Abstract
Many cancer drugs impact cancer cell redox regulatory mechanisms and disrupt redox homeostasis. Pharmacodynamic biomarkers that measure therapeutic efficacy or toxicity could improve patient management. Using immunoblot analyses and mass spectrometry, we identified that serpins A1 and A3 were S-glutathionylated in a dose- and time-dependent manner following treatment of mice with drugs that alter reactive oxygen or nitrogen species. Tandem mass spectrometry analyses identified Cys256 of serpin A1 and Cys263 of serpin A3 as the S-glutathionylated residues. In human plasma from cancer patients, there were higher levels of unmodified serpin A1 and A3, but following treatments with redox active drugs, relative S-glutathionylation of these serpins was higher in plasma from normal individuals. There is potential for S-glutathionylated serpins A1 and A3 to act as pharmacodynamic biomarkers for evaluation of patient response to drugs that target redox pathways.
Introduction
Preclinical and early clinical testing of novel anticancer drugs can be significantly enabled by the inclusion of direct or surrogate biomarkers that correlate with pharmacologic effects (1, 2). Incorporation of such biomarkers can enhance data interpretation, provide an explanation for either positive or negative correlations with efficacy or toxicity, and aid in early decision candidate selection. In complex diseases such as cancer, recent trends in drug development have moved toward targeted therapies to provide maximal therapeutic response while minimizing side effects. Failure of early stage clinical trials incurs great expense and there is merit in including biomarkers, both to estimate treatment efficacy and to identify patients with phenotypic characteristics that might predict response outcomes or toxicities.
In evaluating drug response, a single biomarker can lend itself to quantitative measurements through dose and time response studies. The metabolism of many drugs leads to production of electrophilic species that can generate reactive oxygen or nitrogen species (ROS; RNS). These can directly influence redox balance in both blood and tissue compartments. Biologic sensing of redox changes is most actively monitored through select cysteine residues in various proteins. Although there is debate as to the precise definition of what constitutes redox sensing versus redox signaling (3), there is little doubt that cysteines at various oxidation states are integral. S-glutathionylation is a posttranslational modification that occurs when a cysteine in a low pK environment forms a disulfide bond with GS− . This is reversible and the resultant S-glutathionylation cycle has the potential to selectively regulate the function of numerous enzymes, receptors, structural proteins, transcription factors, and transport proteins. Moreover, posttranslational modification may alter a variety of protein–protein interactions (4). The activities of serine protease inhibitors (serpins) are regulated by modifications of key cysteines (5, 6), in which, for example, S-glutathionylation of serpin A1 results in conformational changes that weaken its affinity for its target protease, thereby reducing its effectiveness in preventing proteolytic activity (7).
Vertebrate serpins are classified into 6 subgroups and represent approximately 2% of the total protein in human plasma (8). Although most are defined by their inhibitory activities, certain serpins have noninhibitory roles as hormone transporters (9), molecular chaperones (10), or tumor suppressors (11). Moreover, serpins can influence myeloproliferation and hematopoetic progenitor cell mobilization. Serpins A1 and A3 have redox-sensitive cysteines and are downregulated in bone marrow during mobilization of hematopoetic progenitor cells into the peripheral bloodstream (12), implicating a regulatory function for S-glutathionylation and redox homeostasis in the marrow compartment. This is consistent with the fact that certain redox active drugs [including the glutathione disulfide mimetic NOV-002 (13) and the GSH peptidomimetic, Telintra (14)] have both preclinical and clinical myeloproliferative effects (15).
In this study, we have identified the S-glutathionylation of plasma serpins A1 and A3 as potential quantifiable response biomarkers for assessing response to 2 different types of drugs that either directly or indirectly impact redox status. NOV-002 is a formulation of disodium glutathione disulfide (GSSG) that in combination protocols is in phase II trials for the treatment of breast cancer. NOV-002 causes a redox regulation of protein thiols that persists in plasma for approximately 4 hours and in rodents and humans stimulates proliferation of bone marrow progenitor cells (13). PABA/NO is a diazeniumdiolate that acts as a direct nitrogen monoxide (NO) donor and is in development as an anticancer drug. We show that plasma serpin S-glutathionylation correlates with drug treatment in vivo and in vitro. Furthermore, analysis of cancer patient plasma samples suggests disease-specific variation in unmodified and S-glutathionylated serpin profiles correlating with drug concentrations.
Materials and Methods
Reagents
Reduced GSH was from Sigma, NOV-002 provided by Novelos Therapeutics, and PABA/NO from Dr. Larry Keefer (National Cancer Institute at Frederick, Frederick, MD; refs. 16, 17).
Animal and human plasma studies
Mice were treated with an intravenous bolus of 25 mg/kg NOV-002 and after 1 hour blood was collected via orbital bleed in heparin-coated tubes. Plasma was separated by centrifugation and proteins separated on SDS-PAGE and transferred for immunoblot analyses. Human blood samples were obtained with informed consent from 8 cancer-free volunteers and 47 oncology patients with various types/stages of cancer: 20 acute myeloid leukemia (AML); 10 acute lymphoblastic leukemia (ALL); 1 multiple myeloma (MM); 4 myelodysplastic syndrome (MDS); 3 chronic lymphoblastic leukemia; 1 chronic myeloid leukemia (CML); 2 lung carcinomas; 2 urothelial carcinomas; 1 leiomyosarcoma; 1 neuroblastoma; 1 spindle cell carcinoma, 1 invasive ductal carcinoma. Eight to 10 cc of blood was collected into Vacutainer tubes containing EDTA. Plasma was separated via centrifugation and treated with NOV-002 or PABA/NO at 37° C.
Immunoblot analysis
Equal amounts of purified recombinant serpins A1/A3 or total plasma protein, as determined by Bradford assay (Bio-Rad Laboratories), were separated on nonreducing SDS-polyacrylamide gels. Proteins were transferred onto poly-vinylidene difluoride membranes (Bio-Rad Laboratories) and probed with a monoclonal anti-GSSG antibody (Virogen) to detect S-glutathionylation, or polyclonal antibodies for serpins A1 and A3 (R&D systems), glutaredoxin1 (Abeam) or GSTP (MBL). Equal loading was estimated by polyclonal rabbit albumin antibody (Abcam). Secondary antibodies were from Amersham Biosciences.
Identification of S-glutathionylated plasma proteins
Plasma S-glutathionylated protein bands were isolated from gels run simultaneously with immunoblots and trypsin digested. Peptide mass was analyzed by matrix-assisted laser desorption/ionization, time-of-flight (MALDI-TOF) mass spectrometry at the Proteomics Core Facility. Protein identification was done using software from the NCBI protein database.
Immunoprecipitation
Human plasma samples (2 mg) were treated with 50 µmol/L of NOV-002 for 60 minutes at 37°C. Biotinylated rabbit polyclonal antibodies to serpins A1 and A3 (Abeam, 5 µg) were used to pull-down respective proteins by overnight incubations at 4°C. Antibody complexes were isolated through incubation with NeutrAvidin agarose resin (Thermo Scientific) for 2 hours. Resin-bound complexes were washed 5 × with immunoprecipitation buffer (20 mmol/L HEPES, 300 mmol/L NaCl, 1% Triton, 10% glycerol) and proteins eluted in nonreducing SDS-sample buffer. Immunoprecipitated proteins were separated by nonreducing SDS-PAGE and S-glutathionylation of serpins A1 and A3 evaluated by immunoblot.
In vitro S-glutathionylation assays
Purified serpin A1 or A3 (50 ng) was incubated at 37°C in 50 mmol/L potassium phosphate buffer (pH 7.2), 1 mmol/L GSH with either NOV-002 or PABA/NO. Samples were run on nonreducing SDS-PAGE gels for subsequent transfer and immunoblot analysis.
Tandem mass spectroscopy
Purified serpins A1 and A3 were treated with 100 µmol/L PABA/NO or 1 mmol/L NOV-002 for 30 minutes at 37°C, digested with Lys-C and analyzed via liquid chromatography (LC)–electrospray ionization (ESI)–tandem mass spectrometry (MS/MS) on a linear ion trap mass spectrometer (LTQ, Thermo Finnigan) coupled to an LC Packings nano LC system. S-glutathionylated serpin A3 peptides required additional trypsin digestion (See Supplementary Data for instrument settings).
