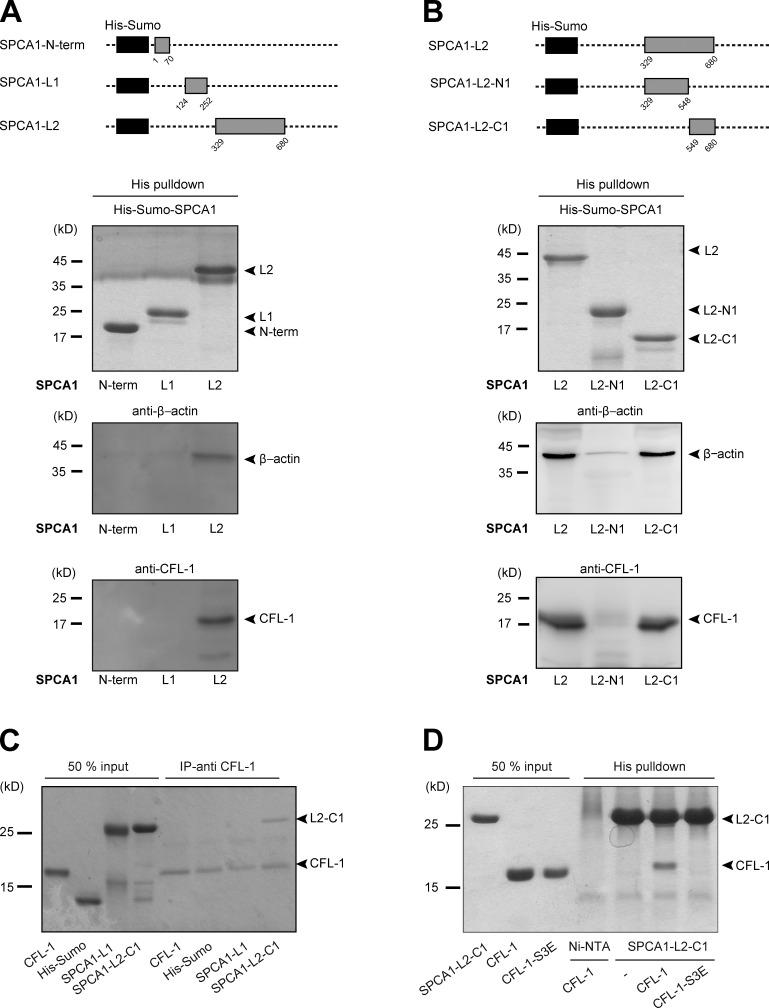
Figure 1.
CFL-1 directly interacts with a small domain of SPCA1 in vitro. (A, top) Schematic representation of the His-Sumo-SPCA1 expression constructs: SPCA1-N-term (aa 1–70), SPCA1-L1 (aa 124–252), and SPCA1-L2 (aa 329–680) comprise the A-, N-, and P-domain of SPCA1, which are the major cytosolic domains responsible for Ca2+ pumping activity. (A, bottom) To perform His pull-downs, E. coli purified His-Sumo-SPCA1 fusion proteins were immobilized on Ni-NTA agarose beads and incubated with equal amounts of HeLa cell lysates. Protein–bead complexes were analyzed by Western blotting using antibodies recognizing His tag, CFL-1, and β-actin. (B, top) To restrict the binding site of actin and CFL-1 to SPCA1-L2 to a smaller region, full-length L2 (aa 329–680) was subdivided into two fragments. The first part is referred to as SPCA1-L2-N1 (aa 329–548), the second part as SPCA1-L2-C1 (aa 549–680). (B, bottom) His pull-downs were performed and analyzed with recombinant His-Sumo-SPCA1-L2 full length, -SPCA1-L2-N1, and -SPCA1-L2-C1 as described in A. (C) His-Sumo-CFL-1 was expressed in E. coli, purified, and the His-tag was subsequently cleaved. CFL-1 was incubated with a CFL-1 antibody and immobilized on agarose beads. Then, His-Sumo tag, His-Sumo-SPCA1-L1, or His-Sumo-SPCA1-L2-C1 were added. Proteins were eluted and separated by SDS-PAGE and stained with Coomassie blue. (D) CFL-1 and mutant CFL-1-S3E constructs were expressed, purified, and cut from His-Sumo tag. Recombinant proteins were incubated with purified His-Sumo-SPCA1-L2-C1 attached to Ni-NTA beads. Proteins were eluted and analyzed by SDS-PAGE and Coomassie blue staining.
To narrow down the binding site of actin and CFL-1 to a smaller domain in SPCA1-L2, we subdivided it into SPCA1-L2-N1 (aa 329–548) and SPCA1-L2-C1 (aa 549–680; Fig. 1 B). After expression and purification, equal amounts of protein were incubated with HeLa cell lysates and processed as described in the Materials and methods. Actin and CFL-1 specifically bound to SPCA1-L2-C1 but not to SPCA1-L2-N1 (Fig. 1 B).
Next, we assessed the presence of a direct interaction between SPCA1 and CFL-1. We expressed and purified His-Sumo-CFL-1 from E. coli. Subsequently, the His-Sumo tag was cleaved, and untagged CFL-1 was separated by gel filtration. The purified protein was then incubated with a CFL-1 antibody and immobilized on agarose beads. The beads were mixed with purified His-Sumo, His-Sumo-fused SPCA1-N-term, SPCA1-L1, and SPCA1-L2-C1. After washing the beads, bound proteins were eluted and separated by SDS-PAGE. Coomassie blue staining of the gel showed that CFL-1 directly interacts with L2-C1 but not with the N-term or L1 (Fig. 1 C).
Active CFL-1 recruits F-actin to SPCA1 in vitro
We have recently reported that the actin-severing activity of CFL-1 is crucial for its localization to the TGN as well as for the facilitation of secretory cargo sorting (von Blume et al., 2009, 2011). Phosphorylation of CFL-1 at Serine 3 (Ser3) by LIM kinase (LIMK) inactivates CFL-1, whereas dephosphorylation reactivates it (Arber et al., 1998). Furthermore, the phosphorylation of CFL-1 at this side impairs its ability to bind to F-actin (Agnew et al., 1995).
To test if inactive CFL-1 binds to SPCA1 in vitro, we expressed and purified a phosphomimetic mutant of CFL-1 (CFL-1-S3E) in E. coli. His-Sumo-SPCA-L2-C1 bound to Ni-NTA beads were then incubated with either recombinant CFL-1 or CFL-1-S3E. Analysis of the bound proteins by SDS-PAGE and Coomassie blue staining revealed that His-Sumo-SPCA1-L2-C1 interacted with CFL-1 but not with inactive CFL-1-S3E (Fig. 1 D). These findings are in line with the observation that active CFL-1 localized to the TGN, whereas the inactive CFL-1 did not (von Blume et al., 2011). These data demonstrate that CFL-1 activity is required for binding to SPCA1 and thus recruitment to the TGN.
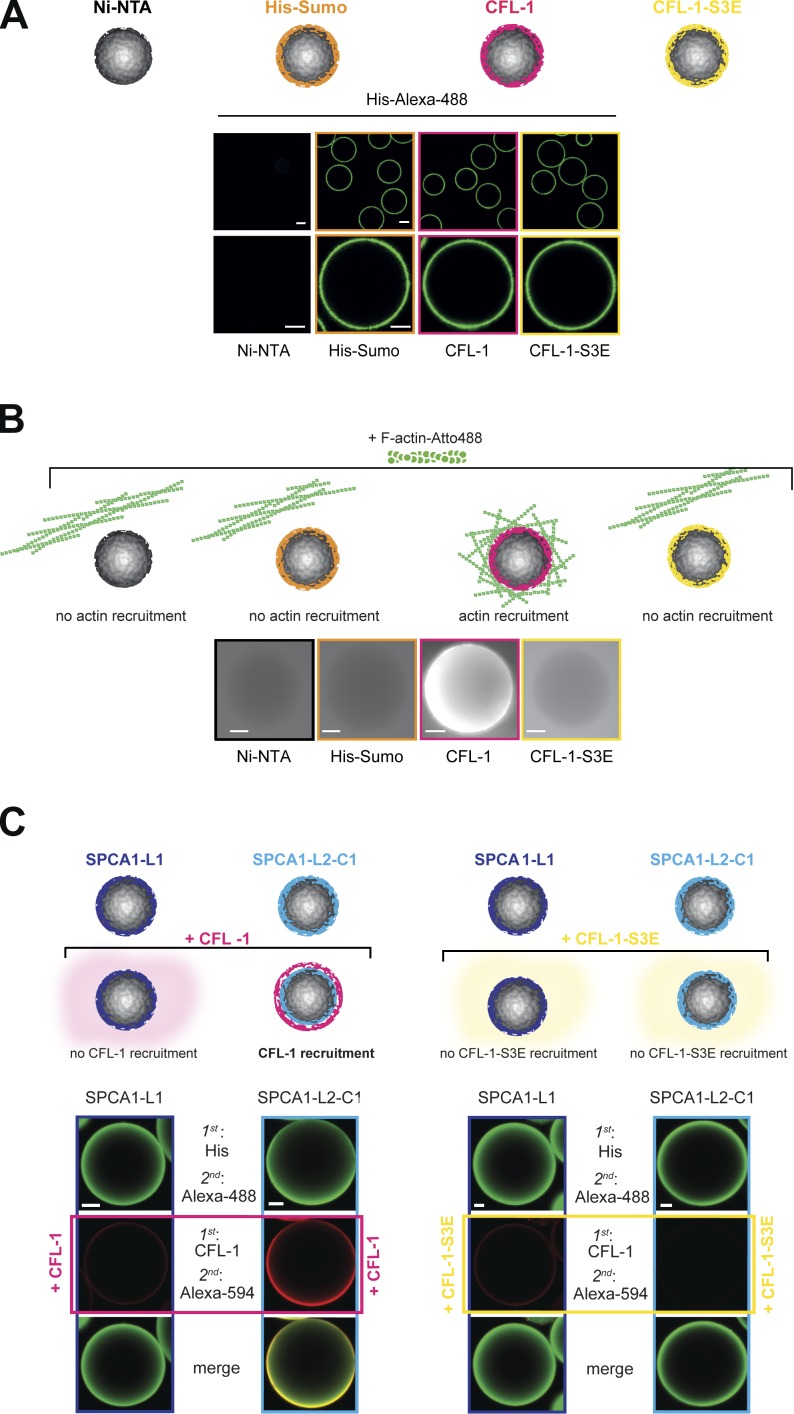
To elucidate how F-actin binds to the SPCA1–CFL-1 complex, we established a reconstitution assay to visualize the process. To set up the assay, we attached His-Sumo, His-Sumo-CFL-1-S3E, and His-Sumo-CFL-1 to Ni-NTA beads. As a negative control, we incubated Ni-NTA beads with BSA. To visualize the proteins, beads were incubated with an anti-His antibody and an Alexa Fluor 488–labeled secondary antibody (Fig. 2 A), and analyzed by fluorescence microscopy. The His-tagged proteins were visible on the beads in equal amounts, whereas the BSA-incubated Ni-NTA beads did not show a fluorescent signal (Fig. 2 A). To test if actin can be recruited to CFL-1 under these conditions, we incubated the beads with fluorescently labeled F-actin and monitored them by fluorescence microscopy. F-actin strongly associated to His-Sumo-CFL-1 beads but not to beads alone, His-Sumo, or His-Sumo-CFL-1-S3E (Fig. 2 B).
Figure 2.
Establishment of an in vitro reconstitution assay. (A) Establishment of the F-actin bead assay with controls: Ni-NTA beads, His-Sumo, His-Sumo-CFL-1, or mutant His-Sumo-CFL-1-S3E protein were attached to Ni-NTA beads (also depicted in the scheme). Immunofluorescence staining was performed using a primary anti-His antibody and a secondary Alexa Fluor 488 antibody to visualize the recombinant proteins on beads. (B) Immobilized proteins on beads were incubated with polymerized Atto-488 actin (also depicted in the scheme), and the actin signal was monitored by epifluorescence microscopy. In case F-actin was recruited, a bright fluorescent signal would have been detectable on the surface of the bead. Only immobilized CFL-1 recruited F-actin to beads. (C) His-Sumo-SPCA1-L1 or His-Sumo-SPCA1-L2-C1 were attached to beads and subsequently incubated with either recombinant CFL-1 or CFL-1-S3E without the His-Sumo tag, as depicted in the scheme. Staining with His or CFL-1, Alexa Fluor 488– or Alexa Fluor 594–specific antibodies, and subsequent confocal imaging revealed that CFL-1 binds SPCA1-L2-C1 exclusively, whereas CFL-1-S3E failed to bind. Bars, 10 µm.
Next, we investigated the interaction between SPCA1-L2-C1 and CFL-1 on the surface of the Ni-NTA beads. To perform the experiment, we incubated His-Sumo-SPCA1-L1 and -SPCA1-L2-C1–coated Ni-NTA beads with recombinant CFL-1 or CFL-1-S3E, respectively, and the proteins were visualized with anti-His and anti–CFL-1 antibodies followed by Alexa Fluor 488 and Alexa Fluor 594 secondary antibody staining. CFL-1 was only detectable on His-Sumo-SPCA1-L2-C1 but not on SPCA1-L1 beads. Furthermore, CFL-S3E was not detectable on His-Sumo-SPCA1-L1 beads nor on His-Sumo-L2-C1 beads (Fig. 2 C). These observations further confirmed our pull-down experiments that showed that SPCA1-L2-C1 directly interacts with CFL-1 but not with CFL-1-S3E.
