Abstract
Structural maintenance of chromosomes (SMC) complexes are essential for maintaining chromatin structure and regulating gene expression. Two the three known SMC complexes, cohesin and condensin, are important for sister chromatid cohesion and condensation, respectively; however, the function of the third complex, SMC5–6, which includes the E3 SUMO-ligase NSMCE2 (also widely known as MMS21) is less clear. Here, we characterized 2 patients with primordial dwarfism, extreme insulin resistance, and gonadal failure and identified compound heterozygous frameshift mutations in NSMCE2. Both mutations reduced NSMCE2 expression in patient cells. Primary cells from one patient showed increased micronucleus and nucleoplasmic bridge formation, delayed recovery of DNA synthesis, and reduced formation of foci containing Bloom syndrome helicase (BLM) after hydroxyurea-induced replication fork stalling. These nuclear abnormalities in patient dermal fibroblast were restored by expression of WT NSMCE2, but not a mutant form lacking SUMO-ligase activity. Furthermore, in zebrafish, knockdown of the NSMCE2 ortholog produced dwarfism, which was ameliorated by reexpression of WT, but not SUMO-ligase–deficient NSMCE. Collectively, these findings support a role for NSMCE2 in recovery from DNA damage and raise the possibility that loss of its function produces dwarfism through reduced tolerance of replicative stress.
Introduction
Primordial dwarfism denotes extreme linear growth impairment from early gestation. When severely reduced head size is also seen, primordial dwarfism is said to be microcephalic. This is generally taken to imply a global problem with either cellular or organismal growth and contrasts with primordial dwarfism in which the head size is relatively preserved, which is more likely to be accounted for by a more tissue-selective problem, affecting, for example, long bone growth (1).
Many human genetic defects have now been described as underlying microcephalic primordial dwarfism. These may affect endocrine growth regulation by impairing production or action of IGF-1 (2, 3); however, most mutations reported to date affect cell-autonomous growth. Some clustering of defects according to the cellular structure or process affected is apparent. For example, microcephalic primordial dwarfism can result from defects in genes involved in the licensing of DNA replication, in centrosome function, or in DNA damage response and repair pathways (reviewed in ref. 4).
In keeping with the known roles of the defective genes, some syndromes feature a characteristic profile of chromosomal instability or impaired DNA damage repair. Some components of the syndromes cannot easily be explained by known functions of the affected gene, however. An example is the severe insulin resistance described in pericentrin deficiency (5). This usually appears during the first decade of life and is associated with severely fatty liver, elevated blood triglyceride levels, and diabetes, recapitulating in severe form the pandemic metabolic syndrome. Similar metabolic abnormalities are seen with milder growth impairment in both Bloom syndrome (6) and Werner syndrome (7), each caused by loss of a DNA helicase, and without growth impairment in patients with a heterozygous polymerase-inactivating mutation in DNA polymerase δ (8). However despite increasing evidence of a link between impaired DNA replication and/or damage responses and severe insulin resistance, the underlying mechanisms linking these remain unclear.
Results
A novel syndrome of primordial dwarfism.
We identified 2 female patients with similar syndromes (details in Supplemental Material and Methods; supplemental material available online with this article; doi:10.1172/JCI73264DS1). The shared features are (a) severe primordial dwarfism, with facial dysmorphism, including a small jaw and prominent midface (Figure 1, A–G and I–K); (b) extremely insulin resistant diabetes, fatty liver, and hypertriglyceridemia developing in childhood; and (c) primary gonadal failure. Both patients had normal karyotypes. Patient 1 (P1) had evidence of a mild excess of chromosome breaks in lymphocyte culture at several stages in her childhood, but diagnostic testing for Bloom syndrome and Fanconi anemia was normal. The degree of chromosome damage reported reduced with time and, by the age of 16.9 years, no excess chromosome damage could be detected. No similar cytogenetic studies of P2 were performed. Neither patient had any history of cancer, bone marrow dysfunction, nor any overt clinical evidence of accelerated ageing. P2 was congenitally blind, reportedly due to bilateral retinal detachment, and died suddenly at 33 years old of a sudden cardiovascular event.
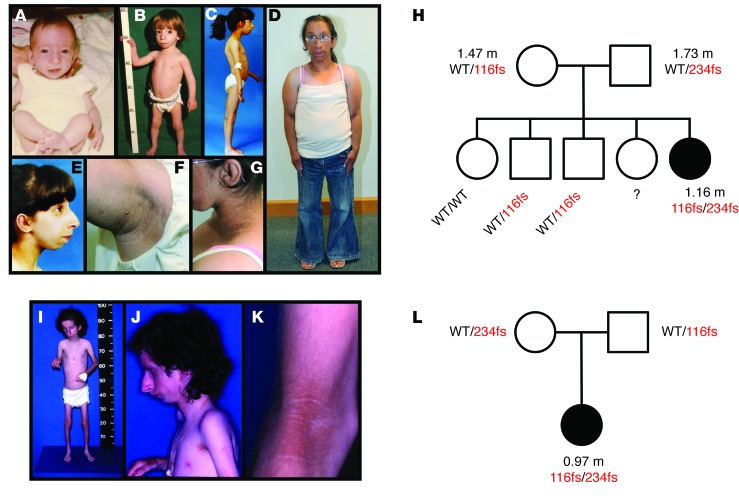
Figure 1. A syndrome of primordial dwarfism and extreme insulin resistance associated with compound heterozygous NSMCE2/MMS21 frameshift mutations.
(A–G) P1 at (A) 4 months, (B) 2 years, (C) 12 years, and (D) 24 years of age. (E) Profile at 12 years showing prominent midface and small lower jaw. (F) Severe axillary acanthosis nigricans with skin tags. (G) Severe acanthosis nigricans on the lateral neck. (H) Pedigree diagram for P1 with adult heights, where available, shown in meters. NSMCE2 genotypes are shown as WT, 116fs (p.Ser116Leufs*18), and 234fs (p.Ala234Glufs*4). (I–K) P2 shown at 27 years old. (I) Whole-body appearance illustrating dwarfism and paucity of adipose tissue and muscle. (J) Profile showing prominent midface and small lower jaw. (K) Detail of arm at antecubital fossa showing severe acanthosis nigricans. (L) Pedigree diagram for P2. Written, informed consent was obtained from patients or their families for publication of these images.
Identification of NSMCE2 mutations.
P1 was born to unrelated parents of European ancestry from Wales and had 4 unaffected siblings. We hypothesized either biallelic, likely compound heterozygous, loss-of-function mutations or a de novo dominant mutation, affecting a critical growth-related gene. Exome-wide sequences of P1 and her parents were analyzed using DeNovoGear (described in Methods) to look for de novo mutations in P1 and to search for genes with compound heterozygous or homozygous rare mutations plausibly affecting protein function (details in Methods and Supplemental Figure 1). No rare de novo variants were identified; however, 3 genes, NSMCE2, RUSC1, and WDR36, harbored compound heterozygous variants. Detailed analysis of these genes in large human populations, and prior functional studies, suggested only the NSMCE2 variants to be plausible candidates to cause the observed phenotype (details in Supplemental Appendix). NSMCE2 contained compound heterozygous frameshift mutations: c.345delT (p.Ser116Leufs*18), inherited from the proband’s mother, and c.700_701insAGGG (p.Ala234Glufs*4), inherited from the patient’s father (Figure 1H). Both mutations were absent from 1,092 individuals from the 1000 Genomes Project Consortium (9) and 4,190 internal control exomes and genomes. The p.Ala234Glufs*4 was also absent from 6,250 individuals from the NHLBI GO Exome Sequencing Project, while the p.Ser116Leufs*18 mutation was found in heterozygous form in 2 of the 6,250 individuals (allele frequency 0.00016) (details in Methods and Supplemental Table 2).