Spectroscopic analysis of serpin A1 and A3 in vitro
The effect of serpin S-glutathionylation on enzyme secondary structure was examined by circular dichroism (CD) measurements carried out on a 202 AVIV Associates CD spectrometer using a semi-micro quartz rectangular 1 × 10 × 40 mm cuvette. Serpins A1 and A3 (~2 mg/mL) were S-glutathionylated by treatment with 40 µmol/L PABA/NO and 1 mmol/L GSH for 30 minutes in 20 mmol/L PB, pH 7.4, at 37°C. Excess PABA/NO and GSH were eliminated using Biospin-6 (Bio-Rad) SEC micro-spin columns.
Purified native and S-glutathionylated serpins A1 and A3 (~95% homogeneous, 40 µmol/L in 20 mmol/L PB, pH 7.4) were maintained at 22°C using a Pelletier element. Spectra were recorded while scanning in the far-ultraviolet region (190–260 nm), with bandwidth of 1.0 nm, step size of 0.5 nm, integration time of 30 seconds. Protein tryptophanyl fluorescence was recorded on a QM-4 spectrofluorometer (PTI) using 10 × 10 × 40-mm quartz cuvette, excitation and 2.5 and 5.0 nm emission slits. To minimize effects of protein tyrosines and phenylalanines, an excitation wavelength of 295 nm was used. Background spectra were subtracted from final emission data.
Statistical analyses
Statistical analyses were carried out using GraphPad Prism (GraphPad Software). P values lower than 0.05 were considered significant. Parametric data were statistically evaluated using t tests and nonparametric data were evaluated using Mann-Whitney or Wilcoxon matched pairs signed rank test tests, based on data distribution. Multiple comparisons were analyzed using ANOVAs with Dunnett’s Multiple Comparison tests or Kruskall–Wallis tests with Dunn's multiple Comparsion test. Corrections were applied based on program recommendation. Relative S-glutathionylation levels induced in cancer were analyzed using 2-way ANOVAs.
Results
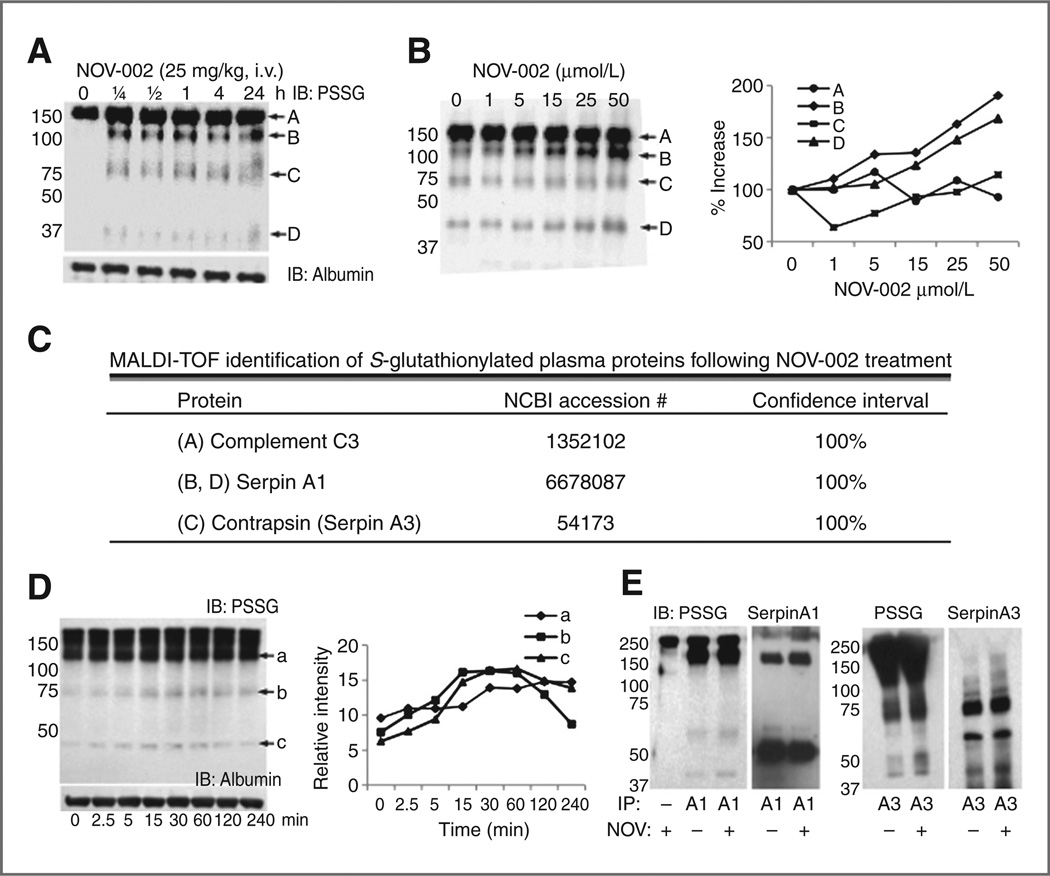
In vivo treatment with NOV-002 leads to S-glutathionylation of a limited subset of plasma proteins in mice (Fig. 1A). Immunoblots from animals detected 4 prevalent S-glutathionylated proteins within 15 minutes of treatment. Excised bands were trypsin digested and peptide mass analyzed by MALDI-TOF. Protein identification (Fig. 1C, Table) used software from the NCBI protein database. Complement C3 appears as a common S-glutathionylated protein throughout the analysis, even before drug treatments. Proteins bands B and D were identified as serpin A1, whereas band C was identified as serpin A3. Murine contrapsin is homologous to human serpin A3. Ex vivo treatment of mouse plasma with various concentrations of NOV-002 showed similar S-glutathionylation induction. Quantification of band densities showed increasing serpin S-glutathionylation with dose exposure (Fig. 1B).
Figure 1.
NOV-002 treatment induces serpin A1 and A3 S-glutathionylation in vivo in mouse plasma and ex vivo in human plasma. Immunoblot analyses of S-glutathionylated proteins (PSSG) from wild-type mice treated in vivo with an intravenous bolus of 25 mg/kg NOV-002 at time points following 1 hour (A) and wild-type mouse plasma treated ex vivo with various concentrations of NOV-002 (B). MALDI-TOF mass spectrometry identified S-glutathionylated mouse plasma proteins (C). Human plasma was treated ex vivo with 40 µmol/L NOV-002 for 0 to 240 minutes and evaluated by immunoblot for S-glutathionylated proteins (D). Albumin immunoblotting determined equal protein loading. For immunoprecipitation (IP), human plasma was treated with 40 µmol/L NOV-002 for 1 hour and biotinylated serpin A1 and A3 antibodies were used to pull-down total unmodified and modified serpins. S-glutathionylated proteins were evaluated by immunoblot (E). Blots were stripped and reprobed with anti-serpin A1 or A3 as a loading control. Due to high homology, there is significant cross reactivity of anti-A1 and A3. IB, immunoblot.
Table 1.
