SUMMARY
The function of the mammalian TIMELESS protein (TIM) has been enigmatic. TIM is essential for early embryonic development, but little is known regarding its biochemical and cellular function. Although identified based on similarity to a Drosophila circadian clock factor, it also shares similarity with a second family of proteins that is more widely conserved throughout eukaryotes. Members of this second protein family in yeast (S.c. Tof1p, S.p. Swi1p) have been implicated in DNA synthesis, S-phase-dependent checkpoint activation and chromosome cohesion—three processes coordinated at the level of the replication fork complex. The present work demonstrates that mammalian TIM and its constitutive binding partner, Tipin (ortholog of S.c. Csm3p, S.p. Swi3p), are replisome-associated proteins. Both proteins associate with components of the endogenous replication fork complex, and are present at BrdU-positive DNA replication sites. Knock-down of TIM also compromises DNA replication efficiency. Further, the direct binding of the TIM-Tipin complex to the 34 kDa subunit of replication protein A provides a biochemical explanation for the potential coupling role of these proteins. Like TIM, Tipin is also involved in the molecular mechanism of UV-dependent checkpoint activation and cell growth arrest. Tipin additionally associates with peroxiredoxin2 and appears to be involved in checkpoint responses to H2O2, a role recently described for yeast versions of TIM and Tipin. Together, this work establishes TIM and Tipin as functional orthologs of their replisome-associated yeast counterparts capable of coordinating replication with genotoxic stress responses, and distinguishes mammalian TIM from the circadian-specific paralogs from which it was originally identified.
Keywords: Timeless, Tipin, DNA replication, checkpoint signaling, chromosome cohesion
INTRODUCTION
Mammalian TIMELESS (TIM) proteins were originally identified and named based on weak similarity to Drosophila TIM 1; 2; 3, a protein essential for circadian rhythms in the fly 4. While it has been difficult to pinpoint the specific role of TIM in the circadian clock, database mining and phylogenetic sequence analysis has suggested that mammalian TIMs may not be the true orthologs of the circadian clock-specific proteins found in insects, since they share even greater similarity to second family of proteins that are more widely conserved in eukaryotes. These include Drosophila TIMEOUT (or TIM-2), S. cerevisiae Tof1p, S. pombe Swi1p, and C. elegans TIM (reviewed in 5). Where studied, these proteins appear to be involved in molecular pathways important for cell viability and/or development. Similarly, knockout of mouse TIM results in embryonic lethality just after blastocyst implantation 6, and down-regulation of TIM disrupts epithelial cell morphogenesis important for tubule formation in the developing mammalian lung and kidney 7; 8.
The molecular pathways through which mammalian TIM exerts its vital functions have not been determined. An initial step in understanding the protein's biochemical roles was made with the identification of its binding partner, Tipin (TIM interacting protein). Isolated by yeast two hybrid screening, Tipin avidly associates directly with TIM, and is co-expressed with Tim in proliferating embryonic and adult tissues 9. Orthologs of Tipin are found throughout eukaryotes and include S. cerevisiae Csm3p and S. pombe Swi3p. Remarkably, yeast versions of Tipin bind directly to orthologs of TIM 10; 11; 12; 13, demonstrating the evolutionary conservation of this interaction.
Rapidly accumulating evidence from yeast and C. elegans has demonstrated that orthologs of TIM and Tipin are DNA replication fork-associated factors that are not only involved in DNA synthesis, but also participate in S phase-dependent checkpoint signaling and chromosome cohesion. Within the replisome, both factors are involved in preventing the collapse of replication forks that have stalled adjacent to DNA damage, natural replication barriers and mating type loci. Both genes were identified in S. pombe for their role in mating type switching. Mutations in either Swi1p or Swi3p are associated with a failure of replisomes to stably stall adjacent to mating-type loci 14; 15 and rDNA sites 16; 17. At the biochemical level, these proteins have been postulated to physically coordinate helicase progression with DNA polymerase activity such that mutations in either gene are associated with the appearance of abnormally large regions of ssDNA adjacent to replication origins under both normal and replication stress conditions 17; 18; 19; 20. TIM and Tipin orthologs may also coordinate leading and lagging strand synthesis as genetic interactions have been observed between genes for these proteins and Polα and Polδ 12.
Yeast orthologs of TIM and Tipin also function in intra-S-phase checkpoint activation, a process dependent upon DNA replication. As replication forks encounter DNA damage or metabolic stress in the form of reduced dNTP levels, checkpoint signaling cascades are activated to stabilize stalled replication forks and to arrest cell cycle progression, which ultimately allows damaged DNA to be repaired or dNTP levels to be restored 21; 22; 23; 24. TIM and Tipin orthologs in yeast are involved in molecular and cellular responses to UV irradiation, methyl methanesulfonate (MMS)-induced DNA damage, or hydroxyurea (HU)-induced depletion of dNTP stores. These roles include checkpoint kinase activation, cell cycle arrest, and maintaining the integrity of replication forks stalled by these treatments 20; 25; 26; 27. Like yeast Claspin (Mrc1p), TIM and Tipin orthologs are likely to mediate these functions by coordinating kinases at the replisome while providing a structural link between helicase unwinding and polymerase mediated elongation activity 18; 19.
Protein factors involved in DNA replication and checkpoint signaling also regulate the initiation and maintenance of chromosome cohesion 28; 29, a process crucial for proper segregation of replicated sister chromatids and recombination-mediated repair of double-strand DNA breaks. Three independent genetic screens identified yeast orthologs of TIM and Tipin as being involved in meiotic and mitotic chromosome segregation 11; 30; 31. In C. elegans, TIM-1 has been found to loosely associate with SMC1 of the Cohesin complex, and is involved in loading Cohesin subunits onto newly replicated chromosomes 32.
Previous uncertainty regarding the evolutionary origin and circadian function of mammalian TIM has clouded the potential relevance of the replication fork-associated functions of yeast and C. elegans orthologs. One recent paper by Ünsal-Kaçmaz et al. (2005) showed human TIM to be involved in HU-induced CHK1 phosphorylation and UV-dependent checkpoint responses, providing a hint into the replisome-associated function of TIM. Here we demonstrate that the TIM-Tipin complex associates with the replisome through a direct interaction with replication protein A (RPA) and is important for efficient DNA synthesis. Like TIM, Tipin also appears to be involved in checkpoint signaling and cell growth arrest occurring in response to UV irradiation and additionally, oxidative stress. These studies demonstrate that mammalian TIM is a functional ortholog of a widely conserved family of proteins that are distinct from the circadian-specific factors restricted to insects.
RESULTS
Tipin and TIM Associate with Replication Protein A
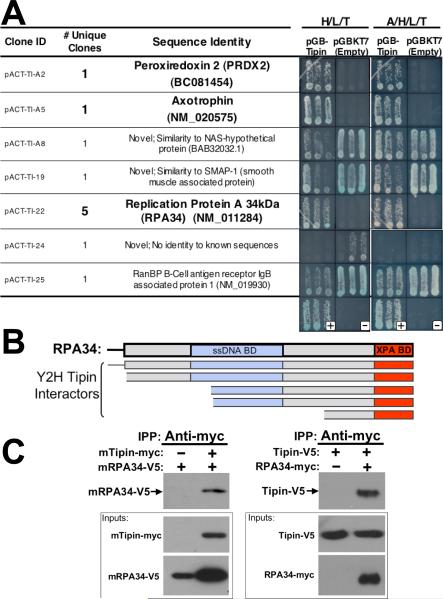
To link the mammalian TIM-Tipin complex with known biochemical pathways, a yeast two-hybrid approach to was used to identify proteins capable of interacting with mouse Tipin. Screening a mouse embryonic cDNA library resulted in the identification of seven unique clones that were classified as true positive interactors (Figure 1(A)). Two of these positive clones encoded portions of the peroxiredoxin 2 and axotrophin proteins. The remaining five positives, which exhibited strong reporter activity, encoded overlapping portions of the same protein: the 34 kDa subunit of replication protein A (RPA34). Examination of the insert sequences encoding these five different RPA34 fragments enabled us to delineate the Tipin binding region contained within the RPA34 protein. The smallest Tipin-interacting clone encoded the C-terminal 84 amino acids of the full-length RPA34 protein (Figure 1(B)), a region containing the 34-residue XPA (xeroderma pigmentosum complementing factor A)-binding domain 33. Conversely, the site on XPA known to bind RPA34 has also been delineated 33, and resembles a 48-residue stretch of the Tipin protein sequence. These observations indicate that Tipin and RPA34 have interaction sites for one another, suggesting that RPA34 is a bona fide Tipin-interacting protein.