P2 was the only child of unrelated parents of European ancestry from the Brittany region of northwest France. Sanger sequencing of NSMCE2 in P2 revealed the same 2 frameshift mutations found in P1. Her mother was heterozygous only for the p.Ala234Glufs*4 mutation, while her father was heterozygous for the p.Ser116Leufs*18 mutation, confirming P2 to be a compound heterozygote (Figure 1L). Although the families of P1 and P2 have no known shared ancestry, genotyping of 21 SNPs and 2 microsatellites within the linkage block encompassing NSMCE2 (chr8: 126104083–1263936) confirmed that both patients inherited the same 2 rare haplotypes within a region greater than 510 kb, one containing the p.Ser116Leufs*18 mutation, the other p.Ala234Glufs*4. This suggests shared common ancestral haplotypes: one, greater than 510-kb long, containing p.Ser116Leufs*18, estimated to have arisen 283 generations (less than 8,500 years) ago, and the other, greater than 339-kb long, containing p.Ala234Glufs*4, estimated to have arisen 150 generations (less than 4,500 years) ago, based on the locus-specific recombination rate. Neither haplotype nor the p.Ser116Leufs*18 nor p.Ala234Glufs*4 mutations were present in a sample of 54 controls from Great Britain (Methods and Supplemental Table 3).
NSMCE2/MMS21 and SMC5–6.
NSMCE2 encodes a protein component of the heteromultimeric structural maintenance of chromosomes 5–6 (SMC5–6) complex (10). It consists of 2 core SMC components (SMC5 and SMC6) and, in humans, 4 non-SMC elements often designated as NSE1-4, among which NSE2 corresponds to NSMCE2/MMS21 (10–12). The NSMCE2 gene product is often named MMS21 due to its original identification in a Saccharomyces cerevisiae screen for mutants sensitive to the alkylating agent methane methylsulphonate (MMS) (13). The Schizosaccharomyces pombe gene is Nse2. Here, we refer to the protein as NSMCE2/MMS21, or NSMCE2 in figures.
The function of SMC5–6, although critical for survival in all organisms studied, is not well understood. Current evidence, particularly from hypomorphic yeast mutants, has defined roles in DNA replication and recombination, including the facilitation of homologous recombination repair (HRR), through aiding of resolution of topologically complex DNA intermediates. Failure to complete HRR may lead to checkpoint activation and apoptosis or to catastrophic failure to untangle highly catenated, interconnected chromosomes.
NSMCE2/MMS21 possesses a Siz/PIAS RING (SP-RING) domain toward its C terminus with SUMO E3-ligase activity. Several targets for this have been documented in various organisms (14–16), including NSMCE2/MMS21 itself, SMC5, and SMC6. Yeast strains with loss of Mms21 SUMO-ligase activity are viable, but exhibit increased genomic instability (14, 17), while SUMO-ligase activity in mammalian cell lines is required for normal responses to some forms of DNA damage (15, 16).
Cellular phenotype associated with NSMCE2 hypomorphism.
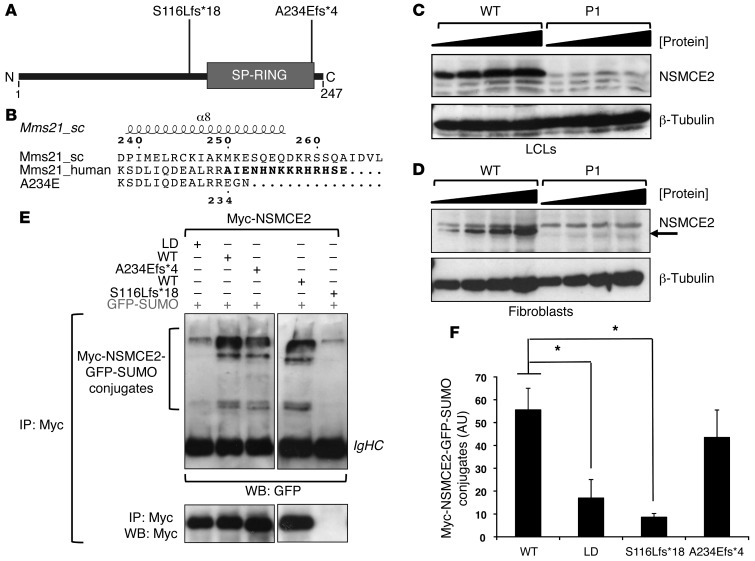
The p.Ser116Leufs*18 mutation occurs between the N-terminal helix bundle and the SP-RING domain and introduces 18 unstructured amino acids, while the p.Ala234Glufs*4 mutation lies at the C terminus of the domain, removing the 14 C-terminal amino acids of the protein (Figure 2A) and thereby disrupting the C-terminal α helix formed by residues 223–240 (ref. 18 and Figure 2B). Western blotting of Epstein Barr virus–transformed lymphoblastoid cells (LCLs) (Figure 2C) and primary dermal fibroblasts (Figure 2D) from P1 revealed severely depressed levels of full-length NSMCE2. Mild underexpression of SMC5 and SMC6 was also observed in whole-cell and chromatin-enriched extracts from P1-derived LCLs compared with WT, suggestive of instability of the core SMC5–6 protein complex in these cells (Supplemental Figure 2, A and B). mRNA levels of NSMCE2/MMS21 were 50% of WT in dermal fibroblasts, although near normal in LCLs. cDNA sequencing showed no evidence of the p.Ser116Leufs*18 species, suggesting that its mRNA is unstable (Supplemental Figure 2, C and D). We did not detect truncated NSMCE2/MMS21 protein. However on heterologous overexpression of a Myc-tagged version of NSMCE2/MMS21 carrying the p.Ser116Leufs*18 mutation in A549 cells, a low level of the truncated protein could be seen on immunoblotting (Supplemental Figure 3A). Although p.Ala234Glufs*4, in contrast, was detected at levels close to those of overexpressed WT NSMCE2/MMS21, its levels decayed more quickly than those of WT NSMCE2/MMS21 after overexpression in HeLa cells treated with cycloheximide, consistent with reduced protein stability (Supplemental Figure 2E).
Figure 2. Expression and auto-SUMOylation of NSMCE2/MMS21 frameshift mutations.
(A) Schematic showing positions of patient mutations with respect to the SP-RING SUMO-ligase domain. (B) Schematic of NSMCE2/MMS21 showing disruption of α helix α8 (residues 223–240) by the p.Ala234Glufs*4 mutation. Alignment of human WT (Mms21_human) and p.Ala234Glufs*4 (A234E) MMS21/NSMCE2 based on the crystal structure of S. cerevisiae Mms21 (Mms21_sc; PDB entry: 3HTK; chain C). The 14 amino acids removed by the mutation are marked in boldface. Numbering and secondary structure of S. cerevisiae Mms21 are displayed. The position of the mutation in human MMS21/NSMCE2 is displayed below the sequences. (C) NSMCE2 expression in dermal fibroblasts from P1 assessed using immunoblotting of increasing amounts of whole-cell extract compared with WT fibroblasts. (D) Similar immunoblotting of whole-cell extract from P1 LCLs. (E) Auto-SUMOylation activity of Myc-tagged WT, SUMO LD, and naturally occurring mutant NSMCE2/MMS21 following coexpression with GFP-SUMO1 (upper panels). Auto-SUMOylation was detected following IP using anti-Myc and blotting (WB, Western blot) using anti-GFP antibodies. IgHC, immunoglobulin heavy chain. The lower panel shows relative amounts of immunoprecipitated Myc-NSMCE2. The p.A234Efs*4 mutation appears not to affect protein gel migration under these conditions, unlike p.S116Lfs*18 (Supplemental Figure 3). (F) Quantification of recovered Myc-NSMCE2-GFP-SUMO conjugates by densitometry. Data represent mean ± SD. (n = 3). *P ≤ 0.05 (unpaired, 2-tailed t test).
