Abstract
Cancer has long been viewed as a genetic disease; however, epigenetic silencing as the result of aberrant promoter DNA methylation is frequently associated with cancer development, suggesting an epigenetic component to the disease. Nonetheless, it has remained unclear whether an epimutation (an aberrant change in epigenetic regulation) can induce tumorigenesis. Here, we exploited a functionally validated cis-acting regulatory element and devised a strategy to induce developmentally regulated genomic targeting of DNA methylation. We used this system to target DNA methylation within the p16Ink4a promoter in mice in vivo. Engineered p16Ink4a promoter hypermethylation led to transcriptional suppression in somatic tissues during aging and increased the incidence of spontaneous cancers in these mice. Further, mice carrying a germline p16Ink4a mutation in one allele and a somatic epimutation in the other had accelerated tumor onset and substantially shortened tumor-free survival. Taken together, these results provide direct functional evidence that p16Ink4a epimutation drives tumor formation and malignant progression and validate a targeted methylation approach to epigenetic engineering.
Introduction
More than 25 years ago, it was proposed that epimutation — mitotically stable gene silencing associated with epigenetic alteration in DNA methylation — can act as 1 of Knudson’s 2 hits required for tumorigenesis (1). In subsequent decades, many promoter CpG island–associated (CGI-associated) genes have been shown to be aberrantly hypermethylated and silenced in various cancers (2). Indeed, recent epigenomic studies revealed that nearly all tumor types harbor hundreds of abnormally hypermethylated promoter CGIs (3), which indicates that epimutations are as common in tumors as genetic mutations. We and others have demonstrated that aberrant promoter CGI methylation is associated with distinct environmental exposures (4), gene mutation patterns (5), cancer prognosis (6), and response to therapy (7). Despite the undisputed importance of DNA methylation in cancer, however, its fundamental role in carcinogenesis remains unclear (8, 9). Most importantly, it remains unknown whether aberrant DNA methylation is a cause of tumorigenesis (8).
Cancer-related promoter CGI hypermethylation originates in normal tissues, which suggests that aberrant methylation could predispose to malignancy (10). p16Ink4a (referred to hereafter as p16) is a tumor suppressor gene that regulates the ability of retinoblastoma protein to control exit from the G1 stage of the cell cycle (11). Inactivation of p16 by promoter CGI methylation is among the most common and earliest epigenetic events in human cancer (12) and is frequently detected in preneoplastic lesions (13–15). Mice provide an apt model in which to study epigenetic dysregulation of p16 in cancer; in a mouse model of chemically induced lung cancer, p16 methylation is a very early event (16). Moreover, p16 age-associated hypermethylation is observed in several normal human and mouse tissues (17–19). Together, these data suggest that epigenetic silencing of p16 in aging cells facilitates early abnormal clonal expansion, driving tumorigenesis (20). Directly testing this hypothesis, however, requires the ability to specifically target methylation to the p16 promoter CGI.
Results and Discussion
Because cis-acting DNA sequences are important for the establishment of genomic patterns of DNA methylation (21, 22), we set out to engineer a cis element to attract DNA methyltransferases during development to achieve targeted de novo DNA methylation in vivo. We built upon our recent demonstration that an exceptional class of promoter CGIs that are methylated and silenced in normal somatic tissues is associated with specific cis-acting DNA motifs (23). To test the function of the identified motifs in vitro, we constructed a 140-bp cis element containing the top 3 motifs in the order and orientation most frequently observed in the normally methylated CGIs (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI76507DS1). We used a stable integrated system to test transgenes targeting 2 human promoter CGIs, one at INSL6 and another at p16, for 80 days in cell culture using both human (LNCaP) and mouse (NIH3T3) cell lines. The cis element induced extensive and progressive de novo methylation throughout the promoter CGIs of the juxtaposed transgenes (Supplemental Figures 2 and 3). These positive results prompted us to generate a mouse model with targeted knockin of the promethylation cis element at the p16 locus.
We introduced the cis element approximately 1 kb upstream of the p16 transcription start site (TSS) in mouse ES cells (mESCs) through homologous recombination (Supplemental Figure 4). The insertion leaves the p16 core promoter intact and avoids affecting the p15Ink4b, p15 antisense (p15AS), and p19Arf promoters, located >27, >12, and >11 kb away, respectively (Supplemental Figure 4, A and B). As a negative control for insertion effects, we compared the effects of cis element against those of an Alu sequence located approximately 1 kb upstream of human p16, since this Alu shows no sequence homology to the cis element, and site-specific integration of this repetitive element does not affect methylation at neighboring genes (24). Germline transmission of both the control and cis elements was achieved and confirmed by Southern blotting, PCR, and DNA sequence analysis (Supplemental Figure 4, C and D).