Relative fold change in protein expression (cancer vs. cancer free) of unmodified serpins and S-glutathionylated serpins
| Protein | Cancer | AML | ALL |
|---|---|---|---|
| Serpin A1 | 1.96 ± 0.12a | 2.02 ± 0.15a | 1.99 ± 0.30a |
| Serpin A3 | 4.16 ± 1.07a | 2.42 ± 0.39a | 5.64 ± 3.30a |
| Glut serpin A1 (PSSGa) | 1.05 ± 0.11 | 1.17 ± 0.15 | 0.35 ± 0.09a |
| Glut serpin A3 (PSSGb) | 0.94 ± 0.19 | 1.00 ± 0.13 | 0.49 ± 0.08 |
| Glut serpin A1 (PSSGc) | 0.70 ± 0.11a | 0.81 ± 0.19 | 0.37 ± 0.07a |
| Glut serpin AVserpin A1 (PSSGa/SerpinA1) | 0.56 ± 0.06a | 0.62 ± 0.09a | 0.19 ± 0.04a |
| Glut serpin AVserpin A3 (PSSGb/SerpinA3) | 0.43 ± 0.06a | 0.56 ± 0.09a | 0.22 ± 0.06a |
| Glut serpin A3/serpin A1 (PSSGc/SerpinA1) | 0.37 ± 0.06a | 0.41 ± 0.10a | 0.19 ± 0.03a |
| GSTp | 0.73 ± 0.05a | 0.86 ± 0.07 | 0.61 ± 0.09a |
| Grx | 2.18 ± 0.32a | 2.95 ± 0.68a | 1.60 ± 0.22 |
NOTE: SDS-PAGE and immunoblot analyses analyzed levels of unmodified and S-glutathionylated serpins A1 and A3. Relative quantities of unmodified serpins and S-glutathionylated serpin A1 (PSSGa and PSSGc), and serpin A3 (PSSGb) were determined by densitometry normalized to both albumin loading controls and internal standards incorporated on all gels, n = 8, cancer free; n = 47, cancer; n = 20, AML; n = 10, ALL. Data are fold change versus cancer-free ± SE.
P < 0.05.
Extending these analyses, purified human plasma samples were similarly treated ex vivo with NOV-002. Anti-PSSG immunoblots showed the induction of several distinct S-glutathionylated protein bands with a profile similar to in vitro and in vivo in mouse plasma assays. Basal levels of S-glutathionylated proteins were detected in both mouse and human untreated samples, most likely the result of ex vivo processing. Treatment with 40 µmol/L NOV-002 caused a time-dependent increase in the relative amount (as normalized to albumin loading control) of S-glutathionylated proteins that peaked between approximately 40 to 60 minutes and gradually decreased with additional time (Fig. 1D). This indicated a relatively rapid turnover rate involving the S-glutathionylation and deglutathionylation of proteins following drugs.
Immunoprecipitation of serpins A1 and A3 from human plasma confirmed that both these proteins are S-glutathionylated (Fig. 1E). Due to high sequence homology (~46%) between human serpins A1 and A3, a certain extent of antibody cross-reactivity may be anticipated. In particular, the immunoprecipitating serpin A3 antibody also pulled down serpin Al, as evidenced in the PSSG blot. Protein bands correspond with the approximate expected molecular weights of serpins A1 and A3 (seen on PSSG immunoblots). Detection of multiple protein bands on immunoblots probed with serpin antibodies is not uncommon. Serpins and serpin/protease complexes undergo gradual proteolysis at the serpin active site producing cleavage products, perhaps explaining the multiple protein bands detected on serpin immunoblots (18).
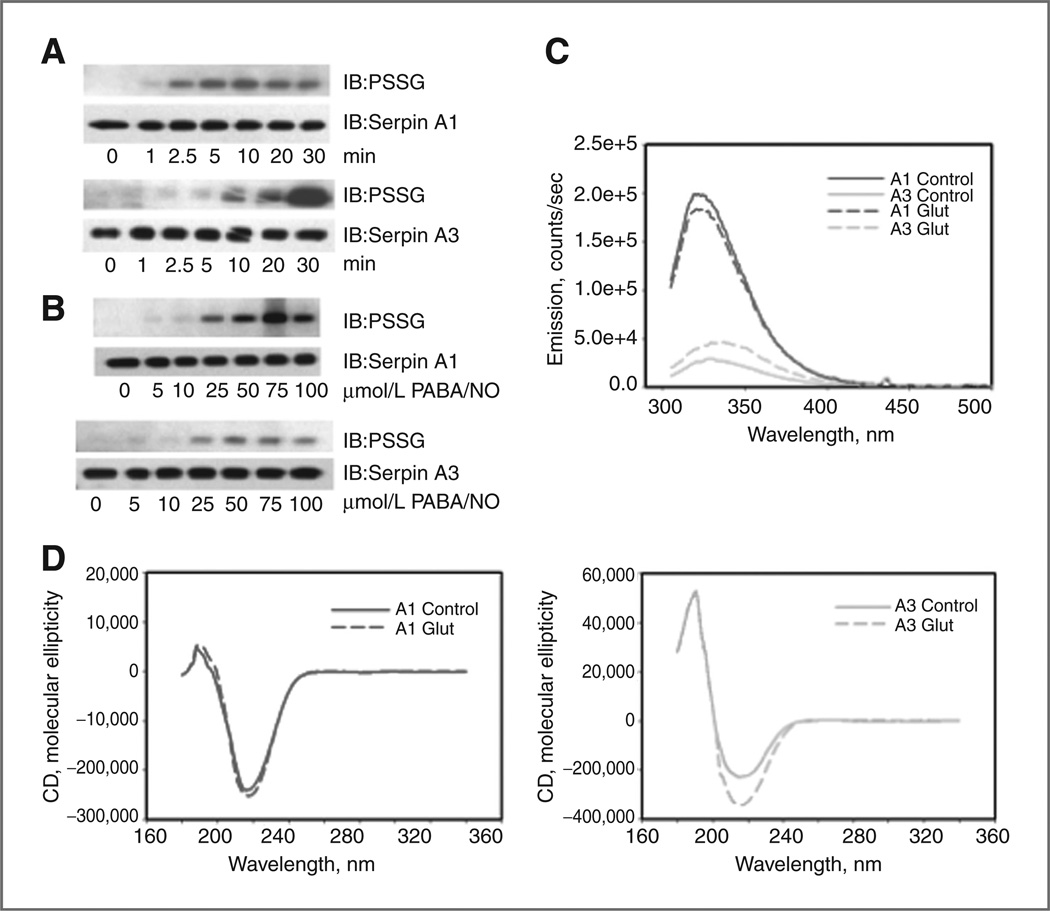
Purified recombinant A1 and A3 protein was treated in vivo and in vitro with agents known to induce protein S-glutathionylation (13, 19). In the presence of PABA/NO and GSH, both serpins A1 and A3 were S-glutathionylated in a time- (Fig. 2A) and concentration-dependent manner (Fig. 2B). Serpin A1 was rapidly S-glutathionylated within 1 minute and peaked at approximately 5 to 10 minutes. Following this, S-glutathionylated protein levels gradually declined. Alternatively, serpin A3 showed a more gradual increase in S-glutathionylation over a 30-minute time course, suggesting unique protein properties despite high sequence homology. Similar drug concentrations were needed to induce minimum (at ~25 µmol/L PABA/NO) and maximum (at ~75 µmol/L PABA/NO) S-glutathionylation levels of both serpins A1 and A3. However, drug-induced S-glutathionylation levels of serpin A3 became relatively saturated between 25 and 50 µmol/L PABA/NO, suggesting that serpin A3 S-glutathionylation may be more time dependent. We evaluated the effects of S-glutathionylation on serpin secondary structure. Tryptophanyl fluorescence of serpin A3 was lower than that of serpin A1 indicating a quenching in the former. Tryptophanyl fluorescence of S-glutathionylated serpin A3 had an increased intensity and a shift of emission maximum from 328 to 336 nm, interpreted as tryptophanyl exposure to a more polar environment, decrease of quenching and structural change (Fig. 2C). The CD spectrum (far UV, 190– 260 nm) of S-glutathionylated serpin A1 was similar to that of native protein, consistent with a minimal increase in the α-helical content (206–220 nm) of the protein (Fig. 2D). This suggested a minor decrease in tryptophanyl fluorescence caused by S-glutathionylation, indicating a reasonably intact tertiary and quaternary structure. In contrast, the CD spectrum of S-glutathionylated serpin A3 was distinct from that of the native protein, consistent with an increase in the β-strand content of the protein and altered protein folding (Fig. 2D). These fluorescent analyses confirmed that S-glutathionylation affects the tertiary structure of serpin A3.
Figure 2.