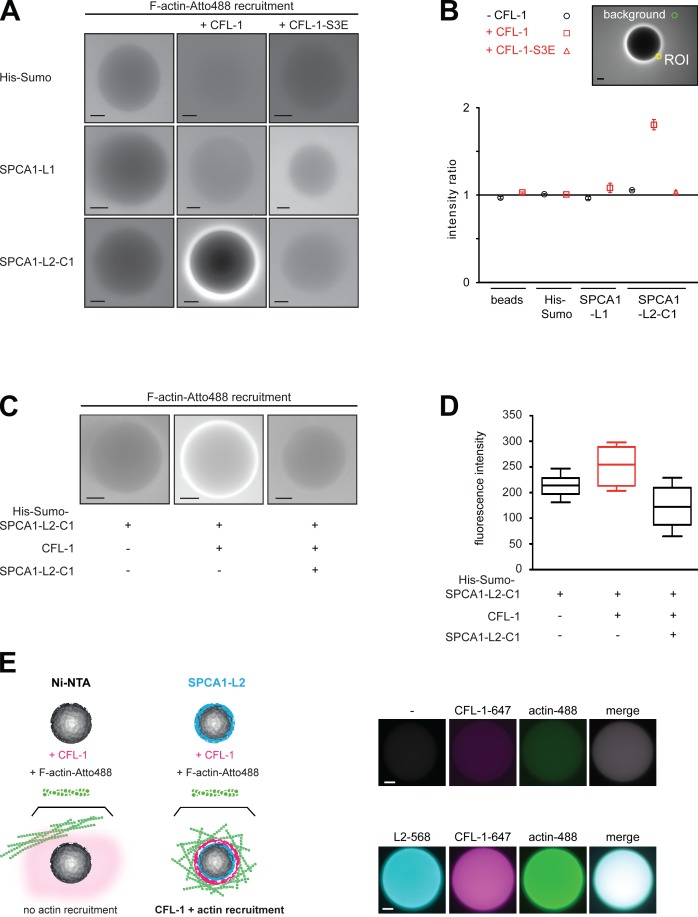
To test the actin recruitment to the SPCA1 domains in the presence or absence of CFL-1 as well as CFL-1-S3E, the proteins were attached to the beads and incubated with fluorescently labeled actin as described in the Materials and methods. Actin did not associate to His-Sumo, or to His Sumo-SPCA1-L1 in either the absence or the presence of CFL-1 or CFL-1-S3E (Fig. 3 A). In contrast, although His-Sumo-SPCA1-L2-C1 did not interact with actin in the absence of CFL-1 or in the presence of CFL-1-S3E, the actin filaments were strongly recruited after recombinant CFL-1 was added (Fig. 3 A). To quantify our results, we examined 10 beads in three independent experiments and quantified the amount of labeled actin recruited to the beads by calculating the fluorescence ratio of the area close to the bead surface to the intensity of the background (Fig. 3 B).
Figure 3.
F-actin is recruited to SPCA1-L2-C1 via CFL-1 in vitro. (A) His-Sumo tag, His-Sumo-SPCA1-L1, and His-Sumo-SPCA1-L2-C1 attached to Ni-NTA beads were incubated with recombinant CFL-1 or CFL-1-S3E in the presence of growing actin filaments labeled with Atto-488. Actin recruitment was assessed by fluorescence microscopy. (B) For the quantification the mean intensity of a circular region of interest (ROI), 12 pixels in diameter adjacent to the rim of the bead and 1 in the background were measured. The ratio of these two intensities is plotted for each experimental condition. A ratio of 1 indicates no increase of fluorescent signal over the background. Error bars represent SEM. n = 20. (C) His-Sumo-SPCA1-L2-C1 on beads was either incubated with CFL-1, actin, and KMEI buffer or with CFL-1 preincubated with untagged SPCA1-L2-C1, actin, and KMEI buffer. (D) The quantification was performed by measuring the absolute fluorescence intensities of a circular region of ROI, 12 pixels in diameter adjacent to the rim of the bead. Error bars represent SEM. n = 20. (E) To visualize the binding of His-Sumo-SPCA1-L2, CFL-1, and actin to the beads (scheme, left), full-length SPCA1-L2 and CFL-1 were labeled with fluorophores. Alexa-568-SPCA1-L2 was immobilized on beads as described earlier and incubated with Alexa-647-CFL-1 and Atto-488 actin induced to polymerize by adding KMEI buffer. As negative controls, Ni-NTA beads were incubated with labeled CFL-1 and actin. Bars, 10 µm.
To further substantiate the mechanism of F-actin recruitment to SPCA1-L2-C1 via CFL-1, we performed a competition experiment. SPCA1-L2-C1 attached to Ni-NTA agarose beads was incubated with a mixture of CFL-1 and soluble untagged SPCA1-L2-C1 and actin filaments as described in the previous paragraph. Actin filaments were strongly recruited to SPCA1-L2-C1 only in the presence of CFl-1, as described previously. This recruitment was strongly impaired when soluble SPCA1-L2-C1 was added to the mixture (Fig. 3, C and D).
To directly visualize the process of CFL1/actin recruitment (Fig. 3 E, scheme), recombinant SPCA1-L2 and CFL-1 were chemically labeled with Alexa Fluor 647 and 568 maleimides, respectively, and the reconstitution assay was performed as described earlier. After incubation of the different components, three additional washing steps were performed to remove the background. As depicted in Fig. 3 E, SPCA1-L2-Alexa-568 was immobilized on Ni-NTA beads, and recombinant CFL-1-Alexa-647 as well as Atto-488-actin were recruited to the beads. Beads incubated with CFL-1-Alexa-647 and Atto-488-actin in the absence of SPCA1-L2-Alexa-568 showed neither CFL-1 nor actin recruitment. These results further illustrate the presence and the order of recruitment of the proteins to form the complex.
Collectively, these data demonstrated that active CFL-1 is required for recruitment of F-actin to SPCA1 in vitro.
Overexpression of SPCA1-L2-C1 in HeLa cells inhibits secretory cargo sorting
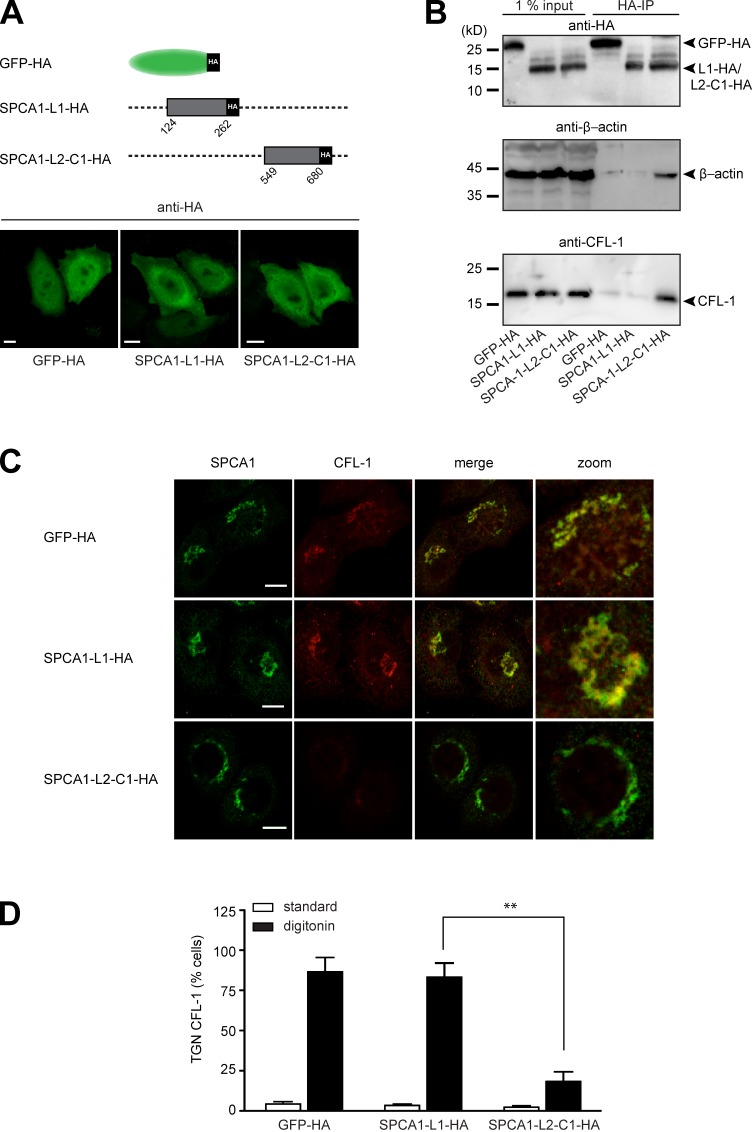
We reported previously that actin and CFL-1 interact with SPCA1 at the TGN to facilitate Ca2+-dependent secretory cargo sorting (von Blume et al., 2009, 2011). Therefore, we aimed to investigate whether the interaction between CFL-1 and SPCA1-L2-C1 is crucial for this process. To address this, we generated stable cell lines transduced with either GFP-HA as a control, SPCA1-L1-HA, or SPCA1-L2-C1-HA (Fig. 4 A, scheme). First, we analyzed the localization of the proteins by immunofluorescence microscopy. The cells overexpressing GFP-HA, L1-HA, or L2-C1-HA were fixed and stained with an antibody against HA. All the HA-tagged proteins showed a comparable localization in the cytosol or the nucleus (Fig. 4 A).
Figure 4.
The interaction of CFL-1 and actin with SPCA1-L2-C1 is crucial for protein sorting at the TGN. (A, top) Schematic representation of GFP-HA (aa 1–239), SPCA1-L1-HA (aa 124–252), and SPCA1-L2-C1-HA (aa 549–680) fusion constructs cloned into the retroviral expression vector pLPCX. (A, bottom) HeLa cells stably transfected with GFP-HA, SPCA1-L1-HA, or SPCA1-L2-C1-HA were visualized with an HA antibody and analyzed by confocal microscopy. (B) HeLa cells stably transfected with GFP-HA, SPCA1-L1-HA, or SPCA1-L2-C1-HA were lysed, and lysates were incubated with µMACS anti-HA magnetic microbeads. GFP-HA–, SPCA1-L1-HA–, and SPCA1-L2-C1-HA–associated proteins were eluted, separated by SDS-PAGE, and analyzed by Western blotting using HA, CFL-1, or β-actin antibodies, respectively. (C) HeLa cells stably expressing GFP-HA, SPCA1-L1-HA, and SPCA1-L2-C1-HA were incubated for 2 h at 20°C and subsequently permeablized with digitonin, washed, and then fixed with formaldehyde before incubation with anti-SPCA1 (green) or anti–CFL-1 (red) antibodies. (D) TGN localization of CFL-1 under different conditions in Triton X-100–permeabilized (images in Fig. S2) versus digitonin-permeabilized cells was determined by counting 100 cells per condition in three independent experiments. Bars, 5 µm.
Next, we attempted to confirm the binding of endogenous CFL-1 and actin to SPCA1-L2-C1-HA. To perform the experiment, lysates of HeLa cells expressing GFP-HA, SPCA1-L1-HA, or SPCA1-L2-C1-HA were incubated with HA antibody–coated magnetic microbeads, and GFP-HA–, SPCA1-L1-HA–, and SPCA1-L2-C1-HA–associated proteins were purified and separated by SDS-PAGE. Proteins were analyzed by Western blotting with anti-HA to confirm the presence of the bait proteins, an anti–β-actin, and an anti– CFL-1 antibody. As demonstrated in Fig. 4 B, GFP-HA, SPCA1-L1-HA, and SPCA1-L2-C1-HA were enriched in the IP fractions. Endogenous actin and CFL-1 coprecipitated with SPCA1-L2-C1-HA but not with SPCA1-L1-HA or GFP-HA. This result further confirmed that SPCA1-L2-C1 also interacts with actin and CFL-1 in living cells.
We reported previously that when secretory cargo is arrested in the TGN by incubating the cells at 20°C for 2 h, a pool of CFL-1 is recruited to the TGN. This pool can be visualized in cells that are permeabilized before fixation (von Blume et al., 2011). We aimed to determine if the overexpression of SPCA1-L2-C1 influences the localization of endogenous, active CFL-1 at the TGN. To perform the experiment, HeLa cells expressing GFP-HA, SPCA1-L1-HA, or SPCA1-L2-HA were incubated at 20°C for 2 h. Cells were subsequently permeabilized with digitonin and washed to remove the cytosolic proteins before fixation procedure. Staining the cells with antibodies against CFL-1 and SPCA1 revealed that CFL-1 colocalized with SPCA1 in cells expressing GFP-HA and SPCA1-L1-HA (Fig. 4 C). Strikingly, CFL-1 did not colocalize with SPCA1 in cells overexpressing HA-SPCA1-L2. The effect of GFP-HA, SPCA1-L1-HA, and SPCA1-L2-C1-HA overexpression on CFL-1 localization in cells that were not permeabilized before fixation (Fig. S2) or digitonin-treated cells was quantified by counting 100 cells in three different experiments (Fig. 4 D).
These findings suggested that cytosolic SPCA1-L2-C1-HA captures the TGN pool of CFL-1 (Fig. 4, C and D) and are in line with the in vitro observation that excess amounts of SPCA1-L2-C1 inhibit the binding of CFL-1 to SPCA1 (Fig. 3 C).