To initially determine whether the knockin cis element specifically induces DNA methylation at the endogenous p16 promoter, we studied mESCs in vitro. Differentiating ESCs recapitulate the earliest stages of embryonic lineage development. We therefore analyzed the progression of DNA methylation before and after induced differentiation (25, 26) in mESCs carrying either knockin allele. To characterize the dynamics of methylation establishment, we performed detailed methylation profiling by quantitative bisulfite pyrosequencing at CpG sites spanning the knockin sequence and the endogenous p16 promoter (–906 to –313 bp relative to TSS). Prior to differentiation, this region was essentially unmethylated in both control (p16+/ctr-neo) and cis-knockin (p16+/cis-neo) mESCs (Figure 1, A–C). Upon differentiation of p16+/ctr-neo mESCs, a few CpG sites within the knockin sequence showed modestly increased methylation, but methylation remained low at the endogenous p16 promoter (Figure 1, A and C). In p16+/cis-neo mESCs, conversely, differentiation induced dramatically increased methylation both at the cis element and at the p16 promoter (Figure 1, B and C). These results clearly demonstrated that the cis element attracts de novo methylation during differentiation, also affecting neighboring endogenous sequence. Bisulfite cloning and sequencing, an independent method of assessing DNA methylation, confirmed the extensive developmentally regulated methylation in p16+/cis-neo mESCs (Figure 1D). Interestingly, methylation at the cis-targeted alleles exhibited clonal heterogeneity. Since our assay was designed to detect methylation at the targeted allele using a forward primer specific for the knockin sequence, our data suggest that the increased methylation occurs in a cell type–specific fashion. Finally, to monitor the functional effects of induced methylation, we assessed p16 expression using quantitative TaqMan real-time RT-PCR. In p16+/ctr-neo cells, p16 expression was upregulated during differentiation (Figure 1E), consistent with its functional role in limiting the replicative capacity of stem cells (27). In p16+/cis-neo cells, however, induced promoter methylation during differentiation led to strong transcriptional repression (Figure 1E). p16 expression was restored by treatment with the DNA hypomethylating agent 5-aza-2′-deoxycytidine (DAC) (Supplemental Figure 5A), providing further validation that p16 silencing is not due to unforeseen side effects of the cis element knockin. Importantly, consistent with previous studies (28, 29), we observed concomitant increases in repressive histone markers (H3K9me2 and H3K27me3), with no changes of active markers (H3K9Ac and H3K4me3), across the p16 promoter (Supplemental Figure 5B), which suggests that cis-mediated epigenetic silencing could be mechanistically reinforced by changes in chromatin configuration.
Figure 1. The cis element specifically induces p16 promoter methylation.
Quantitative DNA methylation profiling in p16+/ctr-neo (A) and p16+/cis-neo (B) mESCs before (Undiff) and after (Diff) differentiation. Schematics of the p16 promoter of targeted alleles, including CpG maps, are shown above. Ex1a, exon 1α. (C) Examples of bisulfite pyrograms of 3 CpG sites between –615 to –589 bp relative to TSS in p16+/ctr-neo and p16+/cis-neo mESCs before and after differentiation. The y axis represents the signal intensity of luminescence as a measure of nucleotide incorporation, and the x axis shows the dispensation order of nucleotide. CpG sites are shaded yellow, and the percentage methylation (blue boxes) was measured based on the signal intensities of C and T (representing methylated and unmethylated cytosines, respectively) in each C of a CpG site. (D) Clonal bisulfite sequencing analysis of 11 CpG sites (white, unmethylated; black, methylated) between –814 to –589 bp relative to TSS. Each row represents an individual clone. Whereas essentially no promoter methylation was observed in undifferentiated p16+/cis-neo mESCs, extensive methylation was found in differentiated p16+/cis-neo mESCs. (E) Quantitative p16 gene expression analysis showed strong transcriptional suppression in differentiated p16+/cis-neo cells compared with controls. Values are mean ± SD. **P < 0.01, Student’s t test.