S-glutathionylation of serpins A1 and A3 is time and dose dependent and impacts protein structure. Recombinant serpin A1 and A3 were treated with 1 mmol/L GSH and 40 µmol/L of PABA/NO for 0 to 30 minutes (A) or 0 to 100 µmol/L of PABA/NO for 30 minutes (B). Immunoblot analyses were used to detect S-glutathionylation (PSSG) and total serpin. Spectroscopic analyses of secondary and tertiary (quaternary) structure of native (control; solid curves) and PABA/ NO + GSH–treated (Glut; dashed curves) A1 (top curves, dark gray) and A3 (bottom curves, light gray) protein were done in vitro using tryptophanyl fluorescence (C) and circular dichroism (D). IB, mmunoblot.
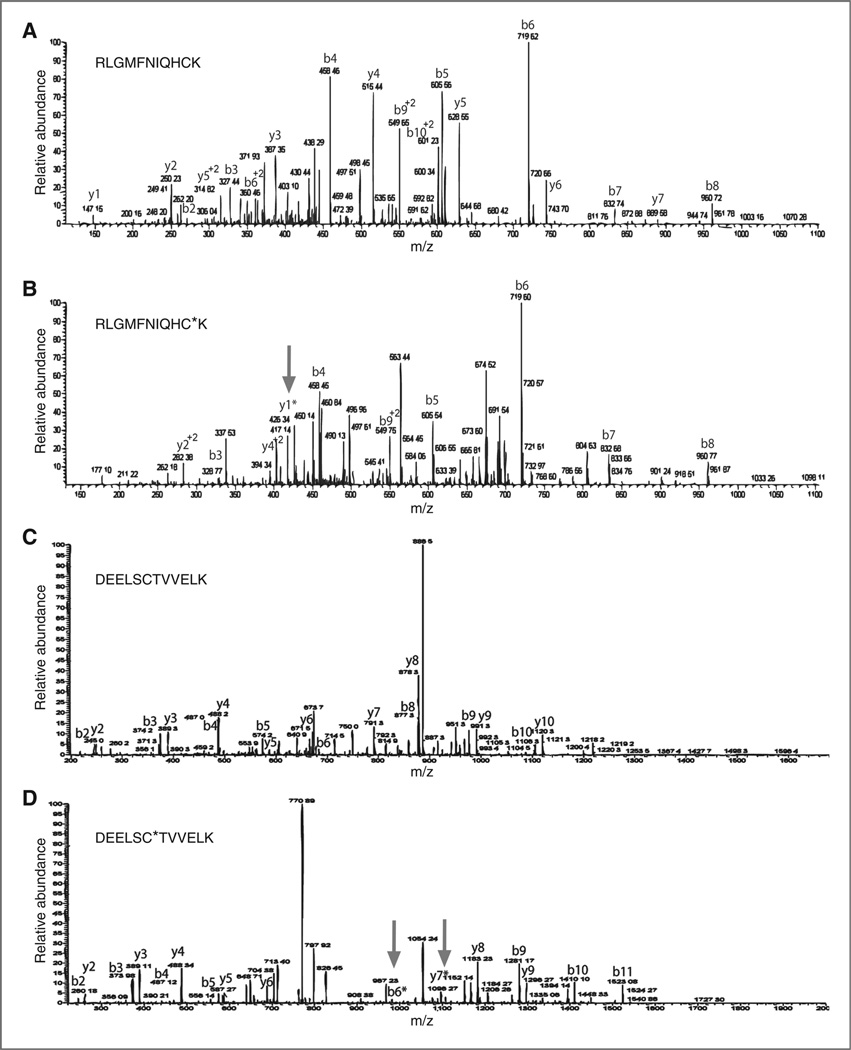
S-glutathionylated recombinant serpins A1 and A3 were analyzed using MS/MS to identify S-glutathionylated cysteines. Purified recombinant serpins A1 and A3 were incubated at 37° C for 30 minutes the presence of 100 µmoI/L PABA/NO and 10 mmol/L GSH. Analyses of (RLGMFNIQHCK) of serpin A1 (Fig. 3A and B) indicated that an additional approximately 305 Da at Cys256, representing GSH, in addition to the Cys alone (~103 Da) and water (~18 Da), was present only in the drug-treated serpin A1 (Fig. 3B). MS/MS analyses similarly identified Cys263 of (DEELSCTWELK; Fig. 3C and D) treated serpin A3 to be modified by the addition of GSH, indicated by an additional approximately 305 Da (Fig. 3D). These data provided a platform to develop analytical methods for quantification of S-glutathionylated serpins in biologic fluids.
Figure 3.
Serpin A1 is S-glutathionylated at Cys256 and serpin A3 is S-glutathionylated at Cys263. Recombinant serpin A1 and A3 proteins were treated with 1 mmol/LGSH and 100 µmol/L PABA/NO for 30 minutes. To identify specific cysteine residues susceptible to S-glutathionylation, control unmodified serpin A1 (A) PABA/NO-treated serpin A1 (B); control unmodified serpin A3 (C) and PABA/NO-treated serpin A3 (D) were digested under nonreducing conditions and analyzed via LC–ESI–MS/MS to identify modification GSH addition. A single S-glutathionylated modification [+305.6] was detected at of Cys256 of serpin A1 (B) and Cys263 of treated serpin A3 (D). Arrows indicate modified peaks. The 2 arrows in D represent both forward and reverse analyses.
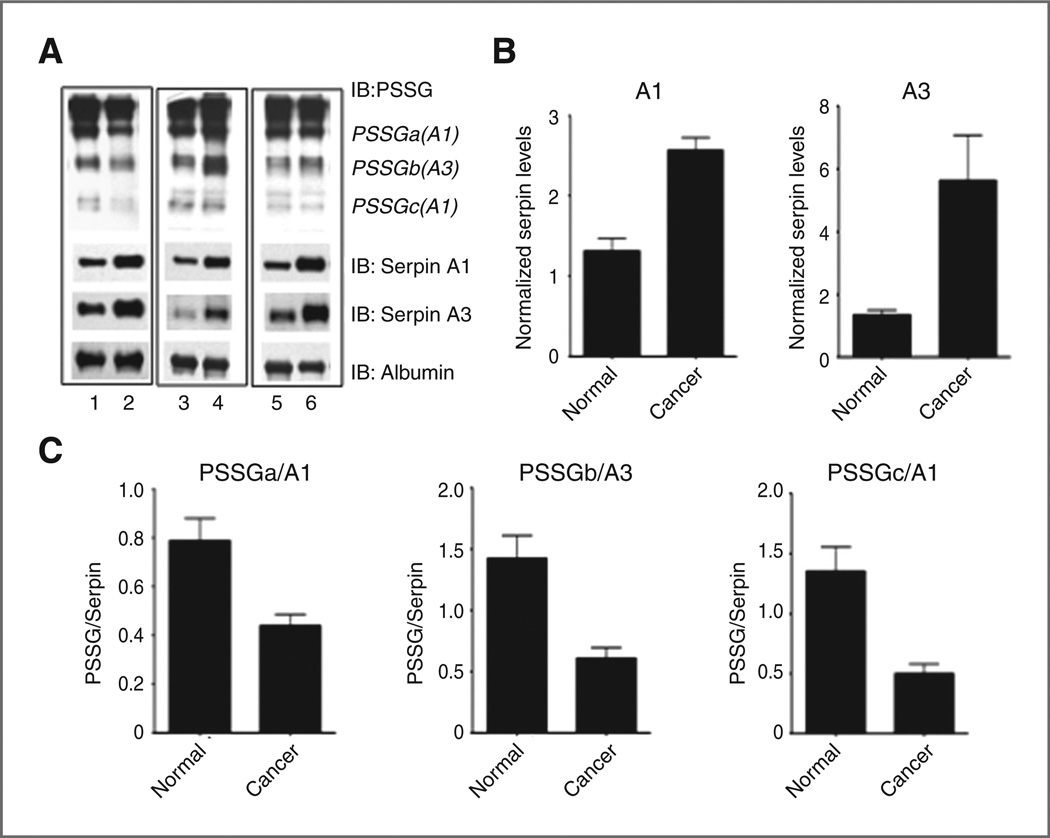
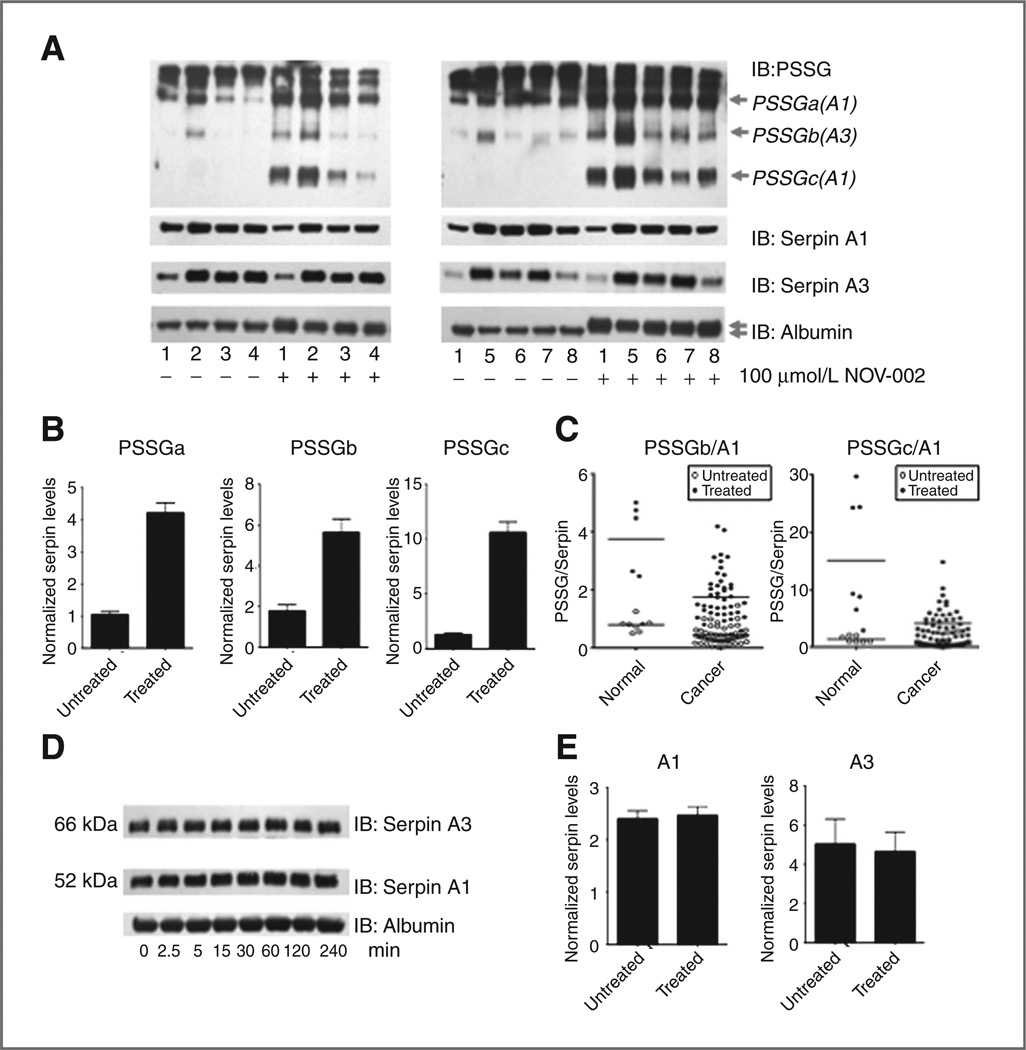
Human plasma samples from 47 cancer patients undergoing chemotherapy treatments and 8 cancer-free patients were analyzed for total and S-gluathionylated serpin A1 and A3 protein. Immunoblot densitometry showed significantly (P < 0.05) elevated amounts of each unmodified serpin in cancer patients (Fig. 4A and B). All protein bands were normalized to an albumin loading control as well as an internal standard, permitting comparison across membranes. Levels of S-glutathionylated serpins A1 and A3 did not correlate with total serpin levels. In fact, serpin S-glutathionylation profiles were distinct between patients, suggesting that individual patients with specific disease and treatment profiles have unique serpin S-glutathionylation. However, when analyzing all cancers in a group, the overall ratio of S-glutathionylated serpin A1 (PSSGa and PSSGc) and A3 (PSSGb) to unmodified serpin A1 and A3, respectively, was significantly decreased (P < 0.05) in cancer patients (Fig. 4C). Thus, in reference to the total amount of serpin, the fraction of serpin protein subject to S-glutathionylation was lower in cancer patients. Table 1 summarizes the relative fold change in protein expression of unmodified serpins and S-glutathionylated serpins in total cancer as compared with cancer-free, in addition to AML and ALL as individual groups. While general trends remain consistent between all cancers, discrepancies in significance may suggest disease-specific profiles. Of particular interest, levels of basal S-glutathionylated serpins were significantly lower in ALL patients as compared with AML and cancers as a group.