To determine the impact of the mutations on the SUMO-ligase activity of NSMCE2/MMS21, we assessed auto-SUMOylation of Myc-tagged NSMCE2/MMS21 ectopically coexpressed with GFP-SUMO1 in A549 cells. NSMCE2/MMS21 species were immunoprecipitated using anti-Myc beads, and the SUMO-modified versions of NSMCE2/MMS21 were detected by Western blotting with anti-GFP (Figure 2, E and F, and Supplemental Figure 3B). A previously validated SUMO-ligase-deficient (LD) version of NSMCE2/MMS21 with mutations in the SP-RING (p.Cys185Ala and p.His187Ala) (19) was used as a negative control. Consistent with its reduced expression and lack of SP-RING domain, auto–SUMO-ligase activity of p.Ser116Leufs*18 was virtually undetectable. As expected, auto-SUMOylation activity of the LD variant was also severely reduced. In contrast, the p.Ala234Glufs*4 mutant exhibited a level of auto-SUMOylation similar to that of WT NSMCE2/MMS21 under these conditions (Figure 2, E and F).
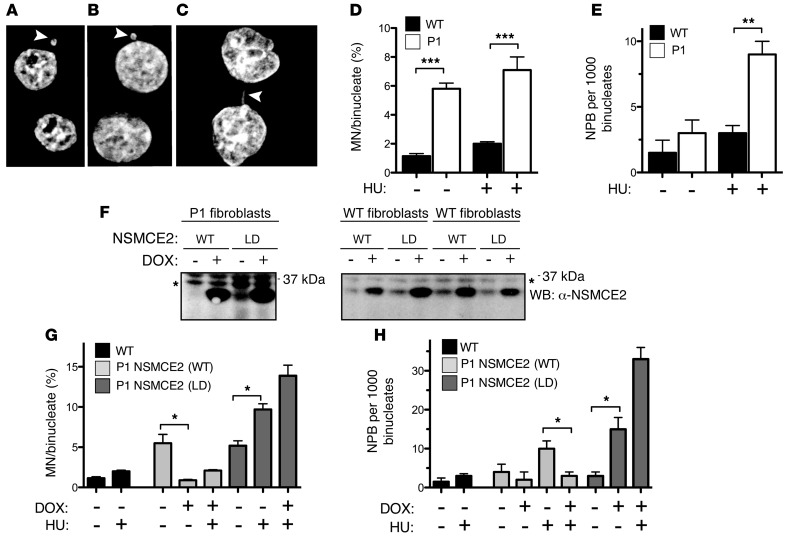
Having confirmed that cells from P1 were severely hypomorphic for NSMCE2/MMS21, we next characterized the associated cellular phenotype. The frequencies of micronuclei (MN) and nucleoplasmic bridges (NPB) in cytokinesis-blocked binucleated cells (Figure 3, A–C) are widely used as indicators of chromosomal damage and instability (20). Typically, MN develop when errors in chromosome segregation during mitosis occur from acentric fragments of chromosomes and/or anaphase-lagging whole chromosomes, while NPB form when centromeres of dicentric chromosomes are drawn to opposite poles at anaphase or by telomere shortening. When the actin polymerization inhibitor cytochalasin B (Cyt-B) is used to inhibit cytokinesis, observed MN are derived from events in the preceding S phase. Given the role suggested for SMC5–6 in resolution of HRR intermediates, we thus assessed the frequencies of MN and NPB in Cyt-B–induced binucleated dermal fibroblasts from P1.
Figure 3. HU-induced nuclear abnormalities in primary fibroblast cells are rescued by expression of WT but not SUMO LD NSMCE2/MMS21.
Typical micrographs of binucleated dermal fibroblasts showing (A and B) MN or (C) NPB. Scale bars: 5 μm. (D) Increased frequency of MN or (E) NPB in Cyt-B–treated dermal fibroblasts from P1 relative to age-matched healthy controls (WT) following a 4-hour block in 1 mM HU. (F) DOX-induced expression of WT or SUMO LD NSMCE2/MMS21 in P1 or control dermal fibroblasts (WT fibroblasts, 2 samples shown) analyzed by immunoblotting with antibodies specific to human NSMCE2/MMS21 (WB: α-NSMCE2). Asterisk indicates a crossreactive band that serves as an internal loading control. (G and H) DOX-induced ectopic expression of WT NSMCE2/MMS21 reduces frequencies of MN (G) and NPB (H) to levels seen in control cells. In contrast, DOX-induced ectopic expression of NSMCE2/MMS21 (LD) significantly increases levels of both MN (G) and NPB (H). Frequencies of MN per 100 or NPB per 1,000 binucleated cells are shown for each experimental condition, as indicated. All bar graphs represent mean ± SEM (n = 4). *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired 1-tailed t test).
The basal frequency of MN was significantly higher in binucleated dermal fibroblasts from P1 relative to WT, although NPB frequencies in these cells were indistinguishable (Figure 3, D and E). We next assessed these following treatment with hydroxyurea (HU), a potent inducer of replication fork stalling and collapse. Following HU, binucleated dermal fibroblasts from P1 showed higher frequencies of both MN and NPB than WT (Figure 3, D and E). Doxycycline-induced (DOX-induced) expression of WT NSMCE2/MMS21 (Figure 3F) significantly reduced the frequency of MN and NPB in P1 binucleates. In contrast, expression of the LD version of NSMCE2/MMS21 markedly increased their frequency (Figure 3, G and H).
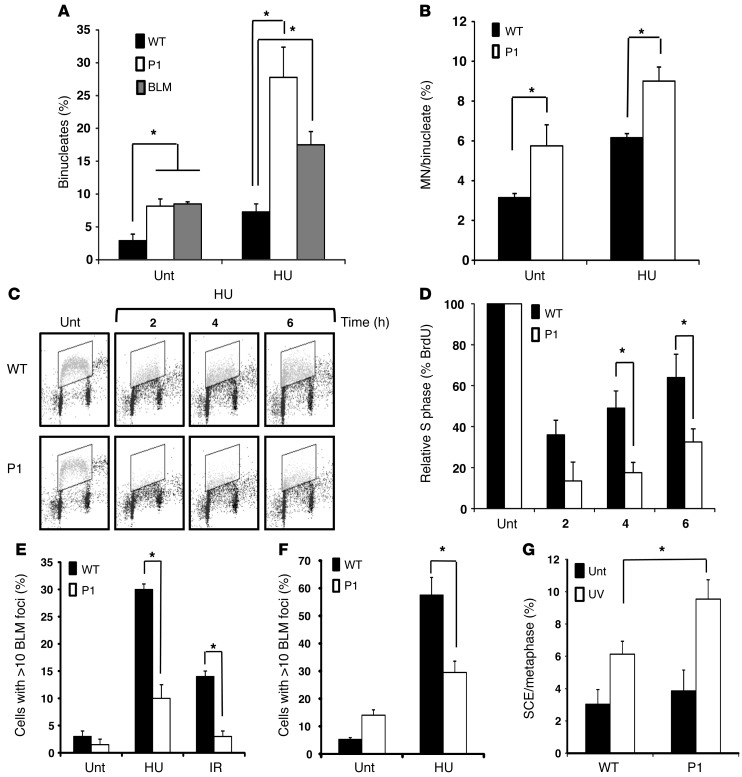
LCLs derived from P1 exhibited a similar phenotype. There was an approximately 1.8-fold increase in spontaneously binucleated cells compared with WT, an increase also observed in Bloom syndrome LCLs. This increase was exacerbated in both P1 and Bloom syndrome LCLs following exposure to repeated treatment with low concentrations of HU (once daily for 4 days) (Figure 4A). Reminiscent of P1 fibroblasts, we also observed modestly elevated spontaneous and HU-induced MN formation in binucleates generated by Cyt-B treatment (Figure 4B).