Having verified that our engineered cis element specifically induced developmentally regulated promoter methylation and transcriptional silencing, we analyzed p16 methylation in multiple tissues from p16cis homozygous (p16cis/cis) mice after excision of the Frt-flanked selection marker (Figure 2A). p16cis/cis mice were viable and fertile and did not display any developmental abnormalities (Supplemental Figure 6). In agreement with our data in differentiating mESCs, we observed initial methylation seeding within the cis element in almost all mouse tissues at birth (P0) (Figure 2B). The notable exception was testis, in which we detected low methylation at all CpG sites. This was remarkably consistent with our previous observations, since the promoter CGIs with which the sequence motifs were originally associated are also highly methylated in most tissues, except testis and sperm (23). With aging, cis element–mediated methylation spread toward the endogenous p16 promoter (Figure 2C). In spleen, liver, and colon, normal age-related increases in DNA methylation at the p16 promoter were significantly accelerated by the cis element, with commensurate reductions in gene expression (Figure 2, D and E, and Supplemental Figure 7). Collectively, these results demonstrated that our approach successfully induced developmentally regulated somatic p16 epimutation, leading to transcriptional repression in vivo.
Figure 2. The cis element induces p16 promoter methylation and age-dependent transcriptional suppression in vivo.
(A) CpG maps and regions assayed for DNA methylation. (B) DNA methylation profiling in multiple tissues from p16cis/cis mice (n = 2) at P0. (C) Comparison of methylation profiling in the spleens obtained from p16cis/cis mice at P0 and 40 weeks of age (n = 2–4 per time point). (D) Significant age-associated increases in DNA methylation in multiple tissues from p16cis/cis mice compared with controls (n = 3–4 per group). Shown are average methylation levels at promoter CpGs between –814 to –589 bp relative to TSS. (E) Significantly reduced p16 mRNA levels in tissues from 40-week-old p16cis/cis mice compared with controls (n = 3–4 per group). Values are mean ± SD. *P < 0.05, **P < 0.01, Student’s t test.
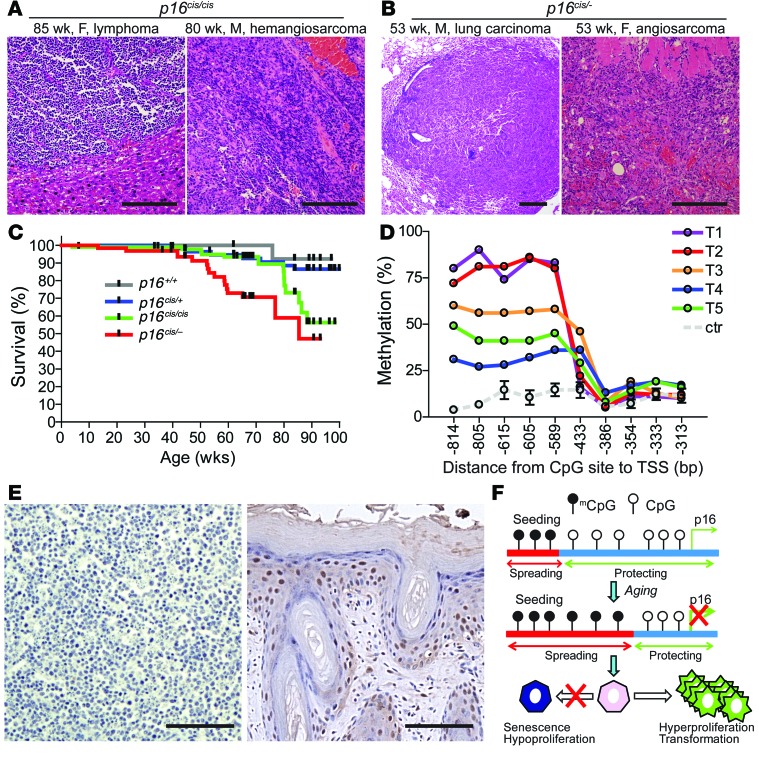
To test whether p16 promoter methylation predisposes animals to tumor development, we used heterozygous intercrossing to generate a cohort of p16+/+, p16+/cis, and p16cis/cis mice (on a mixed 129/C57 genetic background) and monitored tumor development and survival. In mice aged 35–100 weeks, spontaneous tumors developed in 0 of 18 p16+/+ (0%), 3 of 51 p16+/cis (5%), and 6 of 22 p16cis/cis (27%; P = 0.02 vs. p16+/+) mice. Of the 3 p16+/cis mice with spontaneous tumor development, 1 developed lymphoma and 2 developed sarcoma (Supplemental Figure 8, A–C). The malignancies in the 6 p16cis/cis mice were sarcoma (n = 3), lymphoma (n = 2), and lung adenocarcinoma (n = 1) (Figure 3A and Supplemental Figure 8, D–I). Given that p16 deficiency in many human cancers involves mutation of one allele and promoter hypermethylation of the other (30), we next assessed the cooperative tumorigenic effects of combined p16 mutation and epimutation. We bred the p16cis allele into mice on a p16 exon 1α deletion background (31) to generate p16cis/Δexon1α (mixed FVB/129/C57 background; referred to herein as p16cis/–) mice. In 20 p16cis/– mice aged 40–91 weeks, 6 (30%) tumors were found; the malignancies were sarcoma (n = 3), lung