Figure 4.
Unmodified serpin is elevated in certain cancers, whereas the ratio of S-glutathionylated to unmodified serpin is decreased. Human plasma samples from cancer-free patients (n = 8; represented in lanes 1,3,5 of A) and cancer patients undergoing chemotherapeutic treatment (n = 47; represented in lanes 2,4,6 of A) were analyzed by immunoblotting for unmodified serpin A1 and A3 proteinlevels(B)as well as relative levels of S-glutathionylated serpin compared with total unmodified serpin (C). Relative quantities of unmodified serpins and S-glutathionylated serpin A1 (PSSGa and PSSGc), and serpin A3 (PSSGb) were determined by densitometry measurements normalized to both albumin loading controls and an internal standard incorporated on all gels. Data are ± SEM. IB, immunoblot.
Ex vivo treatment of patient plasma with NOV-002 for 60 minutes at 100 µmol/L proportionally enhanced levels of S-glutathionylated A1 and A3 [Fig. 5A and B and Supplementary Fig. S1 (lighter exposures)]. Figure 5B shows the relative quantities of S-glutathionylated serpin A1 (PSSGa and PSSGc) and A3 (PSSGb) after NOV-002 treatment from samples of 47 cancer and 8 cancer-free patients, determined by densitometry measurements normalized to both albumin loading controls and an internal standard incorporated on all gels. S-glutathionylated serpin A1 (PSSGa and PSSGc) and A3 (PSSGb) levels were significantly increased (P < 0.05) in response to NOV-002. Two-way ANOVA analyses show that ex vivo treatment of plasma with 100 µmol/L NOV-002 resulted in significantly higher relative increases (P < 0.05) in serpin A1 S-glutathionylation (PSSGa/A1 and PSSGc/A1) in cancer-free plasma as compared with cancer patients (Fig. 5C). Thus, in response to treatment, serpin S-glutathionylation and disease status are interrelated and showed altered redox homeostasis in cancer patients. NOV-002 treatment also induced S-glutathionylation of albumin, as evidenced by the doublet band detected only at high drug concentrations. Critically, NOV-002 treatment did not alter serpin stability, as indicated by total serpin immunoblotting. Treatment with 40 µmol/L NOV-002 for 0 to 240 minutes did not change levels of unmodified serpin A1 or A3 (P > 0.05) (Fig. 5D). Additional analyses using treated and untreated plasma samples from cancer and cancer-free patients supported these data (Fig. 5E). Treatment with Velcade, vinblastine, Taxol, or tamoxifen did not induce changes in plasma serpin A1 or A3 S-glutathionylation (Supplementary Fig. S2A – D).
Figure 5.
Ex vivo treatment with NOV-002 induces serpin A1 and A3 S-glutathionylation and results in greater relative increases in serpin A1 glutathionylation in cancer-free human plasma. Cancer (samples 2–8) and cancer-free (sample 1) human plasma samples were treated with 100 µmol/L NOV-002 for 60 minutes (A). S-glutathionylation (PSSG), serpin A1, serpin A3, and albumin levels (loading control) were evaluated by immunoblot. Levels of S-glutathionylated serpin A1 (PSSGa and PSSGc) and serpin A3 (PSSGb) after NOV-002 treatment (both cancer and cancer free) were determined by densitometry normalized to both albumin loading controls and an internal standard on all gels (B). Relative S-glutathionylation levels in reference to total unmodified serpin levels induced in response to NOV-002 treatment in cancer patient plasma as compared with those induced in cancer-free plasma were analyzed using 2-way ANOVAs (C). To confirm that S-glutathionylation of serpins does not alter serpin A1 and A3 protein stability, plasma was treated with 40 µmol/L NOV-002 for 0 to 240 minutes (D). The relative quantity of serpins A1 and A3 in treated and untreated samples (both cancer and cancer free) after 100 µmol/L NOV-002 treatment for 1 hour was normalized to both albumin loading controls and an internal standard on all gels(E). Data are mean for 8 cancer-free and 47 cancer samples ± SEM. IB, immunoblot.