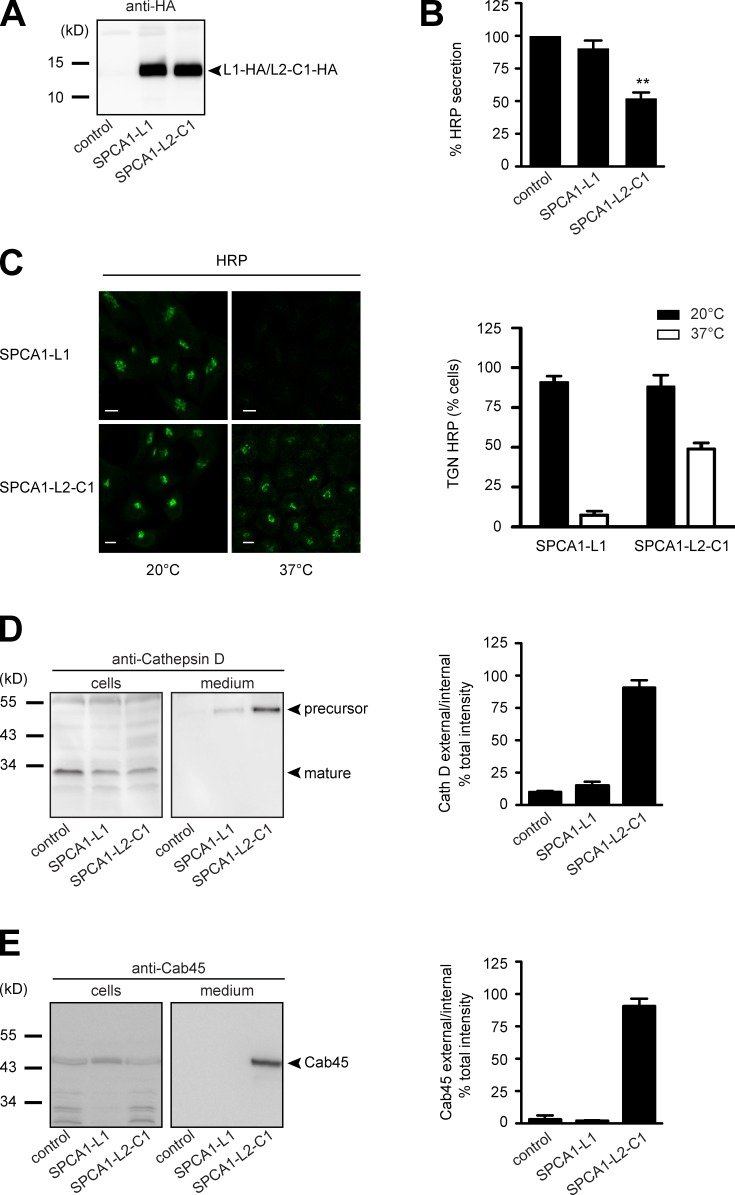
Next, we wanted to know the functional consequence of SPCA1-L2-C1 overexpression for TGN protein sorting in these cells. In recent years we have established robust assays to determine CFL-1– and SPCA1-dependent TGN sorting (von Blume et al., 2009, 2011, 2012). First, HeLa cells stably expressing signal sequence HRP (ss-HRP) were transduced with a control plasmid, SPCA1-L1-HA, or SPCA1-L2-C1-HA. To test the expression of the HA-tagged proteins, lysates were analyzed by Western blotting with an HA antibody. The cells expressed SPCA1-L1-HA and SPCA1-L2-C1-HA at similar levels (Fig. 5 A). The HRP assay was performed by incubating the cells with serum-free medium, and the secretion of HRP was measured by chemiluminescence as described previously (Bard et al., 2006).
Figure 5.
The interaction of CFL-1 and actin to SPCA1-L2-C1 is crucial for protein sorting at the TGN. (A) HeLa cells expressing ss-HRP were transduced with a pLPCX plasmid (control), SPCA1-L1-HA, or SPCA1-L2-C1-HA. Cells were subsequently lysed and proteins were separated by SDS-PAGE. The presence of SPCA1-L1-HA and SPCA1-L2-C1-HA was determined by Western blotting with an anti-HA antibody. (B) Cell culture supernatants of cells described in A were analyzed for HRP activity by chemiluminescence. Error bars show mean ± SD of external HRP activity normalized to internal HRP activity of three independent experiments. Datasets were statistically significant when P < 0.01 (**). (C) HeLa cells stably expressing SPCA1-L1-HA or SPCA1-L2-C1-HA were incubated at 20°C for 2 h in the presence of cycloheximide to accumulate HRP in the TGN. Cells were subsequently shifted to 37°C, and the localization of HRP was analyzed by fluorescence microscopy with an anti-HRP antibody. To quantify the results, 100 cells expressing SPCA1-L1-HA or SPCA-L2-C1-HA in three different experiments at 20°C and 37°C were counted. Bars, 5 µm. (D and E) Media and lysates from the same cells were Western blotted with specific antibodies against Cathepsin D (D) or Cab45 (E). Western blots from three independent experiments were quantified by densitometry using the ImageJ software. Bar graphs represent the densitometry values of external Cathepsin D (D) and Cab45 (E) normalized to internal Cathepsin D and Cab45 values, respectively.
The overexpression of SPCA-L2-C1-HA significantly decreased ss-HRP secretion, whereas control or SPCA1-L1-HA–expressing cells did not show a significant reduction of secretion (Fig. 5 B). To test the localization of HRP under the different conditions, control vector–transduced HeLa cells, or SPCA1-L1-HA– or SPCA1-L2-C1-HA–expressing HeLa cells, were incubated at 20°C to arrest the secretory proteins in the TGN in the presence of cycloheximide. Subsequently, the cells were shifted to 37°C to allow cargo exit from the TGN. Cells were then fixed and stained with an HRP antibody and analyzed by confocal microscopy, as shown previously (von Blume et al., 2009, 2011). At 20°C, HRP was detectable in the TGN in SPCA-L1-HA– and SPCA-L2-C1-HA–expressing cells, whereas at 37°C, HRP was released from the TGN in cells expressing SPCA1-L1-HA but was still accumulated in the TGN in 50% of cells expressing SPCA1-L2-C1-HA (Fig. 5 C). To quantify the results, 100 cells per condition were counted in three independent experiments (Fig. 5 C, histogram).
To monitor sorting defects of endogenous proteins, we investigated cell culture supernatants from control cells, SPCA1-L1-HA–expressing cells, or SPCA1-L2-C1-HA–expressing cells for the presence of the TGN precursor form of Cathepsin D. Cathepsin D is synthesized as a precursor polypeptide of 53 kD, which is subsequently sorted to lysosomes and further converted into a 47-kD intermediate and a 31-kD mature form (Gieselmann et al., 1985; Zaidi et al., 2008). The presence of this protein in the cell culture supernatant indicates a sorting defect, and the sorting of this protein was previously shown to be dependent on SPCA1, actin, and CFL-1 (von Blume et al., 2011). Analysis of the medium by Western blotting showed that HeLa cells overexpressing SPCA1-L2-C1-HA secrete the TGN precursor form of Cathepsin D, whereas control or SPCA1-L1-HA–overexpressing cells did not show aberrant secretion of Cathepsin D (Fig. 5 D). The same samples were also probed for the presence of the Golgi resident protein Cab45. This protein is required for secretory cargo sorting in the TGN and is released from Golgi membranes when Ca2+ in the lumen of the TGN decreases. Cab45 is secreted when SPCA1 is depleted by siRNA (von Blume et al., 2011). The same phenotype was observed when SPCA1-L2-C1-HA was overexpressed but not in the other two cell lines (Fig. 5 E).
These data indicated that overexpressed cytosolic SPCA1-L2-C1 most likely sequesters crucial interaction partners that are required for the pump function of SPCA1.
Overexpression of SPCA1-L2-C1 in HeLa cells impairs TGN Ca2+ uptake
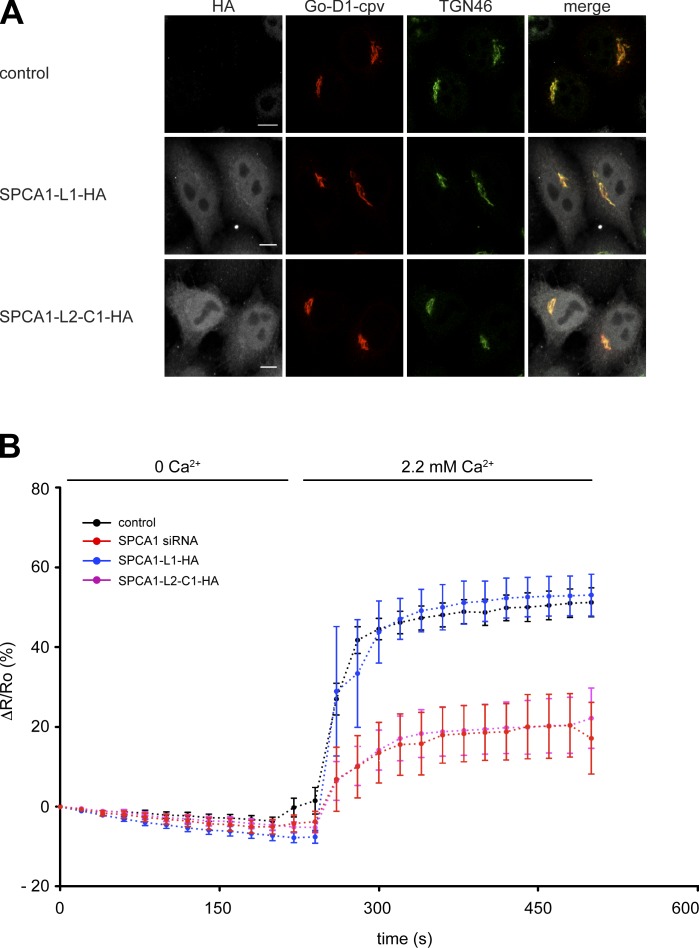
The results of the sorting assays strongly suggested that the overexpression of SPCA1-L2-C1 interferes with SPCA1 function by capturing the necessary interaction partners required for activation and, thus, sorting. To test this hypothesis, we performed Ca2+ measurements with a TGN-targeted Ca2+ FRET sensor (Lissandron et al., 2010). TGN Ca2+ uptake solely relies on SPCA1, as demonstrated before (Lissandron et al., 2010). HeLa cells were transfected with a control plasmid, SPCA1 siRNA, SPCA1-L1-HA, or SPCA1-L2-C1-HA in combination with the Go-D1-cpv sensor to measure the TGN Ca2+ uptake of the TGN, as described previously (von Blume et al., 2011). The coexpression of Go-D1-cpv and SPCA1-L1-HA or SPCA1-L2-C1-HA was confirmed by immunofluorescence microscopy (Fig. 6 A).
Figure 6.
Overexpression of SPC1-L2-C1 inhibits TGN Ca2+ influx. (A) HeLa cells were transfected with the TGN-specific Ca2+ FRET sensor Go-D1-cpv and a control plasmid, SPCA1-HA, or SPCA1-L2-C1-HA. Cells were fixed, stained with anti-HA and anti-TGN46 antibodies, and analyzed by fluorescence microscopy. Bars, 5 µm. (B) HeLa cells were transfected with Go-D1-cpv and a control plasmid, or SPCA1 siRNA and a control plasmid, and either SPCA1-L1-HA or SPCA1-L2-C1-HA, respectively. Ca2+ entry into the TGN was measured in Ca2+-depleted cells transfected with control plasmid (n = 23), SPCA1 siRNA (n = 9), SPCA1-L1-HA, or SPCA1-L2-C1-HA (n = 11). Fluorescent signals reflecting TGN [Ca2+] were presented as ΔR/R0, where R0 is the value obtained before addition of 2.2 mM Ca2+ to the cell’s bathing solution. Data are expressed as the mean ± SEM. Mean maximum values measured after readdition of Ca2+ were statistically different between control/SPCA1-L1-HA and SPCA-L2-C1-HA–transfected cells.
In agreement with previous results (von Blume et al., 2011), the knockdown of SPCA1 significantly reduced the Ca2+ uptake into the TGN compared with control cells (von Blume et al., 2011). Remarkably, a similar result was obtained in cells overexpressing SPCA1-L2-C1-HA. However, there was no significant change in the Ca2+ uptake when SPCA1-L1-HA was overexpressed (Fig. 6 B). These data strongly suggested that the formation of the complex between SPCA1, CFL-1, and actin is required for higher SPCA1 Ca2+ pumping activity at the TGN.
Overexpression of CFL-1-S3E that does not bind to SPCA1 impairs sorting and TGN Ca2+ uptake
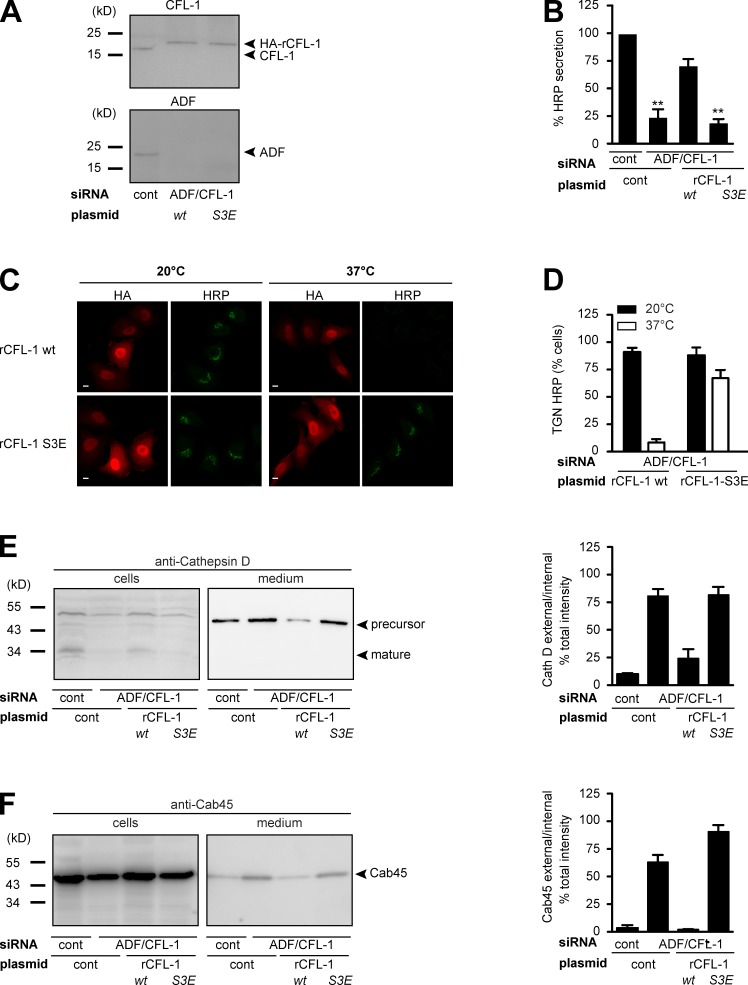
To further substantiate the significance of the formation of the complex for sorting and TGN Ca2+ entry, we aimed at elucidating the consequence of impaired CFL-1 and actin binding to SPCA1. As described previously, SPCA1 binds to active CFL-1, and this binding recruits CFL-1 to the TGN (von Blume et al., 2011). We have also reported previously that inactivation of CFL-1 by overexpression of LIMK, which phosphorylates CFL-1, or knockdown of slingshot, which dephosphorylates CFL-1, interferes with protein secretion (von Blume et al., 2009). To test if an inactive CFL-1 phosphomimetic S3E mutant that does not bind to SPCA1 interferes with Ca2+-dependent protein sorting, HeLa cells were transfected with control or ADF/CFL-1 siRNAs as well as a siRNA-resistant HA-tagged rat CFL-1 (rCFL-1) or rCFL-1-S3E, respectively. To test the knockdown and the expression of CFL-1 and the siRNA-resistant mutants, lysates were analyzed by Western blotting with CFL-1 and ADF antibodies, respectively. ADF/CFL-1 was successfully depleted by siRNAs, and overexpression of HA-rCFL-1 wild type (wt) and S3E in ADF/CFL-1–depleted cells occurred at similar levels (Fig. 7 A).