Figure 1.
Tipin binds the 34 kDa subunit of RPA. A, Secondary screening of representative Tipin-interacting clones identified by yeast two hybrid screening of a mouse embryonic day 11 cDNA library was performed by co-retransforming positive clones into yeast with either the bait construct (pGBTipin) or empty vector (pGBKT7). Three independent transformants were then streaked on media lacking histidine, leucine and tryptophan (moderate stringency, “H/L/T”) or media also lacking adenine (high stringency, “A/H/L/T”). Clones exhibiting growth when co-transformed with the empty vector indicate a false positive. Of 25 initial positives, 11 clones encoded unique insert sequence, including five with overlapping RPA34 sequence. The largest RPA34 clone (pACT-TI-22) was included in this analysis. B, Schematic of the encoded regions of five overlapping RPA34 fragments isolated in the yeast two-hybrid screen for Tipin-interacting proteins relative to the coding region and functional domains of full-length RPA34. (“ssDNA BD”, single strand DNA binding domain; “XPA BD,” xeroderma pigmentosum complementing factor A binding domain). C, Tipin-RPA34 interactions were confirmed in immunoprecipitation experiments of epitope-tagged mouse proteins expressed in NIH3T3 cells (left) and human proteins expressed in HEK293 cells (right). Extracts from cells transfected to express the indicated constructs were subjected to immunoprecipitation with anti-myc antibody (0.5 μg). Western blot exposures of co-precipitated samples probed with anti-V5 antibody are shown above blots of proteins present in 10 μg of input samples (boxed exposures). Augmented RPA34 levels associated with Tipin over-expression is typical (and unavoidable) of experiments done in NIH3T3 cells, and may represent a stabilizing effect of Tipin.
The interaction of RPA34 with Tipin was of particular interest given the known cellular roles for this protein. The RPA holoenzyme, comprised of 17, 34 and 70 kDa subunits, is a single strand DNA (ssDNA) binding protein involved not only in DNA replication, but also DNA repair, cell cycle checkpoint signaling and meiotic recombination of synapsed sister chromatids 34— functions reminiscent of those described for yeast orthologs of TIM and Tipin. The interaction of RPA34 with Tipin was confirmed by co-immunoprecipitation of epitope-tagged proteins expressed in both mouse NIH3T3 and human HEK293 cells. Mouse (m)RPA34-V5 co-precipitated with anti-myc antibody from NIH3T3 cell extracts only when mTipin-myc was expressed (Figure 1(C), left panel). In human HEK293 cells transfected to express human versions of these proteins, anti-myc antibody brought down V5-tagged Tipin only when RPA34-myc was co-expressed (Figure 1(C), right panel).
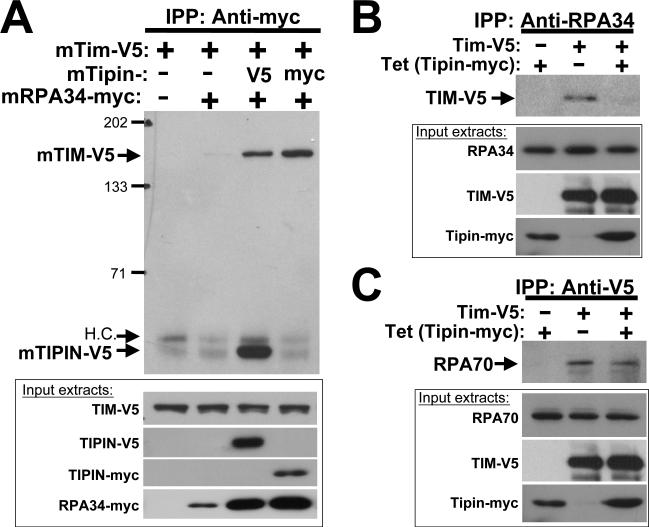
The capacity of the RPA holoprotein to exist in a complex with both Tipin and TIM was also evaluated. In one set of experiments, NIH3T3 cells were transiently transfected to differentially express epitope-tagged versions of mRPA34, mTipin and/or mTIM (Figure 2(A)). Using an anti-myc antibody for immunoprecipitation, TIM-V5 co-precipitated with mRPA34-myc when mTipin-V5 was also expressed in these cells. The lack of an appreciable mTIM-V5 signal in immune complexes brought down with anti-myc in the absence of mTipin expression (either mTipin-V5 with mRPA34-myc or mTipin-myc alone) indicates that TIM's association with RPA34 is dependent upon Tipin. Interactions between human TIM and Tipin with endogenous RPA were further examined in human HEK293 cells stably transfected to express human Tipin-myc in a tetracycline-inducible manner. Both endogenous RPA34 and RPA70 co-precipitated TIM-V5 even in the absence of tetracycline-induced Tipin expression (Figure 1(B and C), second lanes), indicating that endogenous Tipin levels are sufficient to mediate the association of TIM with the RPA holoprotein. In fact, over-expression of Tipin attenuated the association of TIM-V5 with endogenous RPA subunits (Figure 1(B and C), third lanes; see also experiments with endogenous proteins below). This indicates that TIM associates with RPA through Tipin as an intermediary, since elevated levels of both exogenous TIM and Tipin would be expected to favor separate TIM-Tipin and Tipin-RPA interactions over the TIM-Tipin-RPA interaction detected in the presence of endogenous Tipin levels.
Figure 2.
The TIM-Tipin complex binds the RPA holoprotein. A, Co-immunoprecipitation of mouse (m)TIM-V5 and mTipin-V5 with mRPA34-myc in NIH3T3 cells. Extracts were prepared from cells transfected with the indicated constructs and immunoprecipitations done using anti-myc antibody. Immunoprecipitated samples were visualized on blots with anti-V5. Boxed exposures, protein expression in 10 μg of input extracts visualized by anti-V5 and anti-myc western blotting. “H.C.”, cross-reacting heavy chain of the immunoprecipitating antibody. Lane 4, positive control illustrating the TIM-Tipin interaction. Results are representative of over 4 independent trials. B and C, HEK293 cells stably transfected to express hTipin-myc in a tetracycline inducible manner were additionally transfected to express hTIM-V5 as indicated. Complexes precipitated with anti-RPA34 (B) or anti-V5 (C), antibodies were visualized by western blot using anti-V5 and anti-RPA70, respectively (upper exposures, each panel). Protein expression in input extracts was evaluated by western blot using antibodies against endogenous proteins (RPA34, RPA70) or epitope tags (V5, myc).