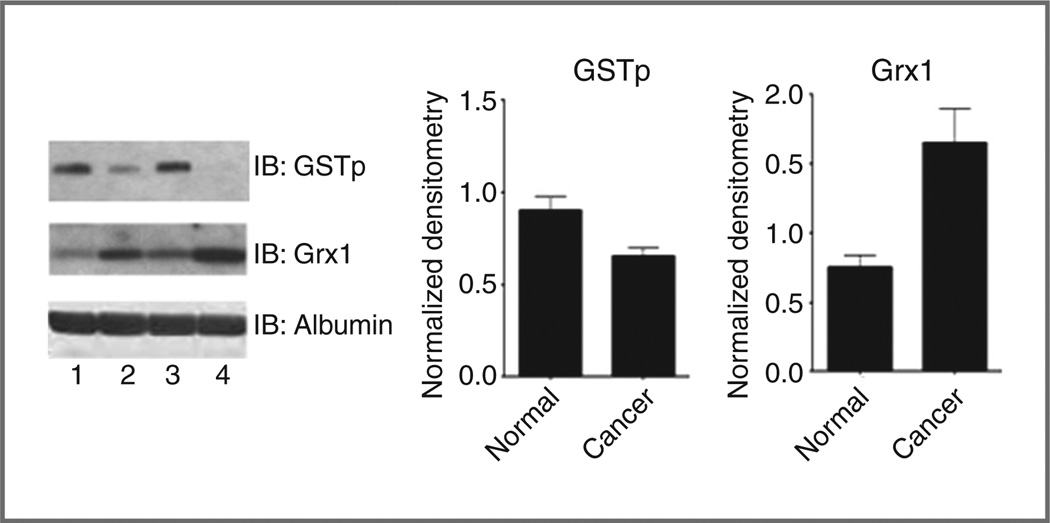
GSTP and Grx1 are involved in the forward and reverse reactions of S-glutathionylation. Relative protein levels were determined via densitometry and then normalized to an albumin loading control, as well as an internal standard incorporated into all gels. Plasma protein levels of GSTP were significantly decreased (P < 0.05) in cancer patients as compared with cancer free. Conversely, plasma Grx1 levels were significantly elevated (P < 0.05) in cancer patients (Fig. 6). These data are consistent with the interpretation that the decreased relative ratio of S-glutathionylated to unmodified serpin in cancer patient plasma may be linked with diminished GSTP levels and a relatively enhanced rate of serpin deglutathionylation, as a consequence of elevated Grx1.
Figure 6.
Cancer patient plasma samples have decreased levels of GSTP protein and elevated levels of Grx1. Human plasma samples from cancer-free patients (represented in lanes 1 and 3) and cancer patients undergoing chemotherapeutic treatment (represented in lanes 2 and 4) were evaluated for GSTP and Grx1 using immunoblots. Relative quantities of GSTP and Grx1 were normalized to both albumin loading controls and an internal standard on all gels. Data are mean for 8 cancer-free and 47 cancer samples ± SEM. IB, immunoblot.
Discussion
There is expanding interest in adopting a redox-modulating platform in drug discovery/development in cancer (20). In this study we used 2 drugs that alter redox homeostasis through distinct mechanisms. NOV-002 is an oxidized glutathione mimetic that modifies extra- and intracellular ratio of GSH: GSSG (13, 21). PABA/NO releases NO to raise levels of RNS and ROS (19). Increased efficacy and limited off-target toxicities could be facilitated by the identification of plausible pharmacodynamic biomarkers (preferably through noninvasive approaches) that serve as predictors of drug response. Initially using mice, we identified S-glutathionylated serpins A1 and A3 as quantifiable response biomarkers. These preclinical observations were extended to human plasma samples in which correlative associations with drug exposure and disease status were identified. Moreover, variations in the expression of unmodified and/or S-glutathionylated serpin profiles were identified and may reflect aberrant redox homeostasis in some cancers.
Because of the variable valence states of sulfur, cysteine-targeted oxidation has evolved as a critical regulator of protein function in a number of signal transduction pathways. Protein S-glutathionylation is cyclical in nature [the forward reaction is catalyzed by GST, the reverse by glutaredoxin (22)], providing the framework for reversible signaling (23). A number of protein clusters with roles in cell survival pathways are characteristically sensitive to posttranslational modification, including enzymes with catalytically important cysteines, signaling proteins, and transcription factors. Serine proteinase inhibitors are susceptible to modulation by redox conditions (7, 24, 25). MS/MS analyses identified Cys256 of serpinA1 and Cys263 of serpin A3 as S-glutathionylation targets. Cys256 of full-length serpin A1 has previously been shown to be redox sensitive (7, 26). However, the present data provided are the first to report a similar redox-sensitive cysteine in serpin A3. Oxidation of cysteine residues in serpins inhibits their activity (7, 24–26). From a functional basis, members of the serpin family are involved in regulation of myeloproliferation and hematopoetic progenitor cell (HPC) mobilization (12, 27). Downregulation of serpins A1 and A3 in bone marrow occurs during progenitor cell mobilization and influences marrow microenvironment and migratory behavior of HPCs (12). Our data show that serpin A1 and A3 S-glutathionylation following NOV-002 or PABA/NO treatment alters their structure, and such factors may alter the role of serpins involved in myeloproliferative pathways. To interrogate the specificity of these effects for ROS/RNS active drugs, we investigated (Supplementary Fig. S2) the effects of Velcade (proteasome inhibitor), vinblastine and Taxol (antimicrotubule drugs), or tamoxifen (estrogen receptor antagonist). Although these drugs may produce low levels of ROS indirectly, the data suggest that there is no dose effect response on plasma serpin S-glutathionylation profiles.
Immunoblots of S-glutathionylated serpins revealed proteins of various molecular weights. Different segments of the A1 molecule may possess unique biologic properties that may or may not be linked to protease inhibitor activity. Low molecular weight fragments of serpin A1, generated by complexing with neutrophil elastase or by macrophage metalloelastase attack, possess chemoattractant properties (28). In this context, serpin S-glutathionylation may have a mechanistic connection with the in vivo myeloproliferative activity of NOV-002 (13). The redox-sensitive serpin, bomapin, is directly involved in the responsiveness of myeloid progenitor cells to their microenvironment (29). Posttranslational regulation of serpins in the peripheral circulation may indicate a role in the control of proteolytic pathways. Whatever the functional association, as the plasma half-life of S-glutathionylated proteins is approximately 4 hours (19), our mouse studies provided evidence that these posttranslationally modified proteins could be viable as pharmacodynamic biomarkers in a clinical setting. There is precedent for using posttranslationally modified proteins as biomarkers. In initial trials of the CML therapeutic imatinib levels of phosphorylated CRKL correlated directly with BCR-ABL inhibition and proved useful in determining appropriate dosing strategies (30, 31). In addition, success of small-molecule inhibitors of epidermal growth factor receptor (gefitinib, erlotinib) and Src kinases (dasatinib) can be linked with phosphorylation of downstream markers such as ERK1/2, FAK, or paxillin (32, 33).