Figure 7.
Expression of CFL-1-S3E that does not bind to SPCA1 and actin causes missorting of secretory cargo. (A) HeLa cells were transfected with control or ADF and CFL-1 siRNA. In parallel, ADF and CFL-1 knockdown HeLa cells were transfected with siRNA-resistant rat HA-tagged CFL-1 (rCFL-1) wt or rCFL-1-S3E plasmids, and analyzed by SDS-PAGE and Western blotting with CFL-1 and ADF antibodies. (B) Cells described in A were transfected with ss-HRP-Flag. Medium from these cells was analyzed for HRP secretion. Error bars indicate the mean SD of HRP activity in the medium normalized by HRP activity in cell lysates collected from three independent experiments. (C) HeLa cells transfected with ADF/CFL-1 siRNA, ss-HRP-Flag, and HA-rCFL-1 wt or rCFL-1-S3E were incubated at 20°C for 2 h in the presence of cycloheximide to arrest HRP in the TGN. Cells were then shifted to 37°C, and the localization of HRP was analyzed with an HRP antibody in confocal microscopy. Bars, 5 µm. (D) 100 cells expressing ss-HRP-Flag and HA-rCFL-1 wt or rHA-CFL-1-S3E were counted in three different experiments at 20°C and 37°C. HeLa cells were transfected with control and ADF/CFL-1 siRNA and incubated for 40 h. Then ADF/CFL-1 knockdown cells were either transfected with a control plasmid or with rCFL-1 wt or rCFL-1-S3E. (E and F) Media and lysates from these cells were Western blotted with Cathepsin D (E) and Cab45 (F) antibodies. Western blots from three independent experiments were quantified by densitometry. Bar graphs represent the densitometry values of external Cathepsin D (E) and Cab45 (F) normalized to internal Cathepsin D and Cab45 values, respectively.
The knockdown of ADF/CFL-1 induces missorting of secretory proteins at the TGN (von Blume et al., 2009). To test the consequence of the overexpression of rCFL-S3E that does not bind to SPCA1 on secretory cargo sorting, HeLa cells were transfected with control or ADF/CFL-1 siRNA and, subsequently, Flag-ss-HRP. In addition, a control plasmid, HA-rCFL-1 wt, or rHA-rCFL-1-S3E were transfected as described in the previous paragraph. Cells and cell culture supernatants were collected and HRP activity was measured as described previously (Bard et al., 2006). Knockdown of ADF/CFL-1 impaired HRP secretion around 75%, which could be successfully rescued by expression of rCFL-1 wt but not rCFL-1-S3E (Fig. 7 B). To test the localization of HRP under the different conditions, HeLa cells treated with ADF/CFL-1 siRNA and transfected with HA-rCFL-1 or HA-rCFL-1-S3E were incubated at 20°C as described earlier. After shifting the cells to 37°C, in order to allow cargo exit from the TGN, cells were fixed and prepared for fluorescence microscopy with anti-HA and anti-HRP antibodies (von Blume et al., 2009, 2011). At 20°C, HRP was arrested in the TGN of cells expressing HA-rCFL-1 and HA-rCFL-1-S3E, respectively. After incubation at 37°C, HRP was released from the TGN in cells expressing HA-rCFL-1 but was still accumulated in the TGN in 70% of cells expressing HA-CFL-1-S3E (Fig. 7 C). To quantify the results, 100 cells per condition were counted in three independent experiments (Fig. 7 D). The same cells were also tested for Cathepsin D secretion (Fig. 7 E). HeLa cells lacking ADF/CFL-1 secreted the precursor of Cathepsin D, and the expression of HA-rCFL-1 rescued the phenotype, whereas HA-rCFL-S3E expression failed to do so. Similar results were obtained when cells and cell culture supernatants were analyzed with an anti-Cab45 antibody (Fig. 7 F). Cab45 was secreted from cells in the absence of ADF/CFL-1, and this was rescued when rHA-CFL-1, but not rHA-CFL1-S3E, was expressed.
These data point out that actin and CFL-1 binding to SPCA1 are indispensable to drive the sorting reaction at the TGN.
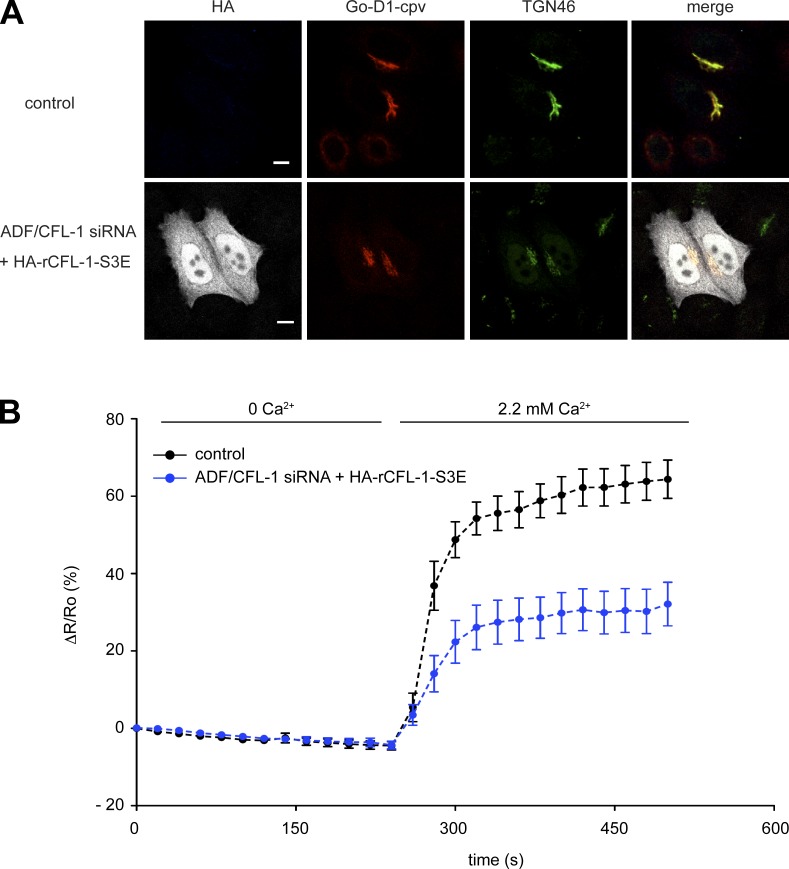
The next question that we aimed to address was whether the expression of the actin and SPCA1 binding–deficient mutant has an effect on TGN Ca2+ uptake. We have shown previously that the knockdown of ADF/CFL-1 significantly impairs TGN Ca2+ uptake. This phenotype could be rescued by the expression of siRNA-resistant rCFL-1 wt (von Blume et al., 2011). To test the effect of CFL-S3E on TGN Ca2+ uptake, HeLa cells were treated with ADF/CFL-1 siRNAs and transfected with either Go-D1-cpv and a control plasmid or HA-rCFL-1-S3E, and processed for immunofluorescence microscopy (Fig. 8 A). The TGN Ca2+ uptake was significantly reduced in ADF/CFL-1–depleted cells expressing HA-rCFL-1-S3E compared with control cells (Fig. 8 B). These data further demonstrate that the binding between SPCA1, actin, and CFL-1 is crucial for Ca2+ pumping at the TGN.
Figure 8.
Expression of CFL-1-S3E inhibits Ca2+ entry into the TGN. (A) HeLa cells were transfected with ADF/CFL-1 siRNA and transfected with Go-D1-cpv and a control plasmid or HA-rCFL-S3E, then processed for immunofluorescence microscopy with anti-HA and anti-TGN46 antibodies. Bars, 5 µm. (B) HeLa cells were transfected with ADF/CFL-1 siRNA and, subsequently, with Go-D1-cpv and a control plasmid or HA-rCFL-S3E. Ca2+ entry into the TGN was measured in Ca2+-depleted cells transfected with control plasmid (n = 23) or rCFL-1-S3E (n = 14). Fluorescent signals reflecting TGN [Ca2+] were presented as ΔR/R0, where R0 is the value obtained before addition of 2.2 mM Ca2+ to the cell’s bathing solution. Data are expressed as the mean ± SEM (error bars). Mean maximum values measured after readdition of Ca2+ were statistically different between control plasmid/HA-rCFL-1-S3E–transfected cells.
Mutation of four conserved amino acids in SPCA1 impairs secretory cargo sorting and TGN Ca2+ uptake
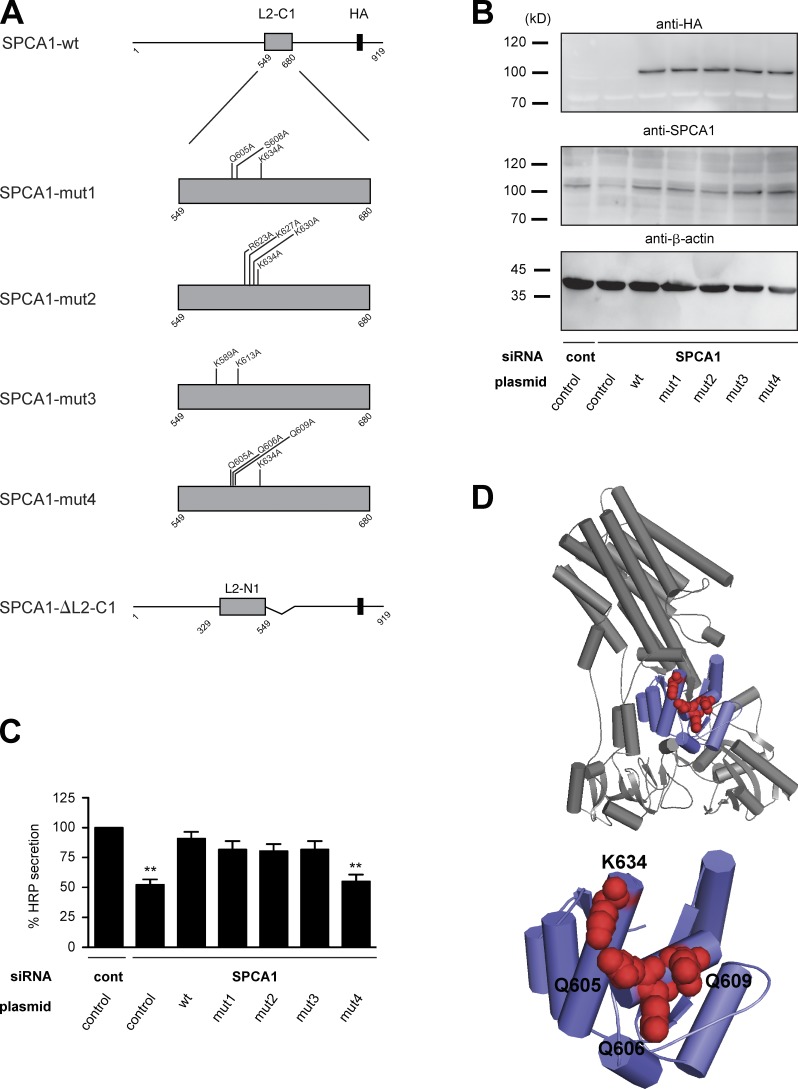
To finally prove that the observed phenotype is directly caused by the loss of CFL-1 binding to SPCA1, we aimed at identifying the crucial amino acids in SPCA1 required for CFL-1 binding. It was reported that CFL-1 binds to and activates the related rat Na+/K+-ATPase (ATP2A2) in a domain that corresponds to L2-C1 in SPCA1 (Lee et al., 2001; Kim et al., 2002). Based on these studies, two residues are required for the functional interaction: D672, which according to the alignment of SPCA1 with ATP2A2 corresponds to S608 in human SPCA1; and R700, which is equivalent to K634 (Fig. S3). However, neither a single nor a double substitution to alanine (S608A/K634A) impaired TGN Ca2+ uptake and secretory cargo sorting (unpublished data). We further reasoned that, potentially, a charged patch rather than a single amino acid could be responsible for CFL-1 binding. We therefore generated a structural model of SPCA1 to analyze the surface accessibility of charged residues in the CFL-1 binding domain (L2-C1). Based on the model structure, we decided to add Q605A to the S608A/K634A mutations (mut1) because this residue is strongly charged and lies in close proximity to S608. In addition, three additional patches were selected for mutagenesis (Fig. 9 A; mut2, R623A/K627A/K630A/K634A; mut3, K589A/K613A; mut4, Q605A/Q606A/Q609A/K634A). Moreover, we generated a L2-C1 deletion mutant (Fig. 9 A). First, stable HeLa cell lines expressing siRNA-resistant SPCA1-wt, -ΔL2-C1, or mutants (-mut1, -mut2, -mut3, -mut4) were generated, and the localization of SPCA1-HA was observed by immunofluorescence microscopy. All the point mutants localized correctly to the TGN, whereas the deletion mutant (ΔL2-C1) accumulated in the ER (Fig. S4). This mutant was therefore not further analyzed.