Cell Cycle-dependent Expression of Endogenous TIM and Tipin
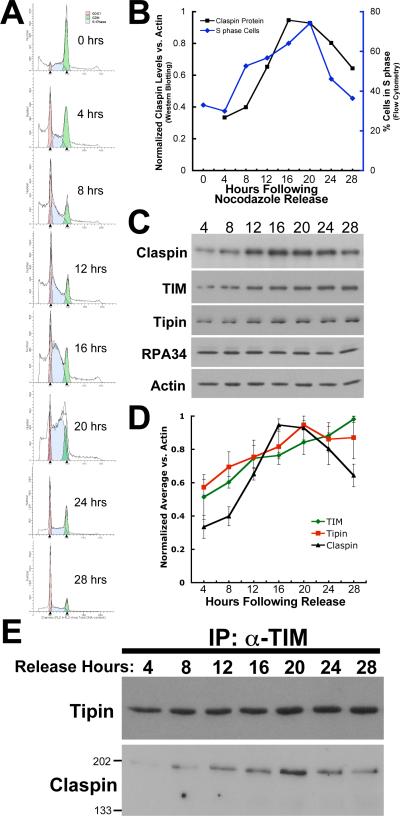
To further understand the cellular function of TIM and Tipin, the cell cycle-dependent expression of these proteins was further assessed. For this purpose, anti-TIM and anti-Tipin antisera were generated in guinea pigs and rabbits, respectively. Affinity purification against their respective antigens resulted in effective reagents that showed little, if any, cross reactivity to other cellular proteins (Supplemental Figures S1 and S2). To assess TIM and Tipin levels during cell cycle progression, HEK293 cells were synchronized by thymidine-nocodazole block and released at 4 hour intervals for 28 hours. Flow cytometry indicates that these cells begin to enter S-phase at approximately 12 hours after release with most cells exiting between 20 and 24 hours (Figure 3(A)). Claspin, a documented S-phase-dependent protein35, peaks in its expression between 16 and 20 hours after release from synchronization, corroborating this S-phase timing (Figure 3(B)). Levels of both TIM and Tipin protein (relative to actin) display a steady increase in expression prior to and during S-phase (Figure 3(C) and 3(D)), identical to a previous description of TIM expression examined at lower resolution 36. This expression pattern of TIM and Tipin is also substantiated by microarray expression analysis of synchronized HeLa cells 37 (http://genomewww.stanford.edu/Human-CellCycle/HeLa/search.shtml) which indicates that Tim and Tipin RNA levels peak just prior to and during S-phase.
Figure 3.
Cell cycle progression of HEK293 cells and TIM and Tipin expression. A, Flow cytometric analysis of thymidine-nocodazole-synchronized HEK293 cells. Percentages of cells in G1, S and G2/M phases were estimated using ModFit LT. Fitted profiles of cell number (y axis) vs. DNA content (FL2-A, x axis) are shown for cells in G1 (red), S (blue) and G1/M (green) at indicated release times. The attenuated G2 phase between 20 and 24 hrs is typical of HEK293-based cells. B, Plot of the percentage of S-phase cells determined by flow cytometry and Claspin protein content relative to actin as determined by western blotting (see below). C, Western blots of extracts from synchronized cells. Endogenous protein levels were assessed relative to actin by re-probing stripped blots with anti-actin. Blots shown are representative of at least three independent experiments. D, Quantitation of TIM, Tipin and Claspin levels during S-phase progression. Endogenous protein levels were assessed by western blot and quantitated by densitometry relative to actin levels measured on blots that were either simultaneously probed (TIM and actin) or stripped and re-probed for actin. Data are the average of three independent experiments. E, Co-immunoprecipitation of Tipin and Claspin with TIM (all endogenous) from synchronized cells released from nocodazole block at the indicated times. Input protein levels are seen in panel C.
The cell cycle-dependent association of TIM with Tipin as well as Claspin was also evaluated in synchronized HEK293 cells. Immunoprecipitation experiments using the anti-TIM antibody to bring-down the endogenous protein, indicated that TIM and Tipin associate with one another constitutively during cell cycle progression, the co-precipitating amount being proportional to their expression level (Figure 3(E)). An association of endogenous TIM with Claspin was also detected. In yeast, orthologs of TIM and Tipin genetically interact with Claspin (Mrc1p) for DNA damage-induced checkpoint activation, chromatid cohesion and DNA repair functions 31. Immunoprecipitation experiments using extracts from these synchronized cells demonstrate that human Claspin also co-precipitates with endogenous TIM in an S-phase-dependent manner proportional to Claspin expression (Figure 3(E), lower panel).
TIM and Tipin Associate With the Endogenous Replication Fork Complex
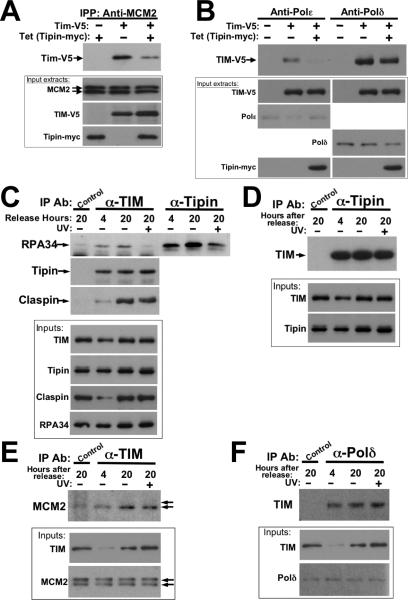
The binding of the human TIM-Tipin complex with RPA, its elevated level of expression during S phase and association with Claspin, suggests that these proteins associate with the replication fork complex. Immunoprecipitation studies of human HEK293 cell extracts demonstrate the interaction of endogenous replisome proteins with both epitope-tagged TIM and Tipin (Figure 4(A) and 4(B)) and the endogenous TIM-Tipin complex (Figure 4(C-F)). In cells stably transfected to express Tipin-myc in a tetracycline inducible manner, MCM2 co-precipitated TIM-V5 even in the absence of Tipin over-expression (Figure 4(A)). Antibodies against replication fork-associated Polε and Polδ DNA polymerases co-precipitated TIM-V5 in a similar manner (Figure 4(B)). When taken with our discovery of a direct interaction of Tipin with RPA, these results suggest that the TIM-Tipin complex may be positioned at the replication fork between the MCM complex, and DNA polymerases by RPA, associating with unwound ssDNA existing between these two complexes 34 (see model of Figure 9). Supporting this assertion is the observation that Tipin over expression attenuates the association of TIM-V5 with MCM2, Polε and Polδ (Figure 4(A) lane 3; 4(B) lanes 3 and 6). An over abundance of Tipin-myc is expected compete with endogenous Tipin to sequester exogenous TIM-V5 from relatively lower levels of RPA at the replisome. Ultimately, this is reflected in a decrease in an observable association of TIM-V5 with proteins present within endogenous replisome complexes as seen in these immunoprecipitation experiments.
Figure 4.
TIM and Tipin interact with the endogenous replication fork complex. A and B, Epitope-tagged TIM and Tipin bind endogenous MCM2 and Polδ/ε. Extracts from HEK293 cells stably transfected to express tetracycline-inducible Tipin-myc were additionally transfected to express TIMV5 where indicated. Antibodies against endogenous MCM2 (A) or Polε and Polδ (B) were used for immunoprecipitation. Co-precipitated TIM-V5 or Tipin-myc was visualized by western blot (upper exposures of each panel). C-F, Endogenous TIM and Tipin interact with endogenous replisome factors. Immunoprecipitation experiments were performed on HEK293 cells released from thymidinenocodazole synchronization at the indicated times as described above. In each experiment, UV treatment was performed on an additional sample released from nocodazole block 19.5 hours prior. C, Association of TIM and Tipin with RPA34 are attenuated by UV irradiation. Proteins co-precipitated with antibodies against TIM, Tipin and an equal concentration of anti-actin antibody (“control”) was visualized on western blots probed with anti-RPA34, Tipin, and Claspin. D, TIM co-precipitation with Tipin. “Control,” 1 μg anti-actin antibody. E, MCM2 co-precipitation with TIM. Arrows indicate alternate forms of MCM2 (as seen in input extracts); TIM preferentially interacts with the lower Mr form. “Control” antibody: anti-HA. F, TIM co-immunoprecipitation with Polδ. “Control” antibody: anti-HA. Each experiment is representative of at least three independent replicates.
Figure 9.