Our patient profile data showed that baseline protein levels of both serpin A1 and A3 were elevated in the plasma of cancer patients. Elevated serpin A1 expression in tumors has been correlated with poor prognosis and highly invasive, metastatic adenocarcinomas (34–36) and with poor response in MM (37). Although the functional importance of serpins A1 and A3 in cancer is not yet clearly defined, serpin A1 has inhibitory activity against cytotoxic T lymphocytes and natural killer cells (38), suggesting that the tumor-promoting effect of serpin A1 could be related to a decreased immune response against malignant cells. Serpin A1 overexpression may also be a useful biomarker in insulinomas (39). In leukemias, in which the cancer is of bone marrow origin, further defining a role of serpin A1 and A3 S-glutathionylation in myeloproliferation may be valuable with regard to development of tailored treatment strategies. Chronic oxidative stress, characterized by excessive cellular ROS levels, occurs in several hematopoietic malignancies including ALL, MDS, CML, and AML (40–42). While ROS is involved in normal hematopoiesis, imbalances of ROS results in redox dysregulation and may influence downstream redox-mediated signaling events, including S-glutathionylation of serpins. Our data suggest that ALL patients have lower basal levels of glutathionylated serpins A1 and A3, perhaps indicating that certain myeloid leukemias might respond preferentially to redox-targeted therapeutics.
Analyses of cancer patients as a group showed that their overall ratio of S-glutathionylated serpins to unmodified serpins was decreased. Furthermore, ex vivo drug-induced S-glutathionylation was blunted in cancer patient plasma, results consistent with a disrupted redox homeostasis associated with cancer. High levels of oxidative stress markers in patient sera often correlate with similar markers within tumors, suggesting the usefulness of patient serum in assessing redox state. Further correlative analysis of patient samples estimated levels of enzymes associated with the forward and reverse reactions of the S-glutathionylation cycle. Levels of GSTP (forward reaction) were significantly decreased in cancer patient plasma compared with cancer free, whereas Grx1 (reverse reaction) levels were elevated. These results are consistent with the relative ratios of S-glutathionylated to unmodified serpin proteins found in cancer patient plasma samples. Modulation of Grx1 levels, either by overexpression or RNAi knockdown, directly influences S-glutathionylation of inhibitory NF-κβ kinase by respectively decreasing or catalyzing its posttranslational modification (43). However, reports indicate a dual role for Grx1, as a glutathionylating enzyme under ROS or as a deglutathionylase when the oxidative signal or stress subsides (44). Earlier results have shown that the glutathione conjugate of cisplatin can inhibit the enzyme activity of Grx (45). However, there were no indications that the parent drug (or related platinum analogs) had inhibitory effects. The drug concentrations used in this study are generally too low to cause enzyme inhibition, either for GSTP (13) or of Grx (45).
This article provides evidence supporting the usefulness of posttranslationally modified serpins as pharmacodynamic biomarkers. Optimization of detection and characterization protocols will provide promising opportunity in a clinical setting in which serpin S-glutathionylation may be directly used to infer patient response to redox-modulating therapeutics. The ability to detect S-glutathionylated serpin peptides, as we have shown in our MS/MS studies, may provide a platform to develop increased sensitivity of detection for quantification of S-glutathionylated serpins in biologic fluids.
Supplementary Material
Acknowledgments
The authors thank the Drug Metabolism and Pharmacokinetics and Proteomics Core Facilities at MUSC.
Grant Support
The work received financial support from the NIH (CA08660 and CA117259); C06 RR015455 from the Extramural Research Facilities Program of NCCR and the South Carolina SmartState program.
Footnotes
Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors’ Contributions
Conception and design: C.L. Grek, D. M. Townsend, J.D. Uys, C. Pazoles, and K.D. Tew
Development of methodology: C.L. Grek, D. M. Townsend, J.D. Uys, Y. Manevich, W. Coker III, and K.D. Tew
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): C.L. Grek, J. D. Uys, Y. Manevich, and W. Coker III.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): C.L. Grek, D. M. Townsend, J.D. Uys, Y. Manevich, and C. Pazoles
Writing, review, and/or revision of the manuscript: C.L. Grek, D. M. Town-send, J.D. Uys, Y. Manevich, W. Coker III, C. Pazoles, and K.D. Tew
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): C.L. Grek and W. Coker III
Study supervision: K.D. Tew
References
- 1.Cummings J, Ward TH, Greystoke A, Ranson M, Dive C. Biomarker method validation in anticancer drug development. Br J Pharmacol. 2008;153:646–656. doi: 10.1038/sj.bjp.0707441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarker D, Workman P. Pharmacodynamic biomarkers for molecular cancer therapeutics. Adv Cancer Res. 2007;96:213–268. doi: 10.1016/S0065-230X(06)96008-4. [DOI] [PubMed] [Google Scholar]
- 3.Jones DP. Redox sensing: orthogonal control in cell cycle and apoptosis signalling. J Intern Med. 2010;268:432–448. doi: 10.1111/j.1365-2796.2010.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Townsend DM. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7:313–324. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyagi SC. Role of oxidative mixed-disulfide formation in elastase-serine proteinase inhibitor (serpin) complex. Biochem Cell Biol. 1996;74:391–401. doi: 10.1139/o96-042. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths SW, King J, Cooney CL. The reactivity and oxidation pathway of cysteine 232 in recombinant human alpha 1 -antitrypsin. J Biol Chem. 2002;277:25486–25492. doi: 10.1074/jbc.M203089200. [DOI] [PubMed] [Google Scholar]
- 7.Tyagi SC, Simon SR. Role of disulfide exchange in alpha 1-protease inhibitor. Biochemistry. 1992;31:10584–10590. doi: 10.1021/bi00158a022. [DOI] [PubMed] [Google Scholar]
- 8.Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–1864. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- 9.Pemberton PA, Stein PE, Pepys MB, Potter JM, Carrell RW. Hormone binding globulins undergo serpin conformational change in inflammation. Nature. 1988;336:257–258. doi: 10.1038/336257a0. [DOI] [PubMed] [Google Scholar]
- 10.Nagata K. Hsp47: a collagen-specific molecular chaperone. Trends Biochem Sci. 1996;21:22–26. doi: 10.1016/0968-0004(96)80881-4. [DOI] [PubMed] [Google Scholar]
- 11.Zou Z, Anisowicz A, Hendrix MJ, Thor A, Neveu M, Sheng S, et al. Maspin, a serpin with tumor-suppressing activity in human mammary epithelial cells. Science. 1994;263:526–529. doi: 10.1126/science.8290962. [DOI] [PubMed] [Google Scholar]
- 12.Winkler IG, Hendy J, Coughlin P, Horvath A, Levesque JP. Serine protease inhibitors serpina1 and serpina3 are down-regulated in bone marrow during hematopoietic progenitor mobilization. J Exp Med. 2005;201:1077–1088. doi: 10.1084/jem.20042299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Townsend DM, He L, Hutchens S, Garrett TE, Pazoles CJ, Tew KD. NOV-002, a glutathione disulfide mimetic, as a modulator of cellular redox balance. Cancer Res. 2008;68:2870–2877. doi: 10.1158/0008-5472.CAN-07-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruscoe JE, Rosario LA, Wang T, Gate L, Arifoglu P, Wolf CR, et al. Pharmacologic or genetic manipulation of glutathione S-transferase P1-1 (GSTpi) influences cell proliferation pathways. J Pharmacol Exp Ther. 2001;298:339–345. [PubMed] [Google Scholar]