Figure 9.
Screen for CFL-1 binding deficient SPCA1 mutants. (A) Schematic representation of wt and SPCA1 mutants (mut1, mut2, mut3, mut4, and ΔL2-C1) used for screening. (B) HeLa cells stably expressing ss-HRP and siRNA-resistant SPCA1-wt or mutants were transfected with control or SPCA1 siRNA, and cell lysates were analyzed by Western blotting with HA, SPCA1, and β-actin antibodies. (C) Medium and cells were analyzed for HRP activity. Error bars indicate the mean SD of HRP activity in the medium normalized by HRP activity in cell lysates collected from three independent experiments. (D) Structural model of human SPCA1 (top). The CFL-1 binding domain (SPCA1-L2-C1) is displayed in purple. The four residues required for CFL1 binding are indicated as red spheres (bottom).
Our first aim was to screen for a mutant that is not able to rescue the SPCA1 knockdown–dependent sorting phenotype. To perform the experiment, cell lines stably expressing ss-HRP, ss-HRP/SPCA1-HA-wt, or ss-HRP with point mutants (-mut1, -mut2, -mut3, -mut4) were treated with control or SPCA1 siRNAs. The HeLa cell lines transduced with siRNA-resistant SPCA1-wt and mutants expressed the HA-tagged proteins at similar levels (Fig. 9 B). Then, the ss-HRP activity in the cell pellets and cell culture supernatants of these cells was measured as described earlier (Fig. 9 C). The secretion defect induced by the SPCA1 knockdown could be rescued by the siRNA-resistant SPCA1-wt as well as mut1, mut2, and mut3, but not mut4. Therefore, we assumed that mut4 lacks the crucial charged amino acids needed for interaction with CFL-1 (Fig. 9). To confirm this hypothesis, lysates of SPCA1-HA-wt and -mut4–expressing HeLa cells were subjected to an HA immunoprecipitation, and associated proteins were analyzed by Western blotting (Fig. S5). CFL-1 coprecipitated with SPCA-HA-wt, as demonstrated earlier. In contrast, SPCA1-HA-mut4 interaction with CFL-1 was strongly reduced, which indicates that we targeted most of the crucial amino acids required for the interaction (Fig. S5).
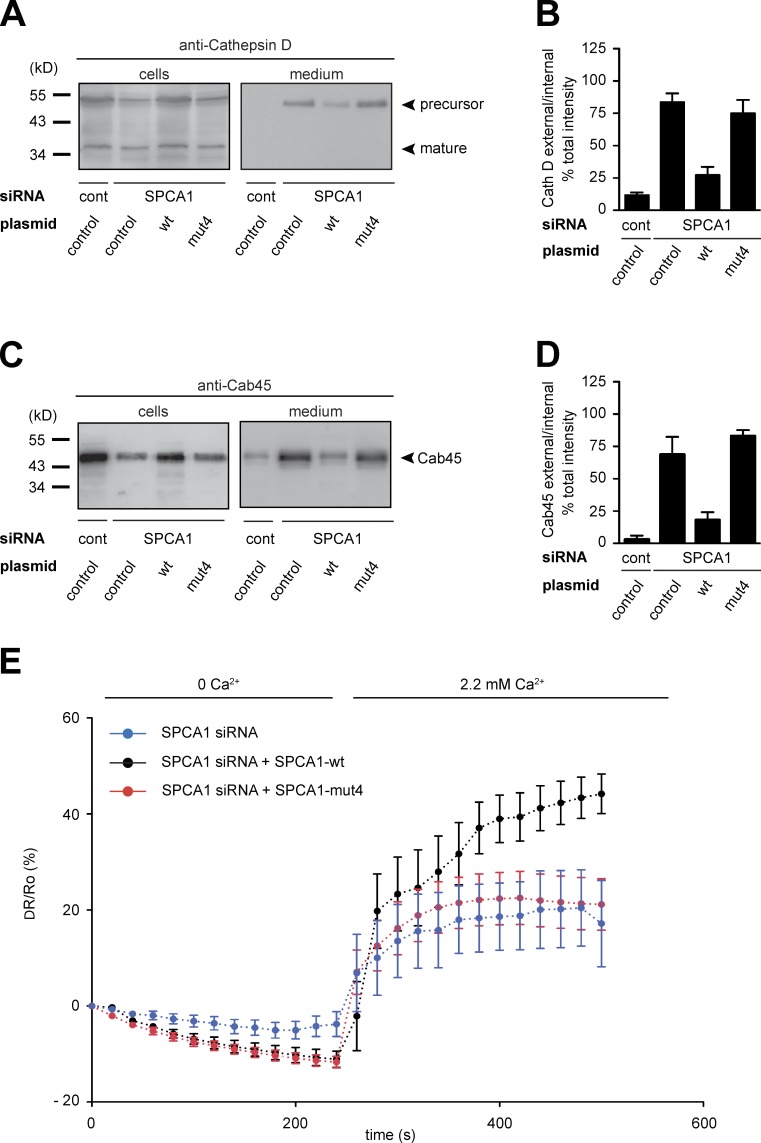
To confirm the ss-HRP secretion phenotype with additional sorting markers, control HeLa cells and HeLa cells expressing siRNA-resistant SPCA1-wt and -mut4 were transfected with control or SPCA1 siRNA, respectively. Cell pellets and supernatants were analyzed for the presence of Cab45 and Cathepsin D by Western blotting. The data clearly showed that SPCA1-wt rescued Cab45 and Cathepsin D sorting, whereas the -mut4 did not (Fig. 10, A–D). Finally, we measured the TGN Ca2+ uptake in HeLa cells stably expressing SPCA1-wt and -mut4 treated with SPCA1 siRNA and transfected with Go-D1-cpv. SPCA1-wt rescued the siRNA-induced defect of Ca2+ uptake, whereas SPCA1-mut4 did not (Fig. 10 E).
Figure 10.
A SPCA1 point mutant impairs TGN Ca2+ uptake and secretory cargo sorting. (A–D) HeLa cells expressing SPCA1-wt and mutants were treated with control and SPCA1 siRNAs. Media and lysates from these cells were Western blotted with Cathepsin D (A) and Cab45 (C) antibodies. Western blots from three independent experiments were quantified by densitometry. Bar graphs represent the densitometry values of external Cathepsin D (B) and Cab45 (D) normalized to internal Cathepsin D and Cab45 values, respectively. (E) HeLa cells expressing either siRNA-resistant SPCA1-wt-HA or -mut4-HA were transfected with SPCA1 siRNA and subsequently with Go-D1-cpv. Ca2+ entry into the TGN was analyzed in wt (n = 8) or mut4 (n = 15)-expressing cells. Fluorescent signals reflecting TGN [Ca2+] were presented as ΔR/R0, where R0 is the value obtained before addition of 2.2 mM Ca2+ to the cell’s bathing solution.
These data finally confirmed that the direct interaction of SPCA1 with CFL-1 and actin is crucial for the activation of SPCA1 and thus Ca2+-dependent cargo sorting at the TGN.
Discussion
In this study we describe how CFL-1 directly interacts with the P-domain of SPCA1 and how this interaction recruits F-actin to the pump. The reaction is required for secretory cargo sorting because the overexpression of the identified domain of SPCA1 impairs TGN Ca2+ uptake and secretory cargo sorting in living cells. Furthermore, the expression of CFL-1-S3E that does not bind to SPCA1 or a CFL-1 binding–deficient SPCA1 mutant shows the same phenotype in HeLa cells. These data demonstrated that the direct binding of SPCA1 and CFL-1 and the recruitment of F-actin are required for the pumping and sorting reaction. These findings provide a new insight into the mechanism of the sorting process and contribute to understanding the action by which actin filaments are recruited to the TGN.
SPCA1 recruits F-actin via activated CFL-1
We have shown previously that active CFL-1 localizes to the TGN (von Blume et al., 2011). Our new findings formally support this hypothesis, as, in vitro, only active CFL-1 binds to SPCA1. This binding is required for the activation of SPCA1 because the expression of CFL-1-S3E impairs TGN Ca2+ uptake and thus the pumping activity. LIMK phosphorylates and inactivates the actin filament severing activity of CFL-1. Dephosphorylation by slingshot phosphatase reactivates CFL-1 (Agnew et al., 1995; Arber et al., 1998). Interestingly, both the kinase (Salvarezza et al., 2009) and the phosphatase (unpublished data) have been localized to the TGN. We could also demonstrate that slingshot is required for the export of ss-HRP from the TGN (von Blume et al., 2009). In fact, LIMK activity has been shown to regulate sorting and protein kinase D (PKD)-dependent export of cargo on the polarized surface of the neurons (Rosso et al., 2004). Furthermore, slingshot is a known target of PKD (Eiseler et al., 2009; Peterburs et al., 2009). Collectively, these findings provide a handle on additional components involved in the regulation of CFL-1 at the TGN to help to spatially and temporally facilitate the association of the SPCA1–CFL-1 and actin complex to allow correct TGN Ca2+ uptake, subsequent packaging of secretory cargo, and ultimately a PKD-dependent fission to generate specific export carriers at the TGN (Campelo and Malhotra, 2012).
The role of the actin cytoskeleton recruited to SPCA1 by CFL-1
Our new findings illustrate that actin does not directly bind to SPCA1 as assumed previously (von Blume et al., 2011). Furthermore, we showed that CFL-1 acts as a connecter between the actin cytoskeleton and the Ca2+ pump. But how do these components support Ca2+ pumping and consequently the sorting of secretory cargo in the lumen of the TGN? Several reports have described the role of actin in modulating the activation of pumps and channels (Mazzochi et al., 2006; Dalghi et al., 2013; Vanagas et al., 2013).
According to our data and the literature, we favor the model in which SPCA1 accumulates in membrane microdomains, and this has an impact on its activity (Simons and van Meer, 1988; van Meer and Simons, 1988; Simons and Ikonen, 1997). These sphingolipid- and cholesterol-rich domains have been reported to occur at the Golgi apparatus, and their major function was found to be the sorting of proteins to the apical plasma membrane in polarized cells and in yeast (van Meer and Simons, 1988; Klemm et al., 2009). In our case, we suggest that the formation of such domains is required for the activation of SPCA1. What is the role of CFL-1, actin, and SPCA1 in the process and how can these proteins support the formation of such a microdomain? It has been proposed that SPCA1 accumulates in cholesterol-rich membrane domains, and if cholesterol is depleted from these domains, the pump is 50% less active (Baron et al., 2010). Our unpublished results show that CFL-1 and SPCA1 colocalize in small domains that partially overlap with TGN46 and Cab45. The actin cytoskeleton is known to organize membrane-associated proteins into functional domains at the plasma membrane (Plowman et al., 2005). Furthermore, the actin attachment to the membrane can also strongly affect phase separation of initially homogenous lipids and stabilize phase-separated domains (Liu and Fletcher, 2006). Therefore, CFL-1 and actin might act in a similar manner at the TGN to collect SPCA1 in such cholesterol-rich domains in which SPCA1 is active. The activation and deactivation of this complex would then be controlled by TGN-localized LIMK and slingshot.
Our former results support this idea and have shown that the treatment of cells with actin depolymerizing (LatA) or hyperpolymerizing (Jasp) agents as well as the knockdown of ADF/CFL-1 inhibit cargo sorting at the TGN (von Blume et al., 2009). The new data showed that inhibition of SPCA1 binding to actin and CFL-1 by the overexpression of the interaction domain (SPCA1-L2-C1), CFL-1-S3E, or a CFL-1 binding-deficient mutant impairs SPCA1 pumping activity and secretory cargo sorting. We assume that under these conditions F-actin is not recruited to the TGN by CFL-1 and, thus, the shaping of the microdomain is impaired. The formation of higher oligomers stabilized by such a domain might have an additional impact on the activity of the ATPase, as demonstrated for several P-Type ATPases such as the Na+/K+-ATPase (Taniguchi et al., 2001) and the H+-ATPase (Chadwick et al., 1987). Therefore, the structure and size of the SPCA1–CFL-1–actin complex at the TGN in the presence or absence of F-actin needs to be further elucidated.
Materials and methods
Antibodies, plasmids, and cell culture
The commercial antibodies used in the performed experiments are as follows: anti-CFL-1 (rabbit polyclonal, ab11062; Abcam), anti-HRP (mouse monoclonal, ab8326; Abcam), anti–β-actin (mouse monoclonal, A5441; Sigma-Aldrich), anti–Cathepsin D (mouse monoclonal; BD), anti-Cab45 (mouse monoclonal, 611065; BD), anti-His (rabbit polyclonal, #2365; Cell Signaling Technology), anti-SPCA1 (mouse monoclonal, H00027032-M01; Abnova), and anti-HA (rat monoclonal, 11867423001; Roche). Western blot antibodies were from Santa Cruz Biotechnology, Inc. (rat sc-2303, rabbit sc-2301, and mouse sc-2314). The HA antibody used for immunofluorescence microscopy was from Covance (mouse monoclonal, MMS-101P) and the TGN46 antibody was from AbD Serotec (sheep polyclonal, AHP500G). Secondary antibodies for immunofluorescence microscopy were from Life Technologies: Alexa Fluor 488 mouse (A21202) and rabbit (A21206), Alexa Fluor 594 mouse (A21203), rabbit (A21207), and sheep A11016; and Alexa Fluor 633 mouse (A21063). Alexa Fluor 568 C5 (A-20341) and 647 C2 (A-20347) maleimides were from Life Technologies. The HRP substrate for detecting chemiluminescence was from EMD Millipore (WBKLS0500). Atto-488–labeled actin was from Hypermol (8153-01). The protease inhibitor cocktail was from Roche (complete, 04693159001). The µMACs HA Tagged Protein Isolation kit was from Miltenyi Biotec (130-091-122). The QuikChange II XL Site-Directed Mutagenesis kit was from Agilent Technologies (#200522).