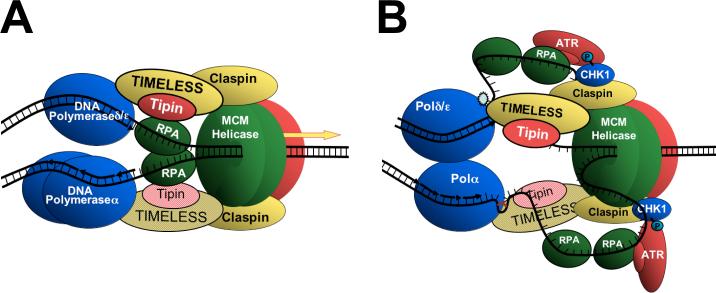
Model for the role of TIM and Tipin at the replisome. A, In progressing replication forks TIM and Tipin associate with the replication fork complex through the Tipin-RPA34 interaction potentially coupling MCM-mediated DNA unwinding with polymerase-mediated DNA synthesis. Claspin is shown near MCM2-7, since its association is dependent upon CDC45 15. B, UV-induced 6-4 photoproducts (upper [leading] strand), MMS-mediated base alkylation (lower [lagging] strand), or insufficient dNTP levels (not depicted) slow polymerase activity resulting in localized increases in RPA-bound ssDNA 21. Light shaded TIM and Tipin [lagging strand] are implied based on their function in yeast to coordinate leading and lagging strand synthesis 12. Other important replication and checkpoint activation factors (e.g. CDC45, PCNA, Rad17, RHR complex) are omitted for simplicity.
We also examined the association of endogenous TIM and Tipin with key replisome factors during S-phase. Figure 4(C) demonstrates that both of these proteins co-immunoprecipitate RPA34 from extracts of synchronized cells. The association of RPA34 with Tipin appears more pronounced, as might be expected for a direct interaction, while the binding of RPA34 to TIM is less robust, presumably because this interaction occurs through Tipin as an intermediary. These associations are seen in extracts from cells collected both during G1 (4 hours after release) and S-phases (20 hours after release), with binding being more obvious under the latter condition. In addition to RPA, endogenous TIM also co-precipitates with both MCM2 and Polδ, which again, is more evident when most of these cells are in S-phase (Figure 4(E) and 4(F)). We were, however, unable to detect co-precipitation of endogenous TIM with Polε above control signals, (not shown), a result that is potentially due to less abundant levels of endogenous Polε (Figure 4(B)). The interaction of TIM with MCM2 is noteworthy in that the former preferentially co-precipitates a more rapidly migrating form of MCM2 (Figure 4(E)), a phosphorylated form of MCM2 typically associated with chromatin 38. Together, these results are consistent with a role for the TIM-Tipin complex at the replisome.
Interestingly, UV irradiation attenuates the amount of RPA34 that co-precipitates with endogenous TIM and Tipin (Figure 4(C), lanes 4 and 7), an effect not seen for the TIM-Tipin interaction, or the association of TIM with Claspin, MCM2 or DNA polymerase δ (Figures 4(C), 4(D), 4(E), 4(F)). UV-dependent disruption of this interaction could be the result of competition between Tipin and XPA (xeroderma pigmentosum complementing factor A) for RPA34 binding. Both Tipin and XPA associate with a similar region of RPA34 (Figure 2(C)), contain similar RPA34-binding sites 9; 33, and compete with one another for RPA34 binding when exogenously expressed in transfected cells (see Supplemental Figure S3). Because RPA34 binds XPA to repair UV-damaged DNA for nucleotide excision repair (NER) 33; 39; 40, XPA may be expected to displace Tipin from RPA in cells exposed to UV irradiation (see Discussion).
TIM and Tipin Co-localize with RPA and Nuclear Replication Sites
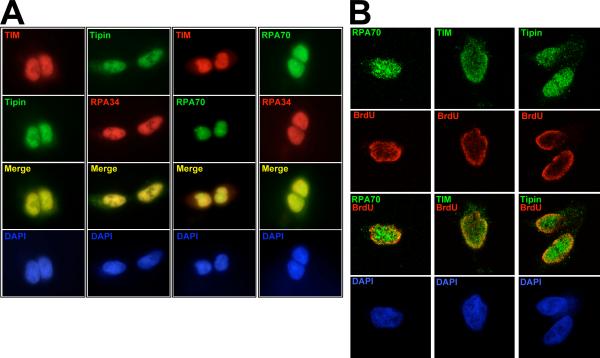
TIM and Tipin also display subcellular localization consistent with a role in processes involving DNA replication. Using the newly generated antibodies described above, these proteins were localized relative to RPA34, RPA70 (Figure 5(A)) and actin (Supplemental Figure S4). In HEK293 cells synchronized to S-phase, endogenous TIM and Tipin are predominantly nuclear with a small, but detectable amount of protein seen in the cytoplasm (Figure 5(A), first column). Both proteins were distributed throughout the nucleus coincident with DAPI localization, suggesting chromatin co-localization. Tipin also co-localizes with RPA34 in the nucleus with the latter being less abundant in the cytoplasm (Figure 5(A), column 2). Similarly, TIM co-localizes with RPA70.
Figure 5.
Endogenous TIM and Tipin exhibit overlapping expression with RPA and DNA replication sites. A, Vertical columns are representative experiments of HEK293 cells synchronized to S-phase (18 hr release from thymidine-nocodazole block) and probed with the indicated affinity-purified primary antibodies. RPA70 exhibits additional punctate staining within the nucleus at sites of attenuated DAPI staining (not seen with the anti-RPA34 antibody), potentially representing localization to promyelocytic leukemia protein (PML) bodies, as observed previously for RPA 61. B, TIM, Tipin and RPA70 localization relative to DNA replication sites. 24 hrs after release from thymidine block, HEK293 cells were incubated in 100 μM BrdU for 10 min, fixed, processed and co-stained with mouse anti-BrdU along with the indicated antibodies. Each column shows representative cells observed in mid-S-phase as defined by BrdU incorporation pattern 41. For both A and B, no appreciable signal was observed in negative control experiments performed with no primary antibody (Supplemental Figure S4).
To assess the presence of TIM and Tipin at DNA replication sites, HEK293 cells, were allowed to incorporate 5’-bromo-2’-deoxyuridine (BrdU) for 10 min prior to processing and coimmunostaining with antibodies against BrdU and either RPA70, TIM or Tipin. In these experiments which evaluated cells 24 hours after thymidine synchronization, BrdU-incorporation took on patterns indicative of all stages of S-phase 41, the most pronounced of which were those in mid-S-phase where BrdU staining was most abundant near the nuclear periphery (not shown, and Figure 5(B)). RPA70 displayed expression throughout the nucleus in addition to the punctate staining similar to that observed in previous experiments. As expected, RPA70 was present at DNA replication sites. Similarly, both TIM and Tipin were also present throughout the nucleus in addition to being present at BrdU-positive sites (Figure 5(B)), indicating that these proteins are present at regions of the nucleus undergoing DNA replication.
TIM Contributes to DNA Replication Efficiency
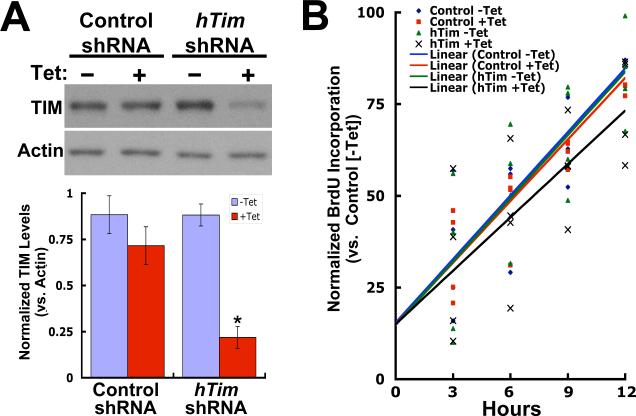
Yeast orthologs of TIM associate with the replisome complex and are thought to coordinate helicase-induced DNA unwinding with polymerase-mediated DNA synthesis. While they are not essential for DNA replication, mutation of these orthologs does effect replication efficiency, presumably due to a lack of coordination between helicase and polymerase activities 18; 19. To evaluate the role of human TIM in DNA replication, a HEK293 cell line was generated in which shRNA-mediated down-regulation of TIM could be controlled in a tetracycline-dependent manner. In these cells, 48 hours of tetracycline induction is accompanied by a 75% reduction in TIM protein levels, while a control cell line, expressing shRNA differing from the human Tim cDNA target sequence by 3 bp, showed no significant reduction (Figure 6(A)). To obtain an indication of the role of human TIM in replication efficiency, these cell lines were used in assays designed to measure the rate of BrdU incorporation into DNA over a 12 hour period. For these experiments, cells grown in the presence or absence of tetracycline for 48 hours were subsequently allowed to incorporate BrdU for 3, 6, 9, and 12 hours followed by colorimetric analysis of BrdU content relative to cell density. In cells expressing control shRNA, the rate of BrdU incorporation was not significantly affected by tetracycline induction (Table 1). In cells expressing Tim shRNA, however, tetracycline-mediated down regulation of TIM significantly reduced the BrdU incorporation rate to 86% of that seen in the same cells grown in the absence of the inducer (p = 0.014, n = 4) (Table 1; shown graphically in Figure 6(B)). Together with our biochemical data, these results are consistent with a conserved replication fork-associated role for mammalian TIM, and further, suggest that mammalian TIM and Tipin may function in intra-S-phase checkpoint signaling similar to their yeast counterparts.