Figure 4. Increased micronucleus formation, impaired S phase progression, and increased SCE in LCLs with NSMCE2/MMS21 mutations.
(A) Binucleates in LCLs from WT, P1, and from a Bloom syndrome patient, either untreated (Unt) or following repeated exposure to HU (50 μM/d for 4 days). (B) HU-induced MN formation in Cyt-B–induced binucleate LCLs. (C) BrdU flow cytometry profiles from WT and P1 LCLs either untreated or following treatment with low-dose HU (250 μM). S phase content (boxed area) following a BrdU pulse 30 minutes prior to each time point is shown. (D) Normalized total S phase content compared with that of untreated cells at each time point. Means ± SD are shown (n = 3). (E) BLM foci formation in P1 LCLs relative to WT following treatment with HU (1 mM for 16 hours) or IR (10 Gy for 16 hours). (F) BLM foci formation in primary dermal fibroblasts from P1 compared with WT following HU exposure (1 mM for 24 hours). (G) UV radiation–induced SCEs in WT and P1 LCLs compared with untreated cells. These effects, although significant, were not as profound as those observed in BLM patient LCLs, which exhibited 23 ± 6 SCEs/metaphase in untreated cells and 38 ± 4 SCEs/metaphase following UV irradiation. *P < 0.05 (unpaired t test).
Yeast SUMO-LD mutants of the NSMCE2/MMS21 ortholog and mutations in smc6 are associated with replication delay and accumulation of unresolved hemi-catenated, X-shaped DNA molecules (21, 22). BrdU pulse chase analysis of asynchronously growing P1-derived LCLs did not reveal a significant difference in S phase progression rate compared with WT (Supplemental Figure 4). However, we did see impaired recovery of DNA replication following acute treatment with a low concentration of HU in P1-derived LCLs compared with WT (Figure 4, C and D).
Bloom syndrome helicase (BLM), which is disrupted in Bloom syndrome, plays a fundamental role in resolution of converging Holliday junctions during HRR. Consistent with synthetic interaction between the BLM ortholog in yeast (Sgs1) and mutation of the NSMCE2/MMS21 ortholog, we observed an impairment in BLM foci formation in P1-derived LCLs both in response to replication stress induced by HU and in response to ionizing radiation (IR) (Figure 4E), in contrast with WT. A similar impairment of BLM focus formation was seen in patient fibroblasts exposed to HU (Figure 4F). Moreover, following UV treatment, another potent means of replication fork stalling, we observed significant elevation in UV-induced sister chromatid exchanges (SCE) in P1 LCLs compared with WT, consistent with the impaired ability to form BLM foci of these cells (Figure 4G). Collectively, these data are consistent with a failure of NSMCE2/MMS21-mutated patient cells to complete cell division effectively following chronic replicative stress. In contrast, P1 LCLs showed no hypersensitivity to camptothecin or mechlorethamine, unlike BLM-deficient cells (Supplemental Figure 5, A–C). As HRR-defective cells are sensitive to killing by both these agents, this suggests that DNA repair per se is not completely deficient as a result of this hypomorphic defect in NSMCE2/MMS2.
Modeling of NSMCE2/MMS21 deficiency in zebrafish.
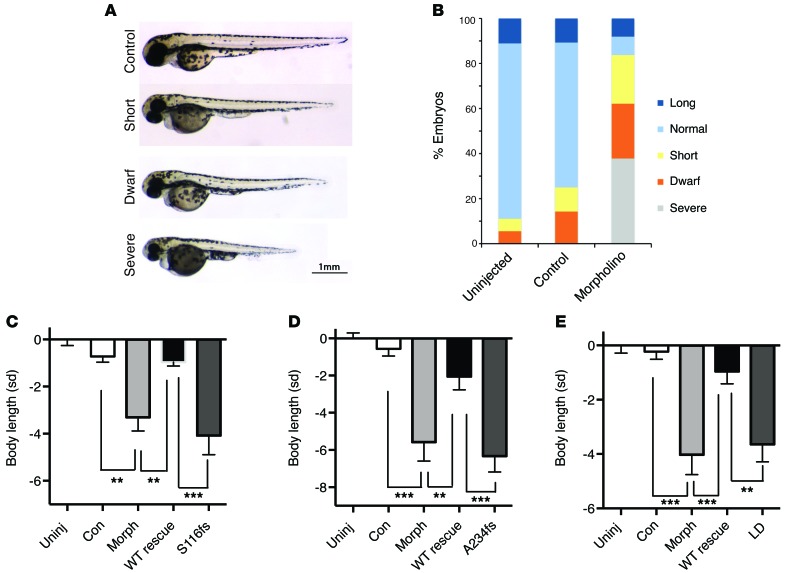
As no Nsmce2 knockout mouse has been reported to date, we used zebrafish to test the consequence of loss of Nsmce2 function in vivo. We utilized a cocktail of 2 splice-site anti-sense morpholinos (MOs) to deplete the zebrafish NSMCE2/MMS21 ortholog, reducing its mRNA levels in the morphant to approximately 20% of those of control MO-injected embryos (Methods and Supplemental Figure 6). This markedly reduced body size compared with uninjected and control MO-injected embryos (Figure 5, A and B). From 5% to 15% of the nsmce2/mms21 MO-injected embryos exhibited an extreme phenotype including body curvature and increased mortality; however, the majority of embryos were grossly normal morphologically. Coinjection of human WT NSMCE2 mRNA with zebrafish nsmce2/mms21-targeting MOs attenuated the dwarf phenotype observed in the nsmce2 morphant alone, with coinjected embryos not significantly different in length from those injected with control MO alone (Figure 5, C–E). In contrast, coinjection of either mRNA variant identified in the patients failed to rescue the nsmce2/mms21 morphant phenotype (Figure 5, C and D).
Figure 5. Modeling of NSMCE2/MMS21 deficiency in zebrafish.
Injection of splice-site MOs targeted to Nsmce2 markedly reduce body size compared with that of control-injected or uninjected WT embryos. (A) Representative images of some arbitrarily defined length categories: normal, –1 to 1 SD; short, –1 to –2 SD; dwarf, –2 to –3 SD; severe, < –3 SD. (B) Quantification of length of MO-injected zebrafish (uninjected, n = 18; control, n = 28; MO, n = 37). (C–E) Coinjection of WT human NSMCE2/MMS21 mRNA (WT rescue) with nsmce2-targeting MOs attenuated the dwarf phenotype observed in the nsmce2 morphant alone (morph). In contrast, coinjection of either (C) human NSMCE2/MMS21 S116Lfs*18, (D) human NSMCE2/MMS21 A234Efs*4, or (E) SUMO LD human NSMCE2/MMS21 mRNAs together with nsmce2-targeting MOs did not affect the severity of the dwarf phenotype. Uninj, uninjected; con, control. Graphs show SD scores relative to those of uninjected WT zebrafish embryos from the same mating. n = 12–15 for each condition. P values were calculated by ANOVA with post-hoc Tukey test; **P < 0.01, ***P < 0.001.