Cell culture and transient/stable transfection
HeLa cells and all stable cell lines were grown in DMEM (Invitrogen) containing 10% FCS at 37°C with 5% CO2. To generate stable cell lines, VSV-G pseudotyped retroviral vectors were produced by transient transfection of HEK293T (human embryonic kidney) cells. Viral particles were concentrated from cell culture supernatant and used for infection.
siRNA transfection
HeLa cells were plated 1 d in advance to ensure 50% confluence on the day of transfection. Functionally siRNA directed against ADF (oligo1, 5′-GCTTTGTATGATGCAAGCTTTGAAA-3′; oligo2, 5′-AGAAGATCTCAATCGGGCTTGTATT-3′), CFL-1 (oligo1, 5′-GGATCAAGCATGAATTGCAAGCAAA-3′), and SPCA1 (oligo1, 5′-CATCGAGAAGTAACATTGCCTTTAT-3′; oligo2, 5′-ATAAAGGCAATGTTAcTTCTCGATG-3′), and a negative control were from Invitrogen. Specific siRNAs (150 nM each) or control siRNA were transfected using HiPerfect (QIAGEN).
Plasmids
The ss-HRP-Flag was generated by PCR from ss-HRP-KDEL (Bard et al., 2006) and cloned into the pEGFP-N1 vector using SmaI and AgeI restriction sites. The EGFP was removed by cutting the backbone with AgeI and NotI. Annealed opposing primers containing the Flag sequence and restriction sites AgeI and NotI replaced the EGFP sequence. The HA-rCFL-1-wt plasmid was generated by PCR from rCFL-pcDNA3 and cloned into the pLPCX (containing CMV promoter) backbone vector using BglII and NotI. HA-rCFL-1-S3E was obtained performing site-directed mutagenesis using the wt plasmid as a template together with sense primer, 5′-GGCCGCTCGATGGCCGAGGGTGTGGCTGTCTCT-3′, and antisense primer, 5′-AGAGACAGCCACACCCTCGGCCATCGAGCGGCC-3′. Substituted nucleotides are indicated with bold letters. Serine 3 of CFL was changed into glutamic acid, rendering the protein constitutively inactive.
The cDNA coding the SPCA1 cytoplasmic domains and CFL-1 constructs were cloned into the pEC-A-His-Sumo vector using a ligase-independent cloning system. Cytoplasmic SPCA1 domains were amplified by PCR using a full-length pcDNA3-SPCA1-Flag plasmid as template with the following primers: N-term (aa 1–70) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGAAGGTTGCACGTTTTC-3′; N-term reverse, 5′-GCAAAGCACCGGCCTCGTTATGGCTCATCTTCACTAATATCAAAC-3′; L1 (aa 124–252) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGTCAGAAAAATCTCTTG-3′; L1 reverse, 5′-GCAAAGCACCGGCCTCGTTACTCTTCTGCTTGCATCATTTTAAAA-3′; L2 (full length, aa 329–680) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGAGGGCCATTGTGAAAAAGCTGCCTATTGTTGAAACTCTG-3′; L2 full length reverse, 5′-GCAAAGCACCGGCCTCGTTAATCATCATCCACTAGGATCATGTCTGCTGCCTCTTTG-3′; L2-N1 (aa 329–548) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGAGGGCCATTGTGAAAAAGCTGCCTATTGTTGAAACTCTG-3′; L2-N1 reverse, 5′-GCAAAGCACCGGCCTCGTTAAGGTGGATCAATGATTCCCACCAAGCCAAG-3′; L2-C1 (aa 549–680) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGAGAACTGGTGTGAAAGAAGCTGTTACAACACTCATTGCC-3′; L2-C1 reverse, 5′-GCAAAGCACCGGCCTCGTTAATCATCCACTAGGATCATGTCTGCT-3′; L3 (aa 751–770) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGGTAGAACCAGTGGATAAAGA-3′; L3 reverse, 5′-GCAAAGCACCGGCCTCGTTAAATGCTGTCTTTCCAGTTGC-3′; L4 (aa 829–840) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGCAGACCAAGTCTGTGTTTGA-3′; L4 reverse, 5′-GCAAAGCACCGGCCTCGTTAACTGCAGAGTCCAATCTCAA-3′; C-term (aa 897–919) forward, 5′-ACCAGGAACAAACCGGCGGCCGCTCGATGAAGGTTGAAAGGAGCAGGGA-3′; C-term reverse, 5′-GCAAAGCACCGGCCTCGTTATACTTCAAGAAAAGATGATG-3′.
Constructs were verified by sequencing procedures.
The CFL-1-S3E mutant was generated by site-directed mutagenesis using CFL-1 wt in pEC-His-Sumo as a template using the sense primer 5′-GGCCGCTCGATGGCCGAGGGTGTGGCTGTCTCT-3′ and the antisense primer 5′-AGAGACAGCCACACCCTCGGCCATCGAGCGGCC-3′ with changed nucleotides (bold letters).
For labeling CFL-1 with Alexa Fluor 647 maleimides, glutamic acid at position 42 was replaced by a cysteine (bold letters) using site-directed mutagenesis primers sense, 5′-CGGTGCTCTTCTGCCTGAGTTGCGACAAGAAGAACATCATCCT-3′, and antisense, 5′-AGGATGATGTTCTTCTTGTCGCAACTCAGGCAGAAGAGCACCG-3′.
GFP-HA was cloned into pLPCX by amplifying GFP from pRetroQ-AcGFP-N1 by PCR using the 5′-CGTGGATCCATGGTGAGCAAGGGCGC-3′ forward primer, introducing a BamHI site, and a reverse primer, 5′-GACGCGGCCGCGAATTCTTAAGCGTAGTCTGGGACGTCGTATGGGTACTTGTACAGCTCATCCAT-3′, introducing HA-tag and a NotI restriction site. The PCR product and the pLPCX backbone were digested with BamHI and NotI and subsequently ligated. The SPCA1-L1-HA construct was cloned into the pLPCX vector, performing PCR using 5′-CTGGTCGACATGTCAGAAAAATCTCTTGAAGAATTGAGTAAACTTG-3′ (including SalI site) as a forward primer and 5′-GCAGAATTCTTAAGCGTAGTCTGGGACGTCGTATGGGTACATGCTCTTCTGCAGAGGG-3′ (including HA-tag sequence and EcoRI site) as a reverse primer from SPCA1 full length (pcDNA3). The PCR product and the pLPCX vector were digested with SalI and EcoRI restriction enzymes, respectively, to create complementary overhangs. Afterward, the pieces were ligated to get SPCA1-L1-HA in pLPCX. The SPCA1-L2-C1-HA construct was cloned into the pLPCX vector, applying the same technique and using 5′-CTGGTCGACATGAGAACTGGTGTGAAAGAAGCTG-3′ as a forward primer and 5′-GCAGAATTCTTAAGCGTAGTCTGGGACGTCGTATGGGTAATCATCCACTAGGATCATGTCTGC-3′ as a reverse primer.
The SPCA1-HA siRNA–resistant mutant was created by introducing silent mutations using the following primers: sense, 5′-CTAATGGAGATCTTGCATCGAGGAGCAATATTGCCTTTATGGGAA-3′; antisense, 5′-TTCCCATAAAGGCAATATTGCTCCTCGATGCAAGATCTCCATTAG-3′. To generate putative SPCA1-Loop2-CFL-1 binding mutants, different clusters of charged amino acids within SPCA1-Loop2-C1 were substituted to alanine (highlighted in bold) using the subsequent primers.
SPCA1-HA_Q605A/S608A/K634A.
Sense Q605A/S608A, 5′-GAGAAGAAATAGATGCAATGGATGTTGCGCAGCTTGCACAAATAGTACCAAAGGTT-3′; antisense Q605A/S608A, 5′-AACCTTTGGTACTATTTGTGCAAGCTGCGCAACATCCATTGCATCTATTTCTTCTC-3′.
We used mutated SPCA1-HA_ Q605A/S608A plasmid as a template for K634A: sense K634A, 5′-GAAAATTATTAAGTCGCTACAGGCGAACGGTTCAGTTGTAGCCATG-3′; antisense K634A, 5′-CATGGCTACAACTGAACCGTTCGCCTGTAGCGACTTAATAATTTTC-3′.
SPCA1-HA_R623A/K627A/K630A/K634A.
Sense, 5′-AGTATTTTACAGAGCTAGCCCAGCGCACAAGATGGCAATTATTGCGTCGCTACAGGCGAACGGTTCAGTTGTAGCCATG-3′; antisense, 5′-CATGGCTACAACTGAACCGTTCGCCTGTAGCGACGCAATAATTGCCATCTTGTGCGCTGGGCTAGCTCTGTAAAATACT-3′.
SPCA1-HA_K589A/K613A.
Sense K589A, 5′-TCGTCTGGGATTGTATTCCGCAACTTCCCAGTCAGTCTCA-3′; antisense K589A, 5′-TGAGACTGACTGGGAAGTTGCGGAATACAATCCCAGACGA-3′.
We used mutated SPCA1-HA_ K589A plasmid as a template for K613A: sense K613A, 5′-GTTCAGCAGCTTTCACAAATAGTACCAGCGGTTGCAGTATTTTACAG-3′; antisense K613A, 5′-CTGTAAAATACTGCAACCGCTGGTACTATTTGTGAAAGCTGCTGAAC-3′.
SPCA1-HA_Q605A/Q606A/Q609A/K634A.
Sense Q605A/Q606A/Q609A, 5′-CAGGAGAAGAAATAGATGCAATGGATGTTGCGGCGCTTTCAGCAATAGTACCAAAGGTTGCAGTATTTTACA-3′; antisense Q605A/Q606A/Q609A, 5′-TGTAAAATACTGCAACCTTTGGTACTATTGCTGAAAGCGCCGCAACATCCATTGCATCTATTTCTTCTCCTG-3′.
We used mutated SPCA1-HA_ Q605A/Q606A/Q609A plasmid as a template for K634A: sense K634A, 5′-GAAAATTATTAAGTCGCTACAGGCGAACGGTTCAGTTGTAGCCATG-3′; antisense K634A, 5′-CATGGCTACAACTGAACCGTTCGCCTGTAGCGACTTAATAATTTTC-3′.
The SPCA1-ΔL2-C1 was generated using primers targeting the sequence before (bold) and after L2-C1: SPCA1_Δ549-680 sense, 5′-GGTGGGAATCATTGATCCACCTGATTTTCAAACCATAATGTCTG-3′; SPCA1_Δ549-680 antisense, 5′-CAGACATTATGGTTTGAAAATCAGGTGGATCAATGATTCCCACC-3′.
All mutants were cloned using the siRNA-resistant mutant as a template.
Purification of recombinant proteins
The constructs for bacterial expression were transformed into E. coli BL21 gold cells (Agilent Technologies), expressed using the auto-induction system for 4 h at 37°C and thereafter overnight at 18°C (Studier, 2005). Bacteria were pelleted by centrifugation and lysed in lysis buffer (20 mM Hepes, 500 mM NaCl, 1 mM DTT, and complete protease inhibitor cocktail) and further sonicated for 10 min (Vs70T sonotrode, 60% amplitude, pulse 1 s/nonpulse 0.5 s). Purification was performed using His-trap columns (5 ml; GE Healthcare) according to the manufacturer’s instructions. After purification, the His-Sumo tag was cut off by SenP2 protease in a ratio of 1:100 for 4 h at 4°C or left with the His-Sumo tag. As a final step, the proteins were purified with the Äkta Gel filtration Sytem (GE Healthcare) using the Superdex 75 column. Fractions were collected and concentrations were measured using Bradford reagent (Carl Roth). The molecular weights of the recombinant proteins were assessed by SDS-PAGE and Coomassie blue staining, respectively.
Pull-down assays
Recombinant His-Sumo fusion proteins were immobilized onto Ni-NTA beads (Macherey-Nagel) for 2 h at 4°C on a rotating wheel. Subsequently, the protein–bead complexes were washed four times using the His elution buffer without imidazole (20 mM Hepes, 400 mM NaCl, and 1 mM DTT) at 4°C. In parallel, confluent HeLa cells (107 cells per condition) were lysed in ice-cold CHAPS buffer (1% CHAPS in PBS supplemented with protease inhibitor cocktail) for 15 min on ice. Subsequently, lysate was cleared by centrifugation. Then, protein–bead complexes were incubated with cell lysate overnight at 4°C. The next day, beads were washed extensively and associated proteins were released by adding SDS sample buffer. Proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane (Millipore). The membranes were further incubated overnight with antibodies against His, CFL-1, and β-actin.
CFL-1 in vitro pull-down assay
Purified CFL-1 was incubated with 2 µg of the CFL-1–specific antibody for 1 h at 4°C, washed with elution buffer (20 mM Hepes, 400 mM NaCl, and 1 mM DTT), and further incubated with 30 µl Protein A/G Agarose (Santa Cruz Biotechnology, Inc.) overnight at 4°C. 40 µm of recombinant SPCA1 proteins was added to the CFL-1–agarose complex for an additional hour at 4°C. Next, beads were extensively washed and proteins released by adding SDS sample buffer. Thereafter, proteins were separated by SDS-PAGE and the gel was incubated with Coomassie blue solution.
His pull-down assay
Ni-NTA beads were incubated with blocking buffer (1% BSA, 20 mM Hepes, and 150 mM NaCl) for 20 min. Then His-Sumo–tagged proteins were added and incubated with beads overnight at 4°C. Afterward, the beads were extensively washed and further incubated with gel-filtrated recombinant proteins for 2 h at 4°C. After extensive washing with 20 mM Hepes and 150 mM NaCl, proteins were eluted by adding SDS sample buffer.