- 15.Grek CL, Townsend DM, Tew KD. The impact of redox and thiol status on the bone marrow: Pharmacological intervention strategies. Pharmacol Ther. 2011;129:172–184. doi: 10.1016/j.pharmthera.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Findlay VJ, Townsend DM, Saavedra JE, Buzard GS, Citro ML, Keefer LK, et al. Tumor cell responses to a novel glutathione S-transferase-activated nitric oxide-releasing prodrug. Mol Pharmacol. 2004;65:1070–1079. doi: 10.1124/mol.65.5.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saavedra JE, Srinivasan A, Buzard GS, Davies KM, Waterhouse DJ, Inami K, et al. PABA/NO as an anticancer lead: analogue synthesis, structure revision, solution chemistry, reactivity toward glutathione, and in vitro activity. J Med Chem. 2006;49:1157–1164. doi: 10.1021/jm050700k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churg A, Dai J, Zay K, Karsan A, Hendricks R, Yee C, et al. Alpha-1-antitrypsin and a broad spectrum metalloprotease inhibitor, RS113456, have similar acute anti-inflammatory effects. Lab Invest. 2001;81:1119–1131. doi: 10.1038/labinvest.3780324. [DOI] [PubMed] [Google Scholar]
- 19.Townsend DM, Findlay VJ, Fazilev F, Ogle M, Fraser J, Saavedra JE, et al. A glutathione S-transferase pi-activated prodrug causes kinase activation concurrent with S-glutathionylation of proteins. Mol Pharmacol. 2006;69:501–508. doi: 10.1124/mol.105.018523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wondrak GT. Redox-directed cancer therapeutics: molecular mechanisms and opportunities. Antioxid Redox Signal. 2009;11:3013–3069. doi: 10.1089/ars.2009.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Townsend DM, Tew KD. Pharmacology of a mimetic of glutathione disulfide, NOV-002. Biomed Pharmacother. 2009;63:75–78. doi: 10.1016/j.biopha.2008.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giustarini D, Rossi R, Milzani A, Colombo R, Dalle-Donne I. S-glutathionylation: from redox regulation of protein functions to human diseases. J Cell Mol Med. 2004;8:201–212. doi: 10.1111/j.1582-4934.2004.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris EC, Dafforn TR, Forsyth SL, Missen MA, Horvath AJ, Hampson L, et al. Murine serpin 2A is a redox-sensitive intracellular protein. Biochem J. 2003;371:165–173. doi: 10.1042/BJ20021567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nawata S, Shi HY, Sugino N, Zhang M. Evidence of post-translational modification of the tumor suppressor maspin under oxidative stress. Int J Mol Med. 2011;27:249–254. doi: 10.3892/ijmm.2010.572. [DOI] [PubMed] [Google Scholar]
- 26.Tyagi SC. Reversible inhibition of neutrophil elastase by thiol-modified alpha-1 protease inhibitor. J Biol Chem. 1991;266:5279–5285. [PubMed] [Google Scholar]
- 27.van Pel M, van Os R, Velders GA, Hagoort H, Heegaard PM, Lindley IJ, et al. Serpina1 is a potent inhibitor of IL-8-induced hematopoietic stem cell mobilization. Proc Natl Acad Sci U S A. 2006;103:1469–1474. doi: 10.1073/pnas.0510192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banda MJ, Rice AG, Griffin GL, Senior RM. Alpha 1-proteinase inhibitor is a neutrophil chemoattractant after proteolytic inactivation by macrophage elastase. J Biol Chem. 1988;263:4481–4484. [PubMed] [Google Scholar]
- 29.Przygodzka P, Ramstedt B, Tengel T, Larsson G, Wilczynska M. Bomapin is a redox-sensitive nuclear serpin that affects responsiveness of myeloid progenitor cells to growth environment. BMC Cell Biol. 2011;11:30. doi: 10.1186/1471-2121-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Druker BJ. Inhibition of the Bcr-Abl tyrosine kinase as a therapeutic strategy for CML. Oncogene. 2002;21:8541–8546. doi: 10.1038/sj.onc.1206081. [DOI] [PubMed] [Google Scholar]
- 31.Druker BJ, Sawyers CL, Kantarjian H, Resta DJ, Reese SF, Ford JM, et al. Activity of a specific inhibitor of the BCR-ABL tyrosine kinase in the blast crisis of chronic myeloid leukemia and acute lymphoblastic leukemia with the Philadelphia chromosome. N Engl J Med. 2001;344:1038–1042. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 32.Serrels A, Macpherson IR, Evans TR, Lee FY, Clark EA, Sansom OJ, et al. Identification of potential biomarkers for measuring inhibition of Src kinase activity in colon cancer cells following treatment with dasatinib. Mol Cancer Ther. 2006;5:3014–3022. doi: 10.1158/1535-7163.MCT-06-0382. [DOI] [PubMed] [Google Scholar]
- 33.Baselga J, Rischin D, Ranson M, Calvert H, Raymond E, Kieback DG, et al. Phase I safety, pharmacokinetic, and pharmacodynamic trial of ZD1839, a selective oral epidermal growth factor receptor tyrosine kinase inhibitor, in patients with five selected solid tumor types. J Clin Oncol. 2002;20:4292–4302. doi: 10.1200/JCO.2002.03.100. [DOI] [PubMed] [Google Scholar]
- 34.Karashima S, Kataoka H, Itoh H, Maruyama R, Koono M. Prognostic significance of alpha-1-antitrypsin in early stage of colorectal carcinomas. Int J Cancer. 1990;45:244–250. doi: 10.1002/ijc.2910450207. [DOI] [PubMed] [Google Scholar]
- 35.Higashiyama M, Doi O, Kodama K, Yokouchi H, Tateishi R. An evaluation of the prognostic significance of alpha-1-antitrypsin expression in adenocarcinomas of the lung: an immunohistochemical analysis. Br J Cancer. 1992;65:300–302. doi: 10.1038/bjc.1992.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allgayer H, Babic R, Grutzner KU, Beyer BC, Tarabichi A, Schildberg FW, et al. Tumor-associated proteases and inhibitors in gastric cancer: analysis of prognostic impact and individual risk protease patterns. Clin Exp Metastasis. 1998;16:62–73. doi: 10.1023/a:1006564002679. [DOI] [PubMed] [Google Scholar]
- 37.Alexandrakis MG, Passam FH, Ganotakis ES, Sfiridaki K, Xilouri I, Perisinakis K, et al. The clinical and prognostic significance of erythrocyte sedimentation rate (ESR), serum interleukin-6 (IL-6) and acute phase protein levels in multiple myeloma. Clin Lab Haematol. 2003;25:41–46. doi: 10.1046/j.1365-2257.2003.00492.x. [DOI] [PubMed] [Google Scholar]
- 38.Laine A, Leroy A, Hachulla E, Davril M, Dessaint JP. Comparison of the effects of purified human alpha 1-antichymotrypsin and alpha 1-proteinase inhibitor on NK cytotoxicity: only alpha 1-proteinase inhibitor inhibits natural killing. Clin Chim Acta. 1990;190:163–173. doi: 10.1016/0009-8981(90)90170-w. [DOI] [PubMed] [Google Scholar]
- 39.de Sa SV, Correa-Giannella ML, Machado MC, Krogh K, de Almeida MQ, Albergaria Pereira MA, et al. Serpin peptidase inhibitor clade A member 1 as a potential marker for malignancy in insulinomas. Clin Cancer Res. 2007;13:5322–5330. doi: 10.1158/1078-0432.CCR-06-1477. [DOI] [PubMed] [Google Scholar]
- 40.Sallmyr A, Fan J, Rassool FV. Genomic instability in myeloid malignancies: increased reactive oxygen species (ROS), DNA double strand breaks (DSBs) and error-prone repair. Cancer Lett. 2008;270:1–9. doi: 10.1016/j.canlet.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Farquhar MJ, Bowen DT. Oxidative stress and the myelodysplastic syndromes. Int J Hematol. 2003;77:342–350. doi: 10.1007/BF02982641. [DOI] [PubMed] [Google Scholar]
- 42.Battisti V, Maders LD, Bagatini MD, Santos KF, Spanevello RM, Maldonado PA, et al. Measurement of oxidative stress and antioxidant status in acute lymphoblastic leukemia patients. Clin Biochem. 2008;41:511–518. doi: 10.1016/j.clinbiochem.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 43.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, et al. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci U S A. 2006;103:13086–13091. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Starke DW, Chock PB, Mieyal JJ. Glutathione-thiyl radical scavenging and transferase properties of human glutaredoxin (thioltransferase). Potential role in redox signal transduction. J Biol Chem. 2003;278:14607–14613. doi: 10.1074/jbc.M210434200. [DOI] [PubMed] [Google Scholar]
- 45.Arner ES, Nakamura H, Sasada T, Yodoi J, Holmgren A, Spyrou G. Analysis of the inhibition of mammalian thioredoxin, .thioredoxin reductase, and glutaredoxin by cis-diamminedichloroplatinum (II) and its major metabolite, the glutathione-platinum complex. Free Radic Biol Med. 2001;31:1170–1178. doi: 10.1016/s0891-5849(01)00698-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.