In keeping with findings in mammalian cells, levels of GFP-tagged mutant NSMCE2/MMS21 protein decayed much more rapidly than those of GFP-tagged WT NSMCE2/MMS21, despite high and equivalent levels of mRNA being detected after injection, suggesting impaired translation and/or instability of the mutants (Supplemental Figure 7). Collectively, these experiments confirm the specificity of the MO effect and suggest that the NSMCE2/MMS21 frameshift mutations identified in the patients lead to loss of function. Interestingly, coinjection of a SUMO LD version of NSMCE2/MMS21 also failed to rescue nsmce2 morphants (Figure 5E) coincident with reduced expression of this GFP-tagged protein, as seen for the other mutants (Supplemental Figure 7).
Discussion
Our findings demonstrate association between compound heterozygosity for rare frameshift mutations in NSMCE2/MMS21 and a syndrome of primordial dwarfism, extreme insulin resistance, and primary gonadal failure. One of the mutations, p.Ser116Leufs*18, removes the whole SP-RING domain, is nearly undetectable in primary cells, has undetectable SUMO autoligase activity, and is unable to rescue the zebrafish morphant phenotype on reexpression. The second mutation, p.Ala234Glufs*4, in contrast, is predicted to remove only 14 amino acids distal to the SP-RING domain, is well expressed from a strong promoter in heterologous cellular systems, and exhibits normal SUMO autoligation after overexpression. The direct evidence that it is a loss-of-function mutation thus rests on the extremely low level of full-length NSMCE2/MMS21 observed in primary fibroblasts and LCLs from P1 and on the failure of the reexpressed mutant to rescue the dwarfism of nsmce2/mms21 morphant zebrafish, which we suggest is due to reduced stability of the mutant protein. These data establish that the patients described are severely hypomorphic for NSMCE2/MMS21 and demonstrate that this results in severe dwarfism, insulin resistance and primary gonadal failure, with mild evidence of spontaneous chromosomal instability.
The SMC5–6 complex is structurally closely related to cohesion (SMC1–3) and condensin (SMC2–4). Although cohesin and condensin have relatively well characterized roles in mitosis, respectively, holding sister chromatids in apposition or mediating the premitotic condensation of chromosomes (23), the function of the SMC5–6 complex is less well understood. The accumulation of aberrant DNA intermediates with hypofunction of the yeast NSMCE2/MMS21 ortholog (21, 22) and synthetic lethality in combination with mutations in the topoisomerase II ortholog (24) argues for a role in resolving topologically complex intermediates of HRR. Inability to resolve these intermediates is particularly harmful in the context of restarting stalled or collapsed DNA replication forks, which appear, for example, when the DNA template is damaged or the nucleotide pool depleted (21, 22, 25).
The phenotype of patient cells, namely impaired ability to recover from replication stress leading to genomic instability both at baseline and in response to induced replication fork stalling, is in keeping with a role for NSMCE2/MMS21 in resolving complex DNA structures and is consistent with findings in yeast, avian, and mammalian cells (e.g., refs. 11, 14–16, 26). We also demonstrate a mild increase in SCE following UV irradiation of patient LCLs; however, this increase is much smaller than that seen in Bloom syndrome. Such an aberrant “hyperrecombination” phenotype has also been reported in chick DT40 cells with NSMCE2/MMS21 deficiency (26).
The precise role of the SUMO-ligase activity of NSMCE2/MMS21 is not fully delineated. Several targets have been documented in various organisms (14–16). As well as NSMCE2/MMS21, SMC5, and SMC6, these include other proteins with important roles in DNA damage sensing and chromosome stability, including KU70, SCC1, and the telomeric proteins TRF1 and TRF2. Studies in avian (26) and mammalian (16, 27) cells have suggested that loss of the SUMO-ligase activity of NSMCE2/MMS21 orthologs leads to genomic instability. Our data now extend these findings. The increased MN and NPB seen after reexpression of a SUMO LD variant of NSMCE2/MMS21 in patient fibroblasts suggest that SUMO-ligase activity may be required for at least some important functions of NSMCE2/MMS21 in higher eukaryotes. Nevertheless, the viability of yeast strains lacking ligase activity and the nonviability of strains wholly lacking the NSMCE2/MMS21 ortholog indicate that NSMCE2/MMS21 has other essential functions that are SUMO-ligase independent.
The question arises as to whether the cellular defects we observe account for the phenotype of the affected patients. Growing cells are subjected to a barrage of endogenously generated DNA damage during every cell division, arising, for example, from generation of reactive oxygen and nitrogen species as a by-product of metabolism (28), so it is plausible that reduced cellular ability to tolerate replicative stress and associated DNA damage is the cause of global growth impairment. This is mechanistically akin to the primordial dwarfism seen when the DNA damage-sensing kinase ATR (29) is mutated in Seckel syndrome.
Both patients with NSMCE2/MMS21 deficiency also exhibit primary ovarian failure. Primary gonadal failure is a consequence of defects in several other human DNA damage response components, including ATM, NBS, BLM and members of the Fanconi anemia complex, presumably due to impaired meiosis and/or mitosis of germ cells in utero. Furthermore, the yeast ortholog of NSMCE2/MMS21 has been shown to be required for meiosis (30). The ovarian failure of the 2 patients we describe is thus potentially explicable through established functions of NSMCE2/MMS21. It remains possible, however, that NSMCE2/MMS21 serves unknown functions that are related to the observed phenotype, as in the case of genetic cohesinopathies, where important components of the clinical syndromes result from perturbed transcriptional regulation, rather than directly to loss of cohesin’s longest established role in sister chromatid cohesion.
The highly insulin resistant diabetes that we report is more difficult to reconcile simply with the observed cellular defects. As our study ascertained patients based on the presence of severe insulin resistance, further work will be needed to determine confidently whether it is an obligate feature of NSMCE2/MMS21 loss of function in humans; however the insulin resistance in both cases was extreme. Although the derangement in insulin and glucose levels was of a similar degree to that seen with loss of insulin receptor function (31), the metabolic phenotype was quite distinct. Both our patients had severe fatty liver and dyslipidemia, which are not seen with insulin receptor dysfunction (32). Instead, the metabolic phenotype closely resembles that seen in primary disorders of adipose tissue development or function (33).
A clue to the origins of the metabolic derangement we report may come from its resemblance to that of patients with Bloom and Werner syndromes (6, 7) or of patients with heterozygous polymerase domain mutations in DNA polymerase δ (8). Bloom and Werner syndromes are caused, respectively, by loss of the BLM (RECQL3) and WRN (RECQL2) DNA helicases, both of which play multiple accessory roles in resolution of complex DNA structures. The yeast phenotype caused by loss of the primordial ancestral DNA helicase that later diverged to produce WRN and BLM resembles the phenotype seen with hypomorphism for the NSMCE2/MMS21 ortholog (22). Our observation of impaired replicative stress–induced BLM foci formation and modestly increased levels of SCE following UV irradiation in NSMCE2/MMS21 patient cells further argues for commonality between Bloom syndrome and the syndrome we describe. On the other hand, our patients did not have cardinal clinical features of either Bloom or Werner syndrome, and the level of increase of SCE was far lower here than in Bloom syndrome. The detailed cellular phenotype of cells with polymerase domain mutations of DNA polymerase δ, the predominant lagging strand DNA polymerase, has yet to be reported.
Although it is tempting to equate whole-body insulin sensitivity to cell-autonomous insulin sensitivity, this is commonly misleading. Maintenance of whole-body insulin sensitivity requires not only intact cellular insulin signaling pathways, but also normal size and function of key insulin-responsive tissues — particularly adipose tissue, muscle, and liver. Indeed, most Mendelian forms of insulin resistance are accounted for by mutations, not in genes involved directly in insulin signaling, but rather those involved in adipose tissue development or function (34). It is thus plausible that genetic defects severely impairing mitosis or response to replication stress ultimately produce insulin resistance through progressive failure to maintain one or more insulin responsive tissues, a notion in keeping with the paucity of adipose tissue and muscle we observe in our patients. This idea remains to be validated; however, there is growing interest in the link between pandemic forms of insulin resistance, oxidative stress, and thereby DNA damage (28), and so in vivo modeling of severe NSMCE2/MMS21 hypomorphism may not only aid understanding of this rare and extreme condition, but may also offer wider, paradigmatic insights into common disease.