Figure 6.
shRNA-mediated TIM down-regulation attenuates DNA replication rate. A (upper panel), Endogenous TIM expression in cells expressing control (mouse Tim sequence with 2 bp mismatches) or and Tim shRNA was determined by western blot of cell extracts prepared 48 hours after tetracycline induction and simultaneously probed with anti-TIM and anti-actin. (Lower panel), TIM levels from three experiments were quantitated densitometrically relative to in-lane actin signals. Normalized TIM/actin levels were averaged (lower panel). *, p < 0.05, unpaired t-test, n=3 (relative to –Tet condition). B, DNA replication rate relative to viable cell density was assessed in control and Tim shRNA-expressing cells grown in the absence or presence of tetracycline (48 hrs) by evaluating time-dependent BrdU incorporation after 3, 6, 9, and 12 hours. Data presented are the results from 4 trials: 4 time points per trial, 6 samples per time point. Each plotted point represents the average of 6 samples for each of 4 trials (normalized to control line -Tet average and y intercept [15%--rates shown do not originate from the origin due to nonlinearity at time points < 3 hrs and compensation for cell density]). Least squares regression lines for each set of averages for both cell lines (−/+ Tet) are shown.
Table 1.
Down regulation of TIM affects DNA replication rate.
| Cell Line (HEK293) | Tetracycline | TIM/Actin Protein Levels (±SEM)a | p Value (t test)c | BrdU Incorporation Rate (±SEM)b | p Value (t test)c |
|---|---|---|---|---|---|
| Control shRNA | − Tet | 0.884 (±0.103) | 100% | ||
| Control shRNA | + Tet | .715 (±0.139) | 0.3850 (vs. −Tet) | 101.60 (±10.70)% | 0.886 (vs. −Tet) |
| Tim shRNA | −Tet | 0.882 (±0.060) | 100% | ||
| Tim shRNA | +Tet | 0.219 (±0.020) | 0.0005 (vs. −Tet) | 86.48 (±3.97)% | 0.014 (vs. −Tet) |
Sub-saturated TIM and actin protein levels were determined by densitometric scanning of western blot signals from three independent cell extract samples (for each cell line and treatment) analyzed on the same film. In-lane TIM/actin levels were calculated, normalized to the single highest value and averaged.
Incorporation rates were determined by comparing the slope of least squares regression lines drawn through data taken at four time points (each trial included 6 samples at each of four time points [3, 6, 9, 12 hrs]). The +Tet slope values from each cell line of each trial were normalized relative to the –Tet condition and these values averaged (n=4).
Two-tailed Student's t test (unpaired).
Tipin is Involved in UV Damage-Induced Responses
The initiation of checkpoint signaling in response to UV irradiation requires DNA replication and factors associated with the replisome 22; 23; 24. UV-induced DNA damage stalls replication fork progression primarily by inhibiting polymerase-mediated DNA synthesis leading to an accumulation of ssDNA 42. ATR (ataxia-telangiectasia and Rad3-related), in a complex with its interacting protein, ATRIP, recognize RPA-bound ssDNA present at stalled replication foci 43. ATR-ATRIP then initiates checkpoint signaling by phosphorylating Chk1 on serine residues 317 and 345 44 for checkpoint activation. Human TIM has recently been demonstrated to be involved in S-phase-dependent cell cycle arrest in response to UV irradiation and to mediate Chk1 phosphorylation in response to metabolic stress 36.
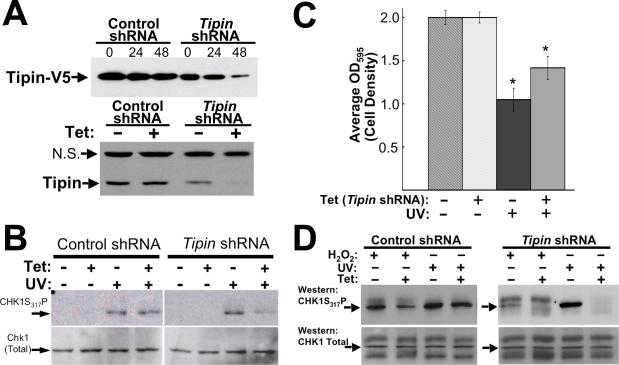
To assess the role of Tipin in UV-dependent Chk1 phosphorylation and cell growth arrest, mouse NIH3T3 cells stably transfected to express control and Tipin shRNA in a tetracyclineinducible manner were generated. Forty-eight hours after tetracycline induction, Tipin shRNA down-regulated both over-expressed Tipin-V5 as well as cells expressing only the endogenous protein (Figure 7(A)). Similar responses were not observed in a control cell line expressing shRNA against human Tipin (2 nucleotide mismatches). UV-dependent Chk1 phosphorylation was assessed in control and Tipin shRNA cell lines by western blot using an anti-CHK1 antibody specific for the S317 phosphorylation site (anti-CHK1S317P), an ATR-dependent modification required for CHK1 activation 44. Thirty minutes after UV irradiation, CHK1 phosphorylation was observed in both cell lines in the absence of tetracycline induction (Figure 7(B), lanes 3, 7). This response was attenuated in Tipin shRNA cells treated with tetracycline (Figure 7(B), lanes 4 and 8), indicating that Tipin is involved in CHK1 activation. As a general measure of the involvement of Tipin in UV-dependent attenuation of cell growth (both cell cycle arrest and apoptosis), cell density of control and Tipin shRNA-expressing lines was assessed 20 hours after either UV or sham treatment, providing a relative measure of cell proliferation occurring after these manipulations. In the absence of Tipin shRNA induction, cell density of UV-exposed cells remained at 52% of the sham treatment (Figure 7(C)). Tetracycline-induced Tipin shRNA expression however, attenuated this UV-dependent response to levels significantly elevated (70.8%) relative to UV-treated cells not grown in the presence of tetracycline (n = 24, p = 0.021, unpaired t test), while no effect of UV was observed in two additional control cell lines (Supplemental Figure S5). The inability of Tipin shRNA expression to completely ablate cell growth arrest in response to UV may be due to either the presence of reduced, yet permissive levels of Tipin protein, or may indicate that Tipin along with TIM are less essential for checkpoint activation as suggested for S. cerevisiae Tof1 17. We did not observe an effect of Tipin shRNA expression on UV-dependent cell cycle arrest of both asynchronous and thymidine-nocodazole synchronized cells analyzed by flow cytometry (not shown). Nevertheless, these observations indicate that Tipin is at least indirectly involved in cell growth arrest in response to UV irradiation.
Figure 7.