carcinoma (n = 2), and lymphoma (n = 1) (Figure 3B and Supplemental Figure 9). Moreover, consistent with the notion of p16 epimutation serving as 1 of Knudson’s 2 hits, p16cis/– mice had accelerated tumor onset and shortened survival (Figure 3C). Our results compare favorably with previous studies that characterized the effects of p16-specific knockout on tumorigenesis (31, 32). Although rigorous comparisons in congenic strains have not yet been performed, the cancer-prone conditions in p16cis/– and p16–/– mice were strikingly similar: tumor spectra were predominantly sarcoma in both, tumor incidence was 30% and 35%, respectively, and tumor latency was 80 and 76 weeks, respectively. In contrast, p16+/– mice developed significantly fewer spontaneous tumors and exhibited significantly longer tumor-free survival. To ascertain the role of p16 promoter methylation in the oncogenic pathway in vivo, we measured p16 promoter methylation and gene expression in the tumor tissues. Hypermethylation of the p16 promoter in tumors was associated with essentially complete loss of protein expression (Figure 3, D and E, and Supplemental Figures 10 and 11). Taken together, our results provide direct evidence for a driving role of p16 epigenetic silencing by promoter hypermethylation in tumor formation and progression (Figure 3F).
Figure 3. The driving role of p16 epimutation in tumorigenesis in vivo.
(A and B) Representative histological appearance of malignancies in p16cis/cis (A) and p16cis/– (B) mice of the indicated age, sex, and tumor type. Scale bars: 200 μm. (C) Kaplan-Meier survival analysis demonstrating shortened lifespan in p16cis/cis mice (P = 0.02 vs. p16+/+, log-rank test) and in p16cis/– mice. (D) p16 promoter methylation significantly increased in the bulk of tumor tissues collected. Tumors 1 and 2 (T1 and T2) were collected from the liver and spleen, respectively, of the same p16cis/cis mouse with metastatic lymphoma; T3 was collected from the p16+/cis mouse; T4 and T5 were collected from 2 individual p16cis/– mice. Values for age-matched control tissues (ctr; mean ± SD, n = 4) are also shown. (E) Immunohistochemical analysis demonstrated that promoter hypermethylation resulted in abrogation of p16 protein expression in tumor cells. Left: Lymphoma with p16 promoter hypermethylation. Right: Positive control skin tissue showing strong immunostaining. Scale bars: 100 μm. (F) Proposed model for a direct role of developmentally regulated p16 methylation in tumorigenesis.
In conclusion, our present study provides the first clear demonstration that p16 epimutation causes tumorigenesis. Potential applications of this novel mouse model include the testing of targeted epigenetic therapies for prevention and treatment of human cancer. Our straightforward approach to epigenetic engineering should be useful in testing the causal role of other epigenetic alterations implicated in carcinogenesis, and broadly applicable toward elucidating epigenetic etiology in a wide range of diseases.
Methods
Further information can be found in Supplemental Methods and Supplemental Tables 1–3.
Statistics.
2-tailed Student’s t test and Fisher exact test were used to determine the significance of differences. Survival analysis was conducted with Kaplan-Meier analysis and log-rank test. A P value less than 0.05 was considered significant.
Study approval.
All animals were treated in accordance with NIH guidelines as approved by the Baylor College of Medicine Animal Care committee.