Methods
Exome sequencing.
Genomic DNA was extracted from blood leukocytes and prepared as described previously (35) for capture. Amplification and sequencing were performed on the Illumina Genome Analyzer II (index case) or Hi-Seq platform (parents) (Illumina) as 54- or 75-bp paired-end reads respectively. BWA software package 0.5.9, Picard v1.46, SamTools v0.1.16 (36), Dindel v1.01 (37), Genome Analysis Toolkit (GATK) v1.1-20 (38), and Variant Effect Predictor (VEP) v2.1 (39) were used to map sequence reads to the genome reference sequence (NCBI build 37), to calculate read quality, to call single nucleotide variants (SNVs) and insertion/deletions, and to annotate against Ensembl build 64. Variants were filtered to include only those with a minimum depth of 4× to a maximum depth of 1200× and were reported in variant calling file (VCF) format v4.1. For the index case, 6.4 Gb of sequence was produced at a mean depth of 60, with 80.4% coverage at a depth greater than or equal to 10, using high mapping quality alignments only (i.e., mapping quality ≥ 30). Equivalent figures for the father and mother were 8.8 Gb, 95% and 86.1%, and 8.9 Gb, 97% and 91.0% respectively. Raw exome sequence data from the trio are available from the European Genome-phenome Archive (EGAN00001101658, EGAN00001001829, EGAN00001001831).
De novo and compound heterozygous mutation analysis.
Exome data for P1 and her parents were merged, and variants with a high posterior probability of being de novo (cutoff value 0.8) were extracted using DeNovoGear v0.5 (https://sourceforge.net/projects/denovogear/) (40). Python scripts were used to extract potential compound heterozygous mutations. Variant annotation has previously been described in detail (41). Potentially causal variants identified were reconfirmed by Sanger sequencing of PCR products using the BigDye Terminator v3.1 Cycle Sequencing Kit according to the manufacturer’s instructions (Applied Biosystems; Life Technologies) and run on ABI 3730 DNA Sequencers (Applied Biosystems). Sequence analysis was performed using the Mutation Surveyor software package v2.3 (LLC; SoftGenetics). For P2, all exons and exon-intron boundaries of NSMCE2 were sequenced using Sanger sequencing as above (for primer sequences, see Supplemental Table 1).
Haplotype analysis.
Twenty-four SNPs were selected from HapMap release 27 (42) to capture the variation within the 2 linkage blocks surrounding the p.Ser116Leufs*18 and p.Ala234Glufs*4 mutations (chr8: 125672673–126874287). These were genotyped along with the microsatellites D8S1179 and D8S266 in both probands, their families and 54 ethnically matched controls predominantly from the west of Great Britain from the 1000 genomes GBR panel (http://www.1000genomes.org/about#ProjectSamples). SNP genotyping was performed using the iPLEX Gold Assay (Sequenom Inc.) according to the manufacturer’s instructions, while microsatellites were amplified by PCR using FAM-labeled forward primers, diluted, mixed with CC5 Internal Lane Size Standard 500 (Promega),and run on an ABI 3730 DNA Analyser (Applied Biosystems). Allelic determination was undertaken using GeneMapper software (Applied Biosystems). Of the 24 SNPs, 3 were monomorphic in our data set.
Analysis was performed on the 21 polymorphic SNPs using SHAPEIT2 (http://www.shapeit.fr/) to phase the genotypes and calculate the most likely haplotypes, and the microsatellite markers were used to further define the haplotypes where ambiguous.
Patient cell culture.
Epstein Barr virus–transformed LCLs were generated from primary lymphocytes using standard procedures and maintained in RPMI 1640 supplemented with l-glutamine, antibiotics (penicillin streptomycin), and 15% FBS. Bloom syndrome LCLs (GM09960) from a patient with a homozygous 6-bp deletion/7-bp insertion (6-bp del/7-bp ins) at nucleotide 2,281 of the open-reading frame of the RECQL3 gene were from Coriell Cell Repository. Dermal fibroblasts were grown from skin biopsies using standard procedures, maintained in MEM supplemented with L-glutamine, antibiotics (penicillin streptomycin), and 15% FBS.
NSMCE2/MMS21 expression analysis.
Endogenous NSMCE2 protein was detected in ureasonicated (15 seconds 30% amplitude using a micro-tip; Sigma-Aldrich) whole-cell extracts (9 M urea, 50 mM Tris-HCl, pH 7.5, 10 mM 2-mercaptoethanol) via immunoblotting with polyclonal rabbit antibody (Ab N2-19) raised against full-length GST-NSMCE2 protein (19).
Generation of mammalian and lentiviral expression vectors.
Expression vectors pCI-Neo-Myc harboring WT or the SUMO LD mutant C185A/H187A (19) were from Elaine Taylor and Alan Lehmann (Genome Damage and Stability Centre). p.S116fsLeufs*18 and p.Ala234Glufs*4 mutants were generated by site-directed mutagenesis of WT Myc-tagged NSMCE2 using the QuikChange XL II protocol (Agilent) and the following mutagenesis primers: p.S116fsLeufs*18: sense primer, 5′-TTTTTGGCTTTACAGAGCAAGAATCTGATGCAGACTTTCAAAATAAT-3′ and antisense primer, 5′-ATTATTTTGAAAGTCTGCATCAGATTCTTGCTCTGTAAAGCCAAAAA-3′; p.Ala234Glufs*4: sense primer, 5′-GGATGAAGCACTTAGAAGGGAGGGCAATTGAGAACCATAACAAG-3′ and antisense primer, 5′-CTTGTTATGGTTCTCAATTGCCCTCCCTTCTAAGTGCTTCATCC-3′. To construct the DOX-inducible lentiviral expression vectors pSLIK-Neo-NSMCE2 (WT) and pSLIK-NSMCE2 (LD), coding regions of human NSMCE2 or the LD mutant were amplified and cloned into the entry vector pEN-Tmcs (MBA-251, LGC Standards; ATCC) and subsequently into pSLIK-Neo (MBA-236, LGC Standards; ATCC) using site-specific recombination (Gateway LR Clonase II Enzyme Mix; Invitrogen).
SUMOylation assay.
4 × 106 A549 cells were cotransfected with 4 μg GFP-SUMO-1 plasmid (pEGFP–SUMO-1) and 10 μg of WT or mutant pCI-Neo-Myc-NSMCE2. Cells were harvested 24 hours after transfection and solubilized with immunoprecipitation (IP) buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 25 mM NaF, 25 mM β-glycerophosphate, 0.1 mM sodium orthovanadate, 0.2% Triton X-100, 0.3% NP40, and Promega protease inhibitors cocktail) containing 20 mM N-ethylmaleimide. Myc-tagged proteins were immunoprecipitated using Red Anti-Myc Affinity Beads (E6654; Sigma-Aldrich). SUMOylation of immunoprecipitated material was detected by immunoblotting using anti-GFP antibody (2555; Cell Signaling).
Establishment of cell lines conditionally expressing WT or ligase-inactive SMCE2/MMS21.