Tipin is involved in checkpoint signaling responses to UV irradiation and oxidative stress. A, Control or Tipin (mTip2) shRNA-expressing NIH3T3 cells were assessed for their ability to down-regulate exogenous Tipin-V5 after treatment with tetracycline (Tet) for 0, 24 or 48 hours, (upper) or endogenous Tipin after 48 hours (lower). Western blots were probed with anti-V5 or anti-Tipin1 antibody. N.S., non-specific band serving as loading control (See Supplemental Fig. S2). B, Control or Tipin shRNA cell lines were cultured for 48 hrs −/+ Tet prior to UV treatment (10 J/m2 at 254 nm, where indicated). Western blots of extracts (10 μg) prepared 15 minutes later were probed with anti-CHK1S317P antibody (top exposures). Stripped blots were re-probed with anti-Chk1 antibody (bottom). Arrows, CHK1S317P band. C, Tipin shRNA-expressing and control cell lines were grown for 48 hours in the presence or absence of tetracycline prior to UV irradiation. Twenty hours after sham or UV treatment, cell density was assessed by MTT assay. OD595 values indicative of cell density were normalized relative to average values in the absence of UV treatment. Error bars, SEM (n=24). *, p=0.021 (unpaired t test). In control cells (control shRNA line and another carrying no construct), no effect of tetracycline on UV-dependent attenuation of cell proliferation was observed (Supplemental Fig. S5). D, CHK1 phosphorylation in cell lines expressing control and Tipin shRNA 30 minutes after treatment with 200 μM H2O2 or UV. Arrows, major UV-dependent band. Differences in immunoreactivity seen in the bottom panels of B and D are due to different anti-human CHK1 antibodies (goat anti-CHK1 [Abcam], rabbit anti-CHK1 [Cell Signaling], respectively).
Tipin Associates with Peroxiredoxin 2
Yeast homologs of TIM and Tipin have recently been described to mediate responses to oxidative stress. In S. cerevisiae, these genes genetically interact with Tsa1, a yeast peroxiredoxin, to prevent oxidative stress-induced synthetic lethality 45. Peroxiredoxins are thiol-dependent enzymes that use thioredoxin as an electron donor to scavenge cellular H2O2 and other reactive oxygen species to protect DNA from oxidative damage 46; 47. Oxidative stress induces CHK1, CHK2 and p53 phosphorylation resulting in cell cycle arrest at G1, intra-S and G2 checkpoints 48.
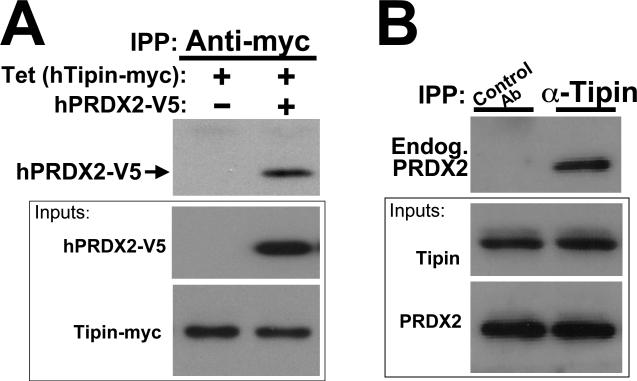
As mentioned earlier, yeast two-hybrid screening for Tipin interacting proteins identified peroxiredoxin 2 (PRDX2), which is, remarkably, the mouse ortholog of S. cerevisiae Tsa1p. To substantiate the yeast two hybrid results, the Tipin-PRDX2 interaction was evaluated in immunoprecipitation experiments of HEK293 cell extracts (Figure 8(A)). In these studies, Tipin and PRDX2 were seen to co-precipitate from extracts of cells transfected to express epitope-tagged proteins (left panel) as well as those in which only endogenous interactions were examined (Figure 7(A), right panel). To evaluate the role of Tipin in signaling responses to oxidative stress relative to UV-induced damage, we examined CHK1S317 phosphorylation in control and Tipin shRNA cell lines (see Figure 7(A)) exposed to H2O2 or UV irradiation. In response to H2O2 exposure, CHK1 undergoes extensive phosphorylation with CHK1S317P signal appearing on at least two different bands, including that which appears in response to UV irradiation (Figure 7(B), arrows). This CHK1S317P signal was attenuated in cells induced to express Tipin shRNA. Additional H2O2-dependent bands not affected by Tipin knock-down are likely to represent further post-translational modifications occurring in response to this broad spectrum DNA damage treatment 47. Oxidative stress also induces double-strand breaks leading to Chk2 phosphorylation 48. In cells manipulated to express Tipin shRNA, however, no H2O2-dependent differences in Chk2T68 phosphorylation were observed in cells grown in the presence or absence of tetracycline (not shown), suggesting that the role of Tipin in these responses may be more specific to ATR-dependent signaling.
Figure 8.
Tipin interacts with peroxiredoxin 2. A, Extracts from HEK293 cells stably-transfected to express Tipin-myc were collected and used in immunoprecipitation experiments after 24 hours of growth in the absence or presence of tetracycline. Anti-myc-precipitated complexes were examined for hPRDX2-V5 (upper exposure). B, Endogenous hPRDX2 co-precipitates with endogenous Tipin. “Control” immunoprecipitating antibody: anti-actin (equal concentration, lane 1).
DISCUSSION
The Role of TIM and Tipin at the Replication Fork
As a ssDNA binding protein, RPA is thought to occupy unwound and unduplicated DNA existing between the MCM helicase and the elongation activity of DNA polymerases 34. As illustrated in the testable model of Figure 9(A), binding of the TIM-Tipin complex to RPA places these proteins in a position advantageous for the coordination of these activities, a function proposed for their yeast counterparts 12; 18; 49. While numerous functional genetic interactions have been described in yeast, the direct biochemical interactions through which those TIM and Tipin orthologs maintain replication fork stability have not. The present work demonstrates that the mammalian proteins not only bind directly to RPA34, but also associate with Claspin, the MCM helicase and DNA polymerases. It should be noted that interactions detected by immunoprecipitation alone do not necessarily indicate a direct protein-protein interaction. It is reasonable to conclude, however, that the TIM-Tipin and Tipin-RPA34 interactions do represent direct binding, since these have been demonstrated in the yeast two-hybrid system 9 (present work), and consistently showed strong immunoprecipitation signals. Claspin also avidly co-precipitated with TIM and may also represent a direct physical interaction. The association of MCM2 and Polδ with TIM in immunoprecipitation experiments, while repeatable, tended to yield weaker signals, suggesting that these may bind through intermediary factors. Indirect interactions mediated by factors like Claspin or CDC45, and PCNA, respectively, would allow TIM to mediate a physical linkage between unwinding and DNA synthesis. A similar biochemical mechanism for yeast orthologs of TIM and Tipin would provide a mechanistic explanation for the role of these proteins in maintaining the integrity of the replisome during stalling adjacent to natural replication fork barriers 16; 17, mating type loci 14; 15 and in the presence of DNA damage or replication stress 12; 18; 19.
Role of TIM and Tipin in Genotoxic Stress Responses
Our current data along with that recently published 36 indicate that mammalian Tipin and TIM have a role in UV-dependent CHK1 phosphorylation and cell growth arrest. In this regard, it is interesting that the peri-implantational embryonic lethality observed in Tim knockout mice is nearly identical to the developmental phenotype resulting from targeted mutagenesis of other checkpoint signaling genes including Rad9, ATR and Chk1 6; 50; 51; 52; 53.
Our results concerning the interaction of the TIM-Tipin complex with RPA, MCM2, Polδ/ε and Claspin suggest that these proteins may facilitate checkpoint activation by maintaining replisome structure. Genotoxic stress in the form of UV-induced DNA damage or reduced dNTP substrate levels preferentially halt polymerase-mediated DNA synthesis rather than DNA unwinding. Even in intact replisomes, these treatments result in an increase in RPA-bound ssDNA that stimulates ATR-dependent checkpoint activation 21. Our data indicate that UV irradiation does not affect the association of the TIM-Tipin complex with the replisome, but does attenuate its association with RPA. These findings suggest a model in which TIM and Tipin may help to maintain the structure of the replication fork complex in the presence of genotoxic stress, while ssDNA is allowed to accumulate in proximity to Claspin, thereby facilitating CHK1 activation (Figure 9(B)). The dissociation of RPA from TIM and Tipin in response to UV is particularly interesting. UV-dependent disruption of the Tipin-RPA34 interaction would also allow XPA to bind RPA for NER processes important for the repair of UV-damaged DNA. In the absence of genotoxic conditions, the TIM-Tipin complex may prevent ATR activation by associating with RPA normally present at the replisome. Upon UV-induced replisome stalling, dissociation of RPA may allow ssDNA to accumulate and make it available for RPA-dependent ATR activation. In support of this mechanism, human TIM has recently been shown to associate with ATRIP and CHK1 in a manner dependent upon hydroxyurea-induced depletion of dNTP stores 36. These interactions are likely to represent recruitment of ATR-ATRIP and CHK1 to the TIM-containing replication fork rather than direct binding to TIM; CHK1 binds directly to Claspin 14, while recruitment of ATRATRIP is dependent upon ssDNA-bound by RPA 22; 23.