Supplementary Material
Acknowledgments
We thank Wei Zhu, Wei Wei, and Jingming Shu for technical assistance and Adam Gillum for assistance with the figures. We acknowledge the shared resources by Baylor College of Medicine ESC and GEM cores (NIH/NCI grant P30CA125123) and by Morphology core at the Texas Medical Center Digestive Disease Center (NIH/NIDDK grant P30DK56338). This work was supported by grants from the Sidney Kimmel Foundation (to L. Shen), USDA (CRIS-6250-51000-055 to L. Shen and R.A. Waterland; CRIS-6250-51000-054 to M.H. Chen), March of Dimes (1-FY-08-392 to R.A. Waterland), and NIDDK (1R01DK081557 to R.A. Waterland).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2014;124(9):3708–3712. doi:10.1172/JCI76507.
References
- 1.Holliday R. The inheritance of epigenetic defects. Science. 1987;238(4824):163–170. doi: 10.1126/science.3310230. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 3.Baylin SB, Jones PA. A decade of exploring the cancer epigenome — biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen L, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94(10):755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 5.Shen L, et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci U S A. 2007;104(47):18654–18659. doi: 10.1073/pnas.0704652104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen L, et al. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007;13(20):6093–6098. doi: 10.1158/1078-0432.CCR-07-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen L, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J Clin Oncol. 2010;28(4):605–613. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylin S, Bestor TH. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell. 2002;1(4):299–305. doi: 10.1016/S1535-6108(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 9.Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;3(4):253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 10.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7(4):536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- 11.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9(1):22–30. doi: 10.1016/S0959-437X(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 12.Herman JG, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55(20):4525–4530. [PubMed] [Google Scholar]
- 13.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61(9):3573–3577. [PubMed] [Google Scholar]
- 14.Kondo Y, Kanai Y, Sakamoto M, Mizokami M, Ueda R, Hirohashi S. Genetic instability and aberrant DNA methylation in chronic hepatitis and cirrhosis — a comprehensive study of loss of heterozygosity and microsatellite instability at 39 loci and DNA hypermethylation on 8 CpG islands in microdissected specimens from patients with hepatocellular carcinoma. Hepatology. 2000;32(5):970–979. doi: 10.1053/jhep.2000.19797. [DOI] [PubMed] [Google Scholar]
- 15.Nuovo GJ, Plaia TW, Belinsky SA, Baylin SB, Herman JG. In situ detection of the hypermethylation-induced inactivation of the p16 gene as an early event in oncogenesis. Proc Natl Acad Sci U S A. 1999;96(22):12754–12759. doi: 10.1073/pnas.96.22.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belinsky SA, et al. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95(20):11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maegawa S, et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20(3):332–340. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishida N, Nagasaka T, Nishimura T, Ikai I, Boland CR, Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47(3):908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waki T, Tamura G, Tsuchiya T, Sato K, Nishizuka S, Motoyama T. Promoter methylation status of E-cadherin, hMLH1, and p16 genes in nonneoplastic gastric epithelia. Am J Pathol. 2002;161(2):399–403. doi: 10.1016/S0002-9440(10)64195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 21.Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43(11):1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 22.Yates PA, Burman RW, Mummaneni P, Krussel S, Turker MS. Tandem B1 elements located in a mouse methylation center provide a target for de novo DNA methylation. J Biol Chem. 1999;274(51):36357–36361. doi: 10.1074/jbc.274.51.36357. [DOI] [PubMed] [Google Scholar]
- 23.Shen L, et al. Genome-wide profiling of DNA methylation reveals a class of normally methylated CpG island promoters. PLoS Genet. 2007;3(10):2023–2036. doi: 10.1371/journal.pgen.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y, Shu J, Si J, Shen L, Estecio MR, Issa JP. Repetitive elements and enforced transcriptional repression co-operate to enhance DNA methylation spreading into a promoter CpG-island. Nucleic Acids Res. 2012;40(15):7257–7268. doi: 10.1093/nar/gks429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bibel M, Richter J, Lacroix E, Barde YA. Generation of a defined and uniform population of CNS progenitors and neurons from mouse embryonic stem cells. Nat Protoc. 2007;2(5):1034–1043. doi: 10.1038/nprot.2007.147. [DOI] [PubMed] [Google Scholar]
- 26.McKinney-Freeman S, Daley G. Derivation of hematopoietic stem cells from murine embryonic stem cells. J Vis Exp. 2007;(2):162. doi: 10.3791/162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esteve PO, et al. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev. 2006;20(22):3089–3103. doi: 10.1101/gad.1463706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamakoshi K, et al. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J Cell Biol. 2009;186(3):393–407. doi: 10.1083/jcb.200904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264(1):42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 31.Sharpless NE, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413(6851):86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 32.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23(2):379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.