10 μg of pSLIK WT or SUMO LD NSMCE2 plasmid, 7.5 μg of each of the packaging plasmids pMDLg/pRRE and pRSREV, and 5 μg of the pseudotyping pVSV-G plasmid were cotransfected into HEK 293T cells on 10-cm dishes using CalPhos Mammalian Transfection Kit (Clontech). The culture medium was replaced 12 hours after transfection with BioWhittaker Ultraculture medium (Lonza). Viral supernatants were harvested at 24, 48, and 72 hours after transfection and concentrated by centrifugation in Centricon Plus-70 Ultracel PL-100 (Millipore). Lentiviral particles were added at low MOI to dermal fibroblasts in the presence of 8 μg/ml of polybrene for 6 hours followed by selection in G418 (600 mg/ml; Sigma-Aldrich) for 3 weeks. Stable cell lines were maintained in the presence of 200 mg/ml G418.
Micronucleus assay.
Dermal fibroblasts were grown to 70% to 80% confluence ± 1 mg/ml of DOX, trypsinized, resuspended, counted, and seeded onto glass coverslips. Twenty hours later, cells were exposed to 1 mM HU or saline for 4 hours, followed by saline washing, and allowed to recover in the presence of 3 μg/ml of cytochalasin-B for 48 hours, fixed with 4% paraformaldehyde for 15 minutes at room temperature, permeabilized in 0.05% Triton X-100, and stained with Actin-stain 488 Phalloidin (Cytoskeleton Inc.) and DAPI (Invitrogen) in PBS. Cells were examined at ×100 magnification using a wide-field fluorescence microscope (BX61; Olympus). Scoring criteria were according to Fenech (20), using 1,000 binucleates per experimental condition.
For micronucleus studies in LCLs, cells were treated with 1 mM HU for 4 hours; HU was washed out and cells cultured in cytochalasin-B for 24 hours prior to harvesting to generate binucleates. Indirect immunofluorescence using anti–β-tubulin antibodies (Santa Cruz Biotechnology Inc.) and counterstaining with DAPI was employed to visualize the cytoplasm and to enable unbiased identification of binucleated cells.
FACS analysis.
LCLS were treated with 250 μM HU for up to 6 hours, and the proportion of cells in S phase was measured using BrdU pulse-mediated flow cytometry. Cells were pulse labeled with 20 μM BrdU 20 minutes prior to harvesting, then fixed in ice-cold 70% ethanol for 24 hours and resuspended in PBS containing 0.5% Tween-20, 10 μg/ml propidium iodide, and 500 μg/ml RNaseA. FITC-conjugated anti-BrdU (clone 44, BD) was used for detection of S phase cells following HCl denaturation. To monitor the rate of S phase progression LCLs were pulse labeled for 30 minutes with BrdU, washed in PBS, and then incubated in complete medium supplemented with Colcemid (100 μg/ml). Cells were harvested at 2-hour intervals and processed as described above. Data were collected using a BD FACS Canto and analyzed with FACS DIVA software.
BLM foci.
LCLs were swollen in 75 mM KCl (10 minutes), immobilized on poly-L-lysine–coated slides by cytospinning (Cytospin; Shandon), fixed with 3% PFA and 2% sucrose in PBS, permeabilized using 0.1% Triton X-100 in 5% BSA-PBS for 2 minutes, blocked (5% BSA-PBS for 10 minutes) and incubated with goat polyclonal anti-BLM (clone C-18, #sc7790; Santa Cruz Biotechnology Inc.) at 1:50 dilution in 5% BSA for 1 hour. Cells were counterstained with the appropriate FITC-conjugated secondary antibody and the nuclei stained with DAPI. Primary fibroblasts were fixed with methanol (–20°C for 20 minutes) and BLM foci detected using anti-BLM antibodies (2742; Cell Signaling). Slides were analyzed using the Zeiss AxioPlan platform and images captured using Simple PCI software.
UV-induced SCE.
LCLs were UV-irradiated (2 J/m2) and differential staining of sister chromatids was achieved by growing LCL in the presence of 25 μM BrdU for 48 hours. After 4 hours of treatment in Colcemid, cells were swollen in 75 mM KCl for 10 minutes and fixed overnight at 4°C with Carnoy’s solution (methanol/acetic acid, 3:1). Chromosomes were spread by dropping cells from 40 cm high onto frozen slides, which were then left to dry for 3 days. Slides were then incubated for 20 minutes in Hoescht (10 μg/ml) in the dark. After washing in SSC buffer (2 M NaCl, 0.3 M tri-sodium citrate, pH 7.0) slides were irradiated for 1 hour under a UV lamp (355 nm), followed by incubation in SCC buffer at 60°C for 1 hour. Slides were finally stained with modified Giemsa stain (GS500; Sigma-Aldrich).
Zebrafish studies.
WT TLF zebrafish (Danio rerio) were maintained and raised using standard protocols (43) in accordance with UK Home Office guidelines. Two splice-blocking MOs, nsmce2 sp1 and nsmce2 sp2, were designed to knock down nsmce2 expression (GeneTools, Philomath) (Supplemental Table 4 and Supplemental Figure 5). nsmce2 sp1 and nsmce2 sp2 were tested independently for effectiveness (data not shown) and then used in combination at a 1:1 ratio (data reported here). One- to two-cell embryos obtained from natural matings were microinjected with 1.8 nl of a 0.55 mM nsmce2 sp1 and 2 cocktail solution or the same concentration of control MO (Supplemental Table 3). Uninjected embryos were kept for comparison.
The efficiency of knockdown was assessed by PCR on cDNA derived from morphant or control embryos collected at day 2. mRNA was extracted using TRIzol (Invitrogen). One microgram of mRNA from each group was transcribed using Superscript II First Strand Synthesis System (Invitrogen). Specific primers were then used to amplify a region across exons 1–3 for nsmce2 sp1 and across exons 2–4 for nsmce2 sp2 as well as control primers for Agpat2 to check equal loading. Primers are detailed in Supplemental Table 3.
Embryos were cultured in egg water at 28.5°C for 48 hours before scoring phenotypes. Live fish images were captured using a Leica M80 stereomicroscope, and body length was calculated by the Leica Application Suite program. Embryos were assigned to categories defined with reference to the SD of the length of a population of uninjected WT fish from the same mating. The experiment was carried out 6 times. Data shown are representative. For rescue experiments capped full-length human NSMCE2 mRNA (either WT, S116Lfs*18, A234Efs*4, or SUMO LD) was transcribed in vitro following the manufacturer’s manual (mMessage mMachine; Invitrogen). RNA was purified using a Megaclear column (Ambion) and quantified spectrophotometrically and the size verified on an agarose gel. 100 picograms of mRNA were comicroinjected with nsmce2/mms21 MO at the 1- to 2-cell stage and the phenotype scored after 48 hours.
Statistics.
All numerical data were analyzed using parametric statistical tests, namely Student’s t test or ANOVA with post hoc Tukey’s testing, as appropriate. All tests used and thresholds for significance are indicated in figure legends.
Study approval.
This study was approved by the UK National Research Ethics Committee. Written informed consent was obtained from all participants or their parents.
Supplementary Material
Acknowledgments
I. Barroso, R.K. Semple, D.B. Savage, and S. O’Rahilly were supported by the Wellcome Trust (grants WT098051, WT098498, WT091551, and WT095515, respectively), the Medical Research Council Centre for Obesity and Related Disorders, and the United Kingdom National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre. M. O’Driscoll is supported by Cancer Research UK, the UK Medical Research Council and Leukaemia Lymphoma Research (United Kingdom). Funding for human studies was also provided by the EU/EFPIA Innovative Medicines Initiative Joint Undertaking (EMIF grant no. 115372). Thanks to Alan Lehmann and Elaine Taylor and Oskar Fernandez-Capatillo for providing antibodies. We are grateful for access to exome sequence data from the CoLaus cohort, sequenced through partnership between the Wellcome Trust Sanger Institute, the CoLaus principal investigators, and GlaxoSmithKline. We also thank the NHLBI GO Exome Sequencing Project and its ongoing studies, which provided exome variant calls for comparison, namely, the Lung GO (HL-102923), the WHI (HL-102924), the Broad GO (HL-102925), the Seattle GO (HL-102926), and the Heart GO (HL-103010) Sequencing Projects. This study also makes use of data generated by the UK10K Consortium derived from samples from TwinsUK and ALSPAC. See Supplemental Appendix for a list of the investigators who contributed to generation of data. UK10K was funded by the Wellcome Trust under award WT091310. Our thanks also to Emma Gray, David Jones (Sample Management), Danielle Walker (Sequencing Pipeline), Carol Scott and Jillian Durham (Variation Informatics), and the staff of the Wellcome Trust Sanger Institute. The authors are grateful for the involvement and assistance of both patients and their parents in this study.