Such indirect mechanisms may explain the inability of Tipin shRNA to completely ablate UV-dependent arrest in cell growth (Figure 5(C)) and cell cycle progression (not shown). In S. cerevisiae, responses to replication stress appear to be mediated primarily through the Claspin homolog, Mrc1p, whereas the role of the TIM ortholog, Tof1p, appears more restricted to the prevention of replication fork collapse 17. Together, the current findings not only provide a basis for further investigation of the role of TIM and Tipin at the replication fork complex and checkpoint signaling, but also illustrate the evolutionary conservation TIM and Tipin function.
The Tipin-PRDX2 interaction
Unlike ATR-mediated checkpoint signaling in response to UV irradiation and metabolic stress, much less is known of the mechanisms through which oxidative stress affects cell cycle progression. Recent evidence from yeast suggests that oxidative stress may be sensed specifically by the PRDX2 ortholog, Tsa1p. Identified as a potent protector of genomic stability, Tsa1p exhibits genetic interactions with factors involved in DNA replication, checkpoint signaling and DNA repair, including S. cerevisiae versions of TIM and Tipin 45; 46. The identification of mammalian PRDX2 as a Tipin interacting protein in our yeast two-hybrid screen, coupled with the co-precipitation of these endogenous proteins, suggests that Tipin has an evolutionarily conserved role in oxidative stress responses. Further, this interaction evokes a biochemical mechanism through which PRDX2 (and Tsa1p in S. cerevisiae) may associate with the replication fork complex to influence checkpoint signaling and/or alleviate the damaging effects of reactive oxygen species.
The interaction of Tipin with PRDX2 is also intriguing given the documented binding of mammalian TIM with the CRY1 and CRY2, proteins that are evolutionarily related to photolyases that have since taken on well documented roles in the circadian clock mechanism 9; 36; 54; 55. Unlike their photolyase ancestors, CRY proteins by themselves lack the ability to respond to light and repair modified DNA in the form of Pyrimidine<>Pyrimidine or (6-4) photoproducts despite retaining methenyltetrahydrofolate (MTHF) and FAD cofactors 56. These proteins do, however, have an affinity for DNA, particularly (6-4) photoproducts 56; 57. In this regard, it is possible that PRDX2, in a complex with TIM-Tipin-CRY, could provide an electron shuttle to facilitate DNA repair activity of CRYs. Conversely, proximity to CRY may facilitate PRDX2 function in alleviating oxidative stress. Interestingly, a recent study has discovered robust circadian oscillations in phosphorylated forms of PRDX6, a related anti-oxidant protein expressed in liver 58. This suggests that the clock mechanism may prime cells to respond to oxidative stress in a time-dependent manner. The specific mechanisms through which TIM might act in this process, however, remain to be determined.
Oxidative stress is also known to affect the circadian clock mechanism. Changes in redox potential modulate the activity of the Clock:BMAL1 transcription factors responsible for clock gene expression and circadian progression 59. Under normal conditions, cyclic changes in Clock:BMAL1 activity are primarily controlled by CRYs, this inhibition being essential to the clock mechanism 55; 60. A potential CRY-TIM-Tipin-PRDX2 complex provides an intriguing mechanism through which redox potential may be sensed and conveyed to the circadian clock mechanism. Down regulation of TIM does effect the circadian clock function 54, but the specific mechanism for this role is presently unclear (for a full review, see Gotter, [2006] 5. If mammalian TIMELESS has a specific function in the circadian clock, these interactions may provide the mechanism. The present studies, however, demonstrate that TIM is a functional ortholog of evolutionarily conserved replisome-associated proteins, distinct from the circadian-specific paralogs from which it was originally identified. For this reason, investigations into the circadian function of TIM will need to adequately control for TIM's primary replication-associated function and its potential role in coordinating DNA synthesis with DNA damage responses.
MATERIALS AND METHODS
Yeast Two-hybrid Screen for Tipin Interacting Proteins
Tipin interacting proteins were identified using a GAL4-based yeast two-hybrid system (Matchmaker system III, BD Biosciences) as described previously 9. Full-length mouse Tipin cDNA was cloned into pGBKT7, and used to screen a commercially available pre-transformed mouse embryonic day 11 cDNA library at both moderate and high stringency plating conditions according to manufacturer's instructions. Screening media included 2 mM 3-AT (3-amino-1,2,4-triazole). Mating efficiency analysis indicated that approximately 3 × 106 library clones were screened against the pGB-mouse Tipin bait construct. From 25 initial positive colonies, DNA was isolated from all 11 colonies exhibiting growth after subsequent patching. These 11 clones were sequenced and retransformed into yeast containing either the empty pGBKT7 vector or the pGB-mTipin bait construct and grown on moderate and high stringency plates to discern true positive from false positives. Positive and negative controls included pGB-p53 co-transformed with pGAD-T and pGB-p53 co-transformed with pGAD-laminin, respectively.
DNA Constructs and Stably-transfected Tetracycline-inducible Cell Lines
Mammalian expression constructs used for transient transfections were generated in pcDNA3.1-myc (His) and pcDNA3.1-V5 (His) (Invitrogen) vectors. Full-length cDNAs for mouse and human Tim, Tipin, RPA34, XPA and human PRDX2 were amplified by RT/PCR from mouse and human brain poly(A)+-selected RNA (BD Biosciences) and cloned into pcDNA3.1-V5 or –myc as previously described 9. Accession numbers: mouse RPA34, NM_011284; mouse XPA, NM_011728; human RPA34, BC001630; human XPA, BC014965; human PRDX2, NM_005809.
NIH3T3 and HEK293 cells were maintained according to supplier guidelines (ATCC, Invitrogen), and transfections done using Lipofectamine PLUS (Invitrogen). HEK293 cells stably-transfected to express hTipin in a tetracycline-dependent manner were generated by transfecting cells with pcDNA4/TO-hTipin-myc followed by selection in 5 μg/ml blasticidin and 125 μg/ml zeocin (Invitrogen's T-Rex system). NIH3T3 cells stably transfected to express control and mTipin shRNA transgenes were generated similarly (Invitrogen's BLOCK-iT system). shRNA constructs were generated by hybridizing forward and reverse oligonucleotides followed by cloning into the pENTR/H1/TO vector (5’ to 3’; target sequences underlined): mTipin2 forward: CACCGCAAAGCTACTGAGTAATAGCCGAAGCTATTACTCAGTAGCTTTGC, mTipin2 reverse: AAAAGCAAAGCTACTGAGTAATAGCTTCGGCTATTACTCAGTAGCTTTGC, Control forward: CACCGCACCCAAGAAACGACAATTGCGAACAATTGTCGTTTCTTGGGTGC, Control reverse: AAAAGCACCCAAGAAACGACAATTGTTCGCAATTGTCGTTTCTTGGGTGC, hTim2 forward: CACCGCACCCAAGAAACGACAATTGCGAACAATTGTCGTTTCTTGGGTGC, hTim2 reverse: AAAAGCACCCAAGAAACGACAATTGTTCGCAATTGTCGTTTCTTGGGTGC, control forward: CACCGGACTCTGATGATGTTCTTGGCGAACCAAGAACATCATCAGAGTCC, control reverse: AAAAGGACTCTGATGATGTTCTTGGTTCGCCAAGAACATCATCAGAGTCC. Tipin and Tim control sequences differ from their respective target cDNAs by 2 and 3 bp, respectively.