Immunofluorescence microscopy
Ni-NTA beads were incubated with blocking buffer (1% BSA, 20 mM Hepes, and 150 mM NaCl), and His-Sumo fusion proteins were attached as described for the His pull-down assay. The beads were then incubated with antibodies against His (rabbit) or CFL-1 (mouse), respectively, for 1 h at room temperature. Samples were then washed three times for 5 min with blocking buffer and further incubated with either Alexa Fluor 488– or Alexa Fluor 594–labeled antibodies, or both, for 1 h in blocking buffer. Next, beads were washed with 20 mM Hepes and 500 mM NaCl, transferred into a glass-bottom cell culture dish containing the wash buffer, and visualized with a confocal laser scanning microscope (LSM 780; Carl Zeiss) using a 40× Plan-Apochromat 1.4 oil objective lens at room temperature. For the detection of Alexa Fluor 594, the 561 nm laser line was used. For Alexa Fluor 488, the 488 nm laser line was used. Pictures were acquired using the Leica software (ZEN 2010) and converted to TIF files in ImageJ (version 1.37).
Immunofluorescence of GFP-HA, L1-HA, and L2-C1-HA in HeLa cells
HeLa cells were fixed with 4% paraformaldehyde for 15 min, washed with PBS, and subsequently permeabilized for 30 min in 0.05% Saponin. Next, cells were incubated with HA antibody for 1 h, washed three times for 5 min, and incubated with secondary antibody conjugated with Alexa Fluor 488. Cells were washed five times for 5 min and mounted using ProLong Gold (Life Technologies). Slides were analyzed using a confocal laser-scanning microscope (LSM 780; Carl Zeiss). A 40× Plan-Apochromat 1.4 oil objective lens was used. For detection of Alexa Fluor 488, the 488-nm laser line was used. Pictures were acquired using Leica software (ZEN 2010) and converted to TIF files in ImageJ (version 1.37).
CFL-1 localization assay
HeLa cells stably expressing GFP-HA, SPCA1-L1-HA, or SPCA1-L2-C1-HA were grown on glass coverslips and incubated at 20°C for 2 h. Cells were then fixed and permeabilized in standard buffer (0.2% Triton X-100, 0.5% SDS, and 4% BSA). Alternatively, cells were washed with KHM buffer (125 mM potassium acetate, 25 mM Hepes, pH 7.2, and 2.5 mM magnesium acetate) at 4°C and permeabilized for 5 min with KHM containing 30 mg/ml digitonin. After washing the cells with KHM for 7 min, cells were fixed and processed for immunofluorescence microscopy. Coverslips from cells permeabilized with the standard procedure and digitonin-permeabilized cells were stained with antibodies against CFL-1 and SPCA1 as well as Alexa Fluor 594/488 secondary antibodies, and processed by immunofluorescence microscopy as described in the previous section.
Protein labeling using Alexa Fluor maleimides
Before the labeling procedure, glutamic acid at position 42 in CFL wt protein was replaced by a cysteine using the QuikChange II site-directed mutagenesis (SDM) kit according to the manufacturer’s instructions. Full-length SPCA1-Loop2 already contains a cysteine in the wt sequence accessible after protein folding. Recombinant proteins (SPCA1-Loop2 and CFL-1-E42C) were purified and gel filtrated as described in “Purification of recombinant proteins.” Purified proteins were either labeled with Alexa Fluor 568 (Loop2) or 647 (CFL-1) fluorophores according to the protocol provided by Life Technologies.
In vitro reconstitution assay
His-sumo tagged SPCA1 proteins (40 µg of each) were immobilized on Ni-NTA beads and extensively washed with His elution buffer without imidazole. 10 µl of the protein–bead complexes were incubated with 20% Atto-488–labeled actin with a final concentration of 23 µM and 3 µl of 10× KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole HCl, pH 7.0) to induce actin polymerization. Additionally, selected samples of protein–bead complexes were incubated with purified CFL-1 (10 µM) and pipetted into self-made flow cells. Flow cells were constructed by placing two strips of parafilm on a glass slide ∼10 mm apart and covering them with an 18 × 18-mm coverslip. When the slide was warmed the parafilm attached the coverslip to it. This resulted in a 20 µl big flow cell. After adding a sample, cells were sealed on both sides using silicone glue (Wacker) to prevent F-actin from drying. Fluorescent F-actin (atto 488) was monitored at room temperature using an epifluorescence microscope (ApoTome Imager.Z1; Carl Zeiss) with a 20× Plan-Apochromat 0.75 NA objective lens (air) and a digital camera for microscopy (AxioCam MRm; Carl Zeiss). Pictures were acquired using the Axio Vision rel. 4.8 (Carl Zeiss) software and converted to TIF files in ImageJ (version 1.37).
For competition experiments, 200 µg of soluble, untagged SPCA1-L2-C1 was incubated with 7 µg of recombinant CFL-1. 10 µl of this mixture was then added to His-Sumo–tagged L2-C1 immobilized on Ni-NTA beads together with 20% Atto-488–labeled actin (final concentration of 23 µM). 3 µl of 10× KMEI buffer was then added to induce actin polymerization. Samples were pipetted in flow cells and monitored by fluorescence microscopy as described in the previous paragraph.
For performing the reconstitution assay with labeled proteins, SPCA1-L2-Alexa-568 was immobilized on beads, then incubated with CFL-1-Alexa-647 and Atto-488 actin together with KMEI buffer as described earlier. The mixture was incubated for 2 h at 4°C on a rotating wheel. Beads with bound protein were washed four to five times before monitoring to remove background signal. As controls, Ni-NTA beads blocked with 1% BSA were incubated with CFL-1-Alexa-647 and Atto-488 actin, respectively. Samples were subjected to epifluorescence microscopy as described in the previous section.
HA pull-down
Confluent HeLa cells stably expressing SPCA1-L1-HA, SPCA1-L2-C1-HA, SPCA1-wt-HA, or SPCA1-mut4-HA were lysed in 1% CHAPS buffer supplemented with protease inhibitor cocktail for 15 min on ice. After centrifugation, lysates were incubated with µMACS anti-HA magnetic microbeads and incubated for 30 min at 4°C. SPCA1-L1-HA and SPCA1-L2-C1-HA or SPCA1-wt-HA and -mut4 associated proteins were eluted from the microbeads using the µMACS anti-HA isolation kit according to the manufacturer's instructions. Eluted proteins were separated by SDS-PAGE and analyzed by Western blotting using antibodies against HA, CFL-1, or β-actin.
ss-HRP transport and localization assay
HeLa cells stably expressing ss-HRP+control, ss-HRP+SPCA1-L1-HA, ss-HRP+SPCA1-L2-C1-HA, ss-HRP+SPCA1-wt-HA, or ss-HRP+SPCA1-mut1-4 were processed for the HRP transport assay. Cells were seeded in 12-well plates and incubated for 24 h. After change of the medium, cells were further incubated for 12 h. Then supernatants were harvested and cells were lysed with 0.5% Triton X-100 in PBS. Finally, 50 µl of the medium and cells were mixed with 50 µl ECL solution, and HRP activity was measured.
For the ss-HRP assay with CFL-1 overexpression, HeLa cells were transfected with control or ADF/CFL-1 siRNAs for 40 h. Then, Flag-ss-HRP and siRNA-resistant HA-rCFL-1 wt or HA-rCFL-1-S3E were cotransfected. 50 µl of medium was harvested 24 h after plasmid transfections. The HRP activity was measured as described in the previous paragraph. For normalization, cells were lysed with 0.5% Triton X-100 in PBS, and internal HRP activity was measured. A second set of HeLa cells transfected with Flag-ss-HRP and siRNA-resistant HA-rCFL-1 wt or HA-rCFL-1-S3E was incubated for 2 h at 20°C in the presence of 100 µg/ml cycloheximide. After the temperature shift to 37°C for 30 min, cells were fixed, permeabilized in standard buffer (0.2% Triton X-100, 0.5% SDS, and 4% BSA), incubated with an HRP antibody, and processed for immunofluorescence microscopy.
Cathepsin D and Cab45 sorting assays
HeLa control cells and cells infected with pLCPX-SPCA1-L1-HA, pLPCX-SPCA1-L2-C1-HA, SPCA1-wt-HA, or SPCA1-mut4-HA were washed five times with serum-free medium and then grown in serum-free medium, which was collected after 4 h. For the CFL-1 overexpression experiments, HeLa cells were transfected with ADF/CFL-1 siRNA for 48 h. Subsequently, cells were either transfected with a control, a rat HA-CFL-1 wt, or an HA-CFL-1-S3E plasmid. Cells were then counted and lysed in PBS supplemented with 1% CHAPS. Media from control cells, SPCA1-L1-HA, SPCA1-L2-HA, rCFL-1 wt, rCFL-1-S3E, or SPCA1-wt-HA– or SPCA1-mut4-HA–expressing cells were filtered using a 0.45-µm filter (EMD Millipore) and centrifuged at 5,000 g for 15 min. The resulting supernatant was then centrifuged at 100,000 g for 2 h. Samples were concentrated using a 3-kD molecular mass cutoff spin column (EMD Millipore). Cell lysates and concentrated media were subsequently analyzed by Western blotting using anti–Cathepsin D and anti-Cab45 antibodies, respectively.
Ca2+ imaging
Ca2+ entry into the TGN was measured as described previously (Lissandron et al., 2010). Measurements of [Ca2+] in the TGN were performed using a fluorescent Ca2+ sensor (Go-D1-cpvcpv) based on the efficiency of FRET between CFP and YFP fluorescent proteins linked by a modified calmodulin and calmodulin-binding domain. This genetically encoded Ca2+ indicator was targeted to the trans-Golgi (Lissandron et al., 2010). HeLa cells, or HeLa cells stable expressing SPCA1-wt-HA or -mut4-HA, were transfected with control, SPCA1, or ADF/CFL-1 siRNAs for 40 h. Then Go-D1-cpv was transfected alone, with a control plasmid, or with SPCA1-L1-HA, SPCA1-L2-HA, or HA-rCFL-1-S3E. Ca2+ entry into the TGN was measured in Ca2+-depleted cells after 1 h of incubation at 4°C in a Ca2+-free solution (20 mM Hepes, Ca2+/Mg2+-free HBSS [Invitrogen], 2 gl−1 glucose, 490 µM MgCl2, and 450 µM MgSO4; 300 mOsmol/liter, pH 7.4) with 2 µM ionomycin (Abcam) and 5 mM EGTA (Palmer and Tsien, 2006). Then cells were washed twice in a Ca2+-free solution. Images were acquired at room temperature using a DeltaVision RT system (Applied Precision) equipped with an inverted base microscope (IX-71; Olympus) using a UPlan-Apochromat 100×/NA 1.35 oil immersion objective lens and a 120-bit monochrome camera (CoolSNAP HQ; Photometrics). To generate the images, the excitation filter (430/24), dual-band Sedat CFP/YFP beam splitter (Chroma Technology Corp.), and the emission filters (535/25 for FRET and 470/24 for CFP) were rapidly changed using an external filter wheel controlled by a motorized unit. Fluorescent signals reflecting TGN [Ca2+] were presented as ΔR/R0, where R0 is the value obtained before addition of 2.2 mM Ca2+ to the cell’s bathing solution. Images were acquired using softWoRx 5.5 software (GE Healthcare), and further image analysis was performed in ImageJ (version 1.47).
Generation of SPCA1 homology model
The multiple sequence alignment of SPCA1 orthologues was generated using MAFFT (Katoh and Standley, 2014). The rat ATPA2 sequence was added manually. The structure of human SPCA1 was predicted using Phyre (Kelley and Sternberg, 2009). The best hit was chosen (2zxe chain A, Squalus acanthias ATP1A1). We displayed the resulting structural model using PyMol (The PyMOL Molecular Graphics System, version 1.5.0.4; Schrödinger, LLC) and used it to select residues for subsequent mutation. The structural model in PyMol was exported as an image and made publication-ready using Illustrator (Adobe).
Statistical analysis
Statistical significance was analyzed in an unpaired Student’s t test using the Graph Pad Prism software. Compared datasets were statistically significant when p-values were <0.01.
Online supplemental material
Fig. S1 illustrates SPCA1 structure und purification of the cytosolic domains from E. coli that are putative CFL-1 and F-actin interaction domains. Fig. S2 shows the cytosolic distribution of CFL-1 in HeLa cells expressing GFP-HA, SPCA1-L1-HA, and SPCA1-L2-C1-HA that were not permeabilized with digitonin before fixation with formaldehyde. Fig. S3 shows a multiple sequence alignment of SPCA1 orthologues. Fig. S4 shows the localization of siRNA-resistant SPCA1-wt, mut1 (Q605A/S608A/K634A), mut2 (R623A/K627A/K630A/K634A), mut3 (K589A/K613A,), mut4 (Q605A/Q606A/Q609A/K634A), and SPCA1-ΔL2-C1 in HeLa cells. Fig. S5 demonstrates that SPCA1-mut4 (Q605A/Q606A/Q609A/K634A) binding to CFL-1 is strongly reduced. Table S1 lists the accession numbers and species abbreviations of human ATP2C1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201311052/DC1.
Supplementary Material
Acknowledgments
We thank Hildegart Reiter for retroviral transfections and Josse van Galen for proofreading.