References
- 1.Klingseisen A, Jackson AP. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 2011;25(19):2011–2024. doi: 10.1101/gad.169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woods KA, Camacho-Hubner C, Savage MO, Clark AJ. Intrauterine growth retardation and postnatal growth failure associated with deletion of the insulin-like growth factor I gene. N Engl J Med. 1996;335(18):1363–1367. doi: 10.1056/NEJM199610313351805. [DOI] [PubMed] [Google Scholar]
- 3.Fang P, Cho YH, Derr MA, Rosenfeld RG, Hwa V, Cowell CT. Severe short stature caused by novel compound heterozygous mutations of the insulin-like growth factor 1 receptor (IGF1R). J Clin Endocrinol Metab. 2012;97(2):E243–E247. doi: 10.1210/jc.2011-2142. [DOI] [PubMed] [Google Scholar]
- 4.Alcantara D, O’Driscoll M. Congenital microcephaly. Am J Med Genet C Semin Med Genet. 2014;166C(2):124–139. doi: 10.1002/ajmg.c.31397. [DOI] [PubMed] [Google Scholar]
- 5.Huang-Doran I, et al. Genetic defects in human pericentrin are associated with severe insulin resistance and diabetes. Diabetes. 2011;60(3):925–935. doi: 10.2337/db10-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diaz A, Vogiatzi MG, Sanz MM, German J. Evaluation of short stature, carbohydrate metabolism and other endocrinopathies in Bloom’s syndrome. Horm Res. 2006;66(3):111–117. doi: 10.1159/000093826. [DOI] [PubMed] [Google Scholar]
- 7.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore). 1966;45(3):177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 8.Weedon MN, et al. An in-frame deletion at the polymerase active site of POLD1 causes a multisystem disorder with lipodystrophy. Nat Genet. 2013;45(8):947–950. doi: 10.1038/ng.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abecasis GR, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Piccoli G, Torres-Rosell J, Aragon L. The unnamed complex: what do we know about Smc5-Smc6? Chromosome Res. 2009;17(2):251–263. doi: 10.1007/s10577-008-9016-8. [DOI] [PubMed] [Google Scholar]
- 11.McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J Biol Chem. 2003;278(46):45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- 12.Sergeant J, et al. Composition and architecture of the Schizosaccharomyces pombe Rad18 (Smc5-6) complex. Mol Cell Biol. 2005;25(1):172–184. doi: 10.1128/MCB.25.1.172-184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prakash S, Prakash L. Increased spontaneous mitotic segregation in MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;87(2):229–236. doi: 10.1093/genetics/87.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25(1):185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci U S A. 2005;102(13):4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25(16):7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rai R, et al. Small ubiquitin-related modifier ligase activity of Mms21 is required for maintenance of chromosome integrity during the unperturbed mitotic cell division cycle in Saccharomyces cerevisiae. J Biol Chem. 2011;286(16):14516–14530. doi: 10.1074/jbc.M110.157149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duan X, Sarangi P, Liu X, Rangi GK, Zhao X, Ye H. Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol Cell. 2009;35(5):657–668. doi: 10.1016/j.molcel.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor EM, Copsey AC, Hudson JJ, Vidot S, Lehmann AR. Identification of the proteins, including MAGEG1, that make up the human SMC5-6 protein complex. Mol Cell Biol. 2008;28(4):1197–1206. doi: 10.1128/MCB.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat Protoc. 2007;2(5):1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 21.Ampatzidou E, Irmisch A, O’Connell MJ, Murray JM. Smc5/6 is required for repair at collapsed replication forks. Mol Cell Biol. 2006;26(24):9387–9401. doi: 10.1128/MCB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Branzei D, et al. Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127(3):509–522. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 23.Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11(6):391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Outwin EA, Irmisch A, Murray JM, O’Connell MJ. Smc5-Smc6-dependent removal of cohesin from mitotic chromosomes. Mol Cell Biol. 2009;29(16):4363–4375. doi: 10.1128/MCB.00377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irmisch A, Ampatzidou E, Mizuno K, O’Connell MJ, Murray JM. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 2009;28(2):144–155. doi: 10.1038/emboj.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kliszczak M, Stephan AK, Flanagan AM, Morrison CG. SUMO ligase activity of vertebrate Mms21/Nse2 is required for efficient DNA repair but not for Smc5/6 complex stability. DNA Repair (Amst). 2012;11(10):799–810. doi: 10.1016/j.dnarep.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Behlke-Steinert S, Touat-Todeschini L, Skoufias DA, Margolis RL. SMC5 and MMS21 are required for chromosome cohesion and mitotic progression. Cell Cycle. 2009;8(14):2211–2218. doi: 10.4161/cc.8.14.8979. [DOI] [PubMed] [Google Scholar]
- 28.Bashan N, Kovsan J, Kachko I, Ovadia H, Rudich A. Positive and negative regulation of insulin signaling by reactive oxygen and nitrogen species. Physiol Rev. 2009;89(1):27–71. doi: 10.1152/physrev.00014.2008. [DOI] [PubMed] [Google Scholar]
- 29.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33(4):497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 30.Pebernard S, McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Nse1, Nse2, and a novel subunit of the Smc5-Smc6 complex, Nse3, play a crucial role in meiosis. Mol Biol Cell. 2004;15(11):4866–4876. doi: 10.1091/mbc.E04-05-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musso C, et al. Clinical course of genetic diseases of the insulin receptor (type A and Rabson-Mendenhall syndromes): a 30-year prospective. Medicine (Baltimore). 2004;83(4):209–222. doi: 10.1097/01.md.0000133625.73570.54. [DOI] [PubMed] [Google Scholar]
- 32.Semple RK, et al. Postreceptor insulin resistance contributes to human dyslipidemia and hepatic steatosis. J Clin Invest. 2009;119(2):315–322. doi: 10.1172/JCI37432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet. 2006;7:175–199. doi: 10.1146/annurev.genom.7.080505.115715. [DOI] [PubMed] [Google Scholar]
- 34.Semple RK, Savage DB, Cochran EK, Gorden P, O’Rahilly S. Genetic syndromes of severe insulin resistance. Endocr Rev. 2011;32(4):498–514. doi: 10.1210/er.2010-0020. [DOI] [PubMed] [Google Scholar]
- 35.Lindhurst MJ, et al. Mosaic overgrowth with fibroadipose hyperplasia is caused by somatic activating mutations in PIK3CA. Nat Genet. 2012;44(8):928–933. doi: 10.1038/ng.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Albers CA, Lunter G, MacArthur DG, McVean G, Ouwehand WH, Durbin R. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21(6):961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2011;43(7):712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payne F, et al. Mutations disrupting the Kennedy phosphatidylcholine pathway in humans with congenital lipodystrophy fatty liver disease. Proc Natl Acad Sci U S A. 2014;111(24):8901–8906. doi: 10.1073/pnas.1408523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.International HapMap Consortium The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 43. Westerfield M. The Zebrafish Book. Eugene, Oregon, USA: University of Oregon Press; 1995. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.