The group of J. von Blume is funded by an Emmy Noether fellowship (project BL 1186/1-1) of the Deutsche Forschungsgemeinschaft (DFG) and by an FP7 Marie Curie Career Reintegration grant. J. von Blume and N. Mizuno are supported by the Max Planck Institute for Biochemistry, and. J. von Blume is promoted by the Department of Prof R. Fässler. A.H. Crevenna acknowledges funding through the SPP 1464 of the DFG.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- A-domain
- actuator domain
- CFL-1
- cofilin-1
- C-term
- C terminus
- LIMK
- LIM kinase
- N-domain
- nucleotide binding domain
- Ni-NTA
- nickel nitrilotriacetic acid
- N-term
- N terminus
- P-domain
- phosphorylation domain
- SPCA1
- secretory pathway calcium ATPase 1
- ss-HRP
- signal sequence horseradish peroxidase
- wt
- wild type
References
- Agnew, B.J., Minamide L.S., and Bamburg J.R.. 1995. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270:17582–17587 10.1074/jbc.270.29.17582 [DOI] [PubMed] [Google Scholar]
- Ang, A.L., Fölsch H., Koivisto U.-M., Pypaert M., and Mellman I.. 2003. The Rab8 GTPase selectively regulates AP-1B-dependent basolateral transport in polarized Madin-Darby canine kidney cells. J. Cell Biol. 163:339–350 10.1083/jcb.200307046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, A.L., Taguchi T., Francis S., Fölsch H., Murrells L.J., Pypaert M., Warren G., and Mellman I.. 2004. Recycling endosomes can serve as intermediates during transport from the Golgi to the plasma membrane of MDCK cells. J. Cell Biol. 167:531–543 10.1083/jcb.200408165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber, S., Barbayannis F.A., Hanser H., Schneider C., Stanyon C.A., Bernard O., and Caroni P.. 1998. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 393:805–809 10.1038/31729 [DOI] [PubMed] [Google Scholar]
- Bard, F., Casano L., Mallabiabarrena A., Wallace E., Saito K., Kitayama H., Guizzunti G., Hu Y., Wendler F., Dasgupta R., et al. . 2006. Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature. 439:604–607 10.1038/nature04377 [DOI] [PubMed] [Google Scholar]
- Barfield, R.M., Fromme J.C., and Schekman R.. 2009. The exomer coat complex transports Fus1p to the plasma membrane via a novel plasma membrane sorting signal in yeast. Mol. Biol. Cell. 20:4985–4996 10.1091/mbc.E09-04-0324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron, S., Vangheluwe P., Sepúlveda M.R., Wuytack F., Raeymaekers L., and Vanoevelen J.. 2010. The secretory pathway Ca2+-ATPase 1 is associated with cholesterol-rich microdomains of human colon adenocarcinoma cells. Biochim. Biophys. Acta. 1798:1512–1521 10.1016/j.bbamem.2010.03.023 [DOI] [PubMed] [Google Scholar]
- Burgos, P.V., Mardones G.A., Rojas A.L., daSilva L.L.P., Prabhu Y., Hurley J.H., and Bonifacino J.S.. 2010. Sorting of the Alzheimer’s disease amyloid precursor protein mediated by the AP-4 complex. Dev. Cell. 18:425–436 10.1016/j.devcel.2010.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campelo, F., and Malhotra V.. 2012. Membrane fission: the biogenesis of transport carriers. Annu. Rev. Biochem. 81:407–427 10.1146/annurev-biochem-051710-094912 [DOI] [PubMed] [Google Scholar]
- Chadwick, C.C., Goormaghtigh E., and Scarborough G.A.. 1987. A hexameric form of the Neurospora crassa plasma membrane H+-ATPase. Arch. Biochem. Biophys. 252:348–356 10.1016/0003-9861(87)90041-5 [DOI] [PubMed] [Google Scholar]
- Curwin, A.J., von Blume J., and Malhotra V.. 2012. Cofilin-mediated sorting and export of specific cargo from the Golgi apparatus in yeast. Mol. Biol. Cell. 23:2327–2338 10.1091/mbc.E11-09-0826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalghi, M.G., Fernández M.M., Ferreira-Gomes M., Mangialavori I.C., Malchiodi E.L., Strehler E.E., and Rossi J.P.F.C.. 2013. Plasma membrane calcium ATPase activity is regulated by actin oligomers through direct interaction. J. Biol. Chem. 288:23380–23393 10.1074/jbc.M113.470542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis, M.A., and Luini A.. 2008. Exiting the Golgi complex. Nat. Rev. Mol. Cell Biol. 9:273–284 10.1038/nrm2378 [DOI] [PubMed] [Google Scholar]
- Dikeakos, J.D., and Reudelhuber T.L.. 2007. Sending proteins to dense core secretory granules: still a lot to sort out. J. Cell Biol. 177:191–196 10.1083/jcb.200701024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doray, B., Bruns K., Ghosh P., and Kornfeld S.. 2002. Interaction of the cation-dependent mannose 6-phosphate receptor with GGA proteins. J. Biol. Chem. 277:18477–18482 10.1074/jbc.M201879200 [DOI] [PubMed] [Google Scholar]
- Eiseler, T., Döppler H., Yan I.K., Kitatani K., Mizuno K., and Storz P.. 2009. Protein kinase D1 regulates cofilin-mediated F-actin reorganization and cell motility through slingshot. Nat. Cell Biol. 11:545–556 10.1038/ncb1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölsch, H., Ohno H., Bonifacino J.S., and Mellman I.. 1999. A novel clathrin adaptor complex mediates basolateral targeting in polarized epithelial cells. Cell. 99:189–198 10.1016/S0092-8674(00)81650-5 [DOI] [PubMed] [Google Scholar]
- Gieselmann, V., Hasilik A., and von Figura K.. 1985. Processing of human cathepsin D in lysosomes in vitro. J. Biol. Chem. 260:3215–3220 [PubMed] [Google Scholar]
- Katoh, K., and Standley D.M.. 2014. MAFFT: iterative refinement and additional methods. Methods Mol. Biol. 1079:131–146 10.1007/978-1-62703-646-7_8 [DOI] [PubMed] [Google Scholar]
- Kelley, L.A., and Sternberg M.J.E.. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- Kienzle, C., and von Blume J.. 2014. Secretory cargo sorting at the trans-Golgi network. Trends Cell Biol. 10.1016/j.tcb.2014.04.007 [DOI] [PubMed] [Google Scholar]
- Kim, M., Jung J., Park C.S., and Lee K.. 2002. Identification of the cofilin-binding sites in the large cytoplasmic domain of Na,K-ATPase. Biochimie. 84:1021–1029 10.1016/S0300-9084(02)00004-4 [DOI] [PubMed] [Google Scholar]
- Klemm, R.W., Ejsing C.S., Surma M.A., Kaiser H.-J., Gerl M.J., Sampaio J.L., de Robillard Q., Ferguson C., Proszynski T.J., Shevchenko A., and Simons K.. 2009. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 185:601–612 10.1083/jcb.200901145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld, S., and Mellman I.. 1989. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5:483–525 10.1146/annurev.cb.05.110189.002411 [DOI] [PubMed] [Google Scholar]
- Kühlbrandt, W.2004. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 5:282–295 10.1038/nrm1354 [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., and Hoflack B.. 1997. Mannose 6-phosphate receptors regulate the formation of clathrin-coated vesicles in the TGN. J. Cell Biol. 137:335–345 10.1083/jcb.137.2.335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K., Jung J., Kim M., and Guidotti G.. 2001. Interaction of the alpha subunit of Na,K-ATPase with cofilin. Biochem. J. 353:377–385 10.1042/0264-6021:3530377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissandron, V., Podini P., Pizzo P., and Pozzan T.. 2010. Unique characteristics of Ca2+ homeostasis of the trans-Golgi compartment. Proc. Natl. Acad. Sci. USA. 107:9198–9203 10.1073/pnas.1004702107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, A.P., and Fletcher D.A.. 2006. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys. J. 91:4064–4070 10.1529/biophysj.106.090852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzochi, C., Benos D.J., and Smith P.R.. 2006. Interaction of epithelial ion channels with the actin-based cytoskeleton. Am. J. Physiol. Renal Physiol. 291:F1113–F1122 [DOI] [PubMed] [Google Scholar]
- Mellman, I., and Nelson W.J.. 2008. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9:833–845 10.1038/nrm2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiaen, L., Dode L., Vanoevelen J., Raeymaekers L., and Wuytack F.. 2007. Calcium in the Golgi apparatus. Cell Calcium. 41:405–416 10.1016/j.ceca.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Palmer, A.E., and Tsien R.Y.. 2006. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1:1057–1065 10.1038/nprot.2006.172 [DOI] [PubMed] [Google Scholar]
- Peterburs, P., Heering J., Link G., Pfizenmaier K., Olayioye M.A., and Hausser A.. 2009. Protein kinase D regulates cell migration by direct phosphorylation of the cofilin phosphatase slingshot 1 like. Cancer Res. 69:5634–5638 10.1158/0008-5472.CAN-09-0718 [DOI] [PubMed] [Google Scholar]
- Pfeffer, S.R.2011. Entry at the trans-face of the Golgi. Cold Spring Harb. Perspect. Biol. 3:a005272 10.1101/cshperspect.a005272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowman, S.J., Muncke C., Parton R.G., and Hancock J.F.. 2005. H-ras, K-ras, and inner plasma membrane raft proteins operate in nanoclusters with differential dependence on the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 102:15500–15505 10.1073/pnas.0504114102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman, M.L., and Kornfeld S.. 1981. Lysosomal enzyme targeting. N-Acetylglucosaminylphosphotransferase selectively phosphorylates native lysosomal enzymes. J. Biol. Chem. 256:11977–11980 [PubMed] [Google Scholar]
- Rockenbauch, U., Ritz A.M., Sacristan C., Roncero C., and Spang A.. 2012. The complex interactions of Chs5p, the ChAPs, and the cargo Chs3p. Mol. Biol. Cell. 23:4402–4415 10.1091/mbc.E11-12-1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso, S., Bollati F., Bisbal M., Peretti D., Sumi T., Nakamura T., Quiroga S., Ferreira A., and Cáceres A.. 2004. LIMK1 regulates Golgi dynamics, traffic of Golgi-derived vesicles, and process extension in primary cultured neurons. Mol. Biol. Cell. 15:3433–3449 10.1091/mbc.E03-05-0328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvarezza, S.B., Deborde S., Schreiner R., Campagne F., Kessels M.M., Qualmann B., Caceres A., Kreitzer G., and Rodriguez-Boulan E.. 2009. LIM kinase 1 and cofilin regulate actin filament population required for dynamin-dependent apical carrier fission from the trans-Golgi network. Mol. Biol. Cell. 20:438–451 10.1091/mbc.E08-08-0891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, K., and Ikonen E.. 1997. Functional rafts in cell membranes. Nature. 387:569–572 10.1038/42408 [DOI] [PubMed] [Google Scholar]
- Simons, K., and van Meer G.. 1988. Lipid sorting in epithelial cells. Biochemistry. 27:6197–6202 10.1021/bi00417a001 [DOI] [PubMed] [Google Scholar]
- Studier, F.W.2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 10.1016/j.pep.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Taniguchi, K., Kaya S., Abe K., and Mårdh S.. 2001. The oligomeric nature of Na/K-transport ATPase. J. Biochem. 129:335–342 10.1093/oxfordjournals.jbchem.a002862 [DOI] [PubMed] [Google Scholar]
- Van Baelen, K., Dode L., Vanoevelen J., Callewaert G., De Smedt H., Missiaen L., Parys J.B., Raeymaekers L., and Wuytack F.. 2004. The Ca2+/Mn2+ pumps in the Golgi apparatus. Biochim. Biophys. Acta. 1742:103–112 10.1016/j.bbamcr.2004.08.018 [DOI] [PubMed] [Google Scholar]
- van Meer, G., and Simons K.. 1988. Lipid polarity and sorting in epithelial cells. J. Cell. Biochem. 36:51–58 10.1002/jcb.240360106 [DOI] [PubMed] [Google Scholar]
- Vanagas, L., de La Fuente M.C., Dalghi M., Ferreira-Gomes M., Rossi R.C., Strehler E.E., Mangialavori I.C., and Rossi J.P.F.C.. 2013. Differential effects of G- and F-actin on the plasma membrane calcium pump activity. Cell Biochem. Biophys. 66:187–198 10.1007/s12013-012-9467-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume, J., Duran J.M., Forlanelli E., Alleaume A.-M., Egorov M., Polishchuk R., Molina H., and Malhotra V.. 2009. Actin remodeling by ADF/cofilin is required for cargo sorting at the trans-Golgi network. J. Cell Biol. 187:1055–1069 10.1083/jcb.200908040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Blume, J., Alleaume A.-M., Cantero-Recasens G., Curwin A., Carreras-Sureda A., Zimmermann T., van Galen J., Wakana Y., Valverde M.A., and Malhotra V.. 2011. ADF/cofilin regulates secretory cargo sorting at the TGN via the Ca2+ ATPase SPCA1. Dev. Cell. 20:652–662 10.1016/j.devcel.2011.03.014 [DOI] [PubMed] [Google Scholar]
- von Blume, J., Alleaume A.-M., Kienzle C., Carreras-Sureda A., Valverde M., and Malhotra V.. 2012. Cab45 is required for Ca2+-dependent secretory cargo sorting at the trans-Golgi network. J. Cell Biol. 199:1057–1066 10.1083/jcb.201207180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakana, Y., van Galen J., Meissner F., Scarpa M., Polishchuk R.S., Mann M., and Malhotra V.. 2012. A new class of carriers that transport selective cargo from the trans Golgi network to the cell surface. EMBO J. 31:3976–3990 10.1038/emboj.2012.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C.-W., Hamamoto S., Orci L., and Schekman R.. 2006. Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 174:973–983 10.1083/jcb.200605106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, N., Maurer A., Nieke S., and Kalbacher H.. 2008. Cathepsin D: a cellular roadmap. Biochem. Biophys. Res. Commun. 376:5–9 10.1016/j.bbrc.2008.08.099 [DOI] [PubMed] [Google Scholar]
- Zanolari, B., Rockenbauch U., Trautwein M., Clay L., Barral Y., and Spang A.. 2011. Transport to the plasma membrane is regulated differently early and late in the cell cycle in Saccharomyces cerevisiae. J. Cell Sci. 124:1055–1066 10.1242/jcs.072371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.