For cell cycle analysis and S-phase-dependent association of endogenous TIM and Tipin with replication fork factors, thymidine-nocodazole synchronization was carried out using standard methodology: HEK293 cells were grown in 2mM thymidine for 24 hours followed by a 4 hr release before mitotic arrest in 100 ng/ml nocodazole for 12 hrs. Cells were then released at 4 hr intervals such that all samples were collected simultaneously. For flow cytometry, cells were propidium iodide stained (BD Cycletest, BD Biosciences), and analyzed on a FACSort instrument (BD Biosciences; University of Pennsylvania's flow cytometry core facility).
Generation of Anti-human TIM and Tipin Antibodies
Antigen against the human TIM protein was nickel affinity purified from bacteria expressing the C-terminal 154 amino acids of the human protein from the pET101 vector (Invitrogen). Guinea pig antisera (Cocalico Biologicals Inc., Reamstown, PA) were analyzed on western blot strips for its ability to recognize both epitope-tagged and endogenous TIM in cultured cell extracts. Final guinea pig anti-hTIM reagent was generated by affinity purifying antibodies using bacterially-expressed antigen coupled to NHS-activated Sepharose (Pharmacia). Rabbit antisera against human Tipin were generated similarly: The N-terminal 143 amino acids of the human protein were expressed in bacteria, nickel affinity purified and used as antigen in multiple animals. Anti-Tipin activity was identified in two different rabbits was affinity purified against the bacterially expressed Tipin antigen coupled to NHS-activated Sepharose. Anti-TIM and anti-Tipin antibody characterization is shown in Supplemental Figure S1 and S2.
Immunoprecipitation
Cell extract preparation, immunoprecipitation (IP) and western blot experiments were performed at 4°C as previously described 9 with modifications. NIH3T3 and HEK293 cell extracts were prepared by washing cells in ice-cold PBS, homogenization in IP buffer (20mM HEPES [pH 7.4], 150 mM KCl, 2mM EDTA, 1mM dithiolthreitol 0.1% (v/v) Triton X-100, 10 Mm β-glycerophosphate, 1 mM Na3VO4, 1 mM NaF, 5% glycerol, protease inhibitors [Complete®, Roche Molecular Biochemicals]) with pestles (Kontes) fitting 1.5ml microfuge tubes, pelleting cell debris at 1000g for 5 min, and analysis of protein content (Bradford). 50 μg of fresh extracts were used for IP of exogenous epitope tagged proteins (125 μg for endogenous protein IPs). Extracts were first cleared of non-specific binders with application to 15 μl of protein G Sepharose (Amersham) for 1 hr. Separately, 10 μl of protein G Sepharose was equilibrated with the indicated IP antibody for 30 min followed by the application of 50 μg of BSA (125 μg for endogenous IPs) for another 30 min to further reduce non-specific binding. Cleared extracts were then combined with antibody-charged protein G Sepharose for 2.5 hrs (2-6 hrs for endogenous IPs) followed by 3 brief washes in 750 μl of IP buffer. SDS PAGE sample buffer was added to IP resin and precipitated complexes analyzed by western blot. IP of endogenous proteins required using antibodies of alternate species for the pull-down vs. western blotting, to avoid background from the IP antibody otherwise obscuring western blot signals of the co-precipitated protein. Equal concentrations of control antibodies (rabbit anti-actin or mouse anti-HA) were used for comparison to relevant experimental IP antibodies.
Commercial antibodies used for immunoprecipitation and western blotting [W]: Rabbit anti-myc (Santa Cruz Biotechnology, IP: 0.6μg, W: 1:10,000); Mouse anti-V5 (Invitrogen, IP: 0.5μg, W: 1:20,000); rabbit anti-actin (Sigma, IP: 1ug, W: 1:5000) goat anti-Chk1 (Abcam, W: 1:7,500); rabbit anti-Chk1S317P (Abcam, W: 1:7,500); rabbit anti-Chk2T68P (Cell Signaling Technology, W: 1:2000); mouse anti-ORC2 (Calbiochem, IP: 1 μg, W: 1:2000), rabbit anti-MCM2 (Abcam, IP: 2 μg, W: 1:5000); anti-Polδ (Abcam, IP: 1 μg W: 1:5000); mouse anti-Polε (Abcam, IP: 1 μg, W: 3 mg/ml); mouse anti-RPA34 ([RPA34-19] Abcam, W: 1:1000); rabbit anti-RPA70 (Abcam, W: 1;6000).
Immunofluorescent Microscopy
HEK293 cells thymidine-nocodazole synchronized to S-phase were fixed for 15 min in 4% paraformaldehyde, permeabilized, immuno-stained and treated with 4’,6-diamidino-2-phenylindole (DAPI) as previously described 9. All experiments examined endogenous protein localization with the following antibodies: Guinea pig anti-hTIM, rabbit anti-hTipin2, rabbit anti-RPA70, mouse anti-RPA34 and rabbit anti-actin were used at 1:500, 1:1000, 1:200, 1:100, and 1:50, respectively. Secondary antibodies (Molecular Probes) included Alexafluor594-conjugated goat anti-guinea pig, Alexafluor488 donkey anti-rabbit and Alexafluor594 donkey anti-mouse (1:1,000).
Localization of DNA replication sites relative to TIM, Tipin and RPA70 was visualized under confocal microscopy (Pathology Core, Children's Hospital of Philadelphia) after processing HEK293 cells using the BrdU Labeling and Detection Kit (Roche) with empirically-derived modifications to the manufacturer's protocol: After 10 min of BrdU labeling (100 μM), cells were washed in PBS, fixed for 15 min in 4% paraformaldehyde, and blocked/permeabilized for 30 min. Antigen retrieval of BrdU-labeled DNA was performed in 100 mM citric acid (pH 6.0) at 89°C for 10 min followed by a 20 min cooling period. Primary antibodies were diluted in mouse anti-BrdU/nuclease cocktail followed by secondary antibody application and DAPI staining as described above. HEK293 cells were examined 24 hrs after release from 2 mM thymidine block to obtain a spread of cells in S-phase stages. Cells displaying a BrdU staining pattern indicative of mid-S-phase 41 were chosen for analysis and comparison.
BrdU Incorporation Rate Measurements
HEK293 cells expressing either control or Tim shRNA in a tetracycline-dependent manner were grown for 48 hours −/+ tetracycline (96 well platform). Cells were then grown in the presence of BrdU for 3, 6, 9, or 12 hours (BrdU Cell Proliferation Assay, Oncogen™). Cells were processed according manufacturer's protocol and BrdU incorporation relative to viable cell density was measured colormetrically (readings at 450nm - 540nm were taken, effectively controlling for any effect of shRNA expression on cell viability). Each trial evaluated 6 samples for each time point. Incorporation over time for each trial was determined by least squares regression analysis of each cell line and treatment (Microsoft Excel). These slopes were normalized relative to the – tetracycline condition for each cell line. For Table 1, values from four trials were averaged and analyzed statistically.
Cellular Responses to UV Irradiation and H2O2
Cell growth following treatment with UV irradiation was assessed in either untransfected NIH3T3 cells, or those stably transfected to express control or Tipin shRNA. Cells plated at 1×104 cells/well, were maintained in normal media or that containing 1 μg/ml tetracycline for 48 hours. Cultures received either sham or UV treatment (cells exposed to 10 J/m2 at 254 nm irradiation followed by media replacement). Cell density was analyzed by MTT assay (3-(4,5-dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide) (ATCC) in separate cultures (n=24) at the time of UV or sham treatment, and 20 hours later. Cell density in the presence or absence of tetracycline was compared for each treatment and for each cell line. Chk1S317 phosphorylation following UV and H2O2 treatment was evaluated in NIH3T3 control and Tipin shRNA-expressing cells. Thirty minutes after UV, 200 μM H2O2 or sham treatment, protein extracts were prepared in IP buffer and 10 μg analyzed with rabbit anti-Chk1S317P antibody. Blots were then stripped and re-probed with anti-Chk1 antibody.
Supplementary Material
ACKNOWLEDEMENTS
Technical assistance of Julia Brown and David Tompkins is greatly appreciated. These studies were supported by CA39926 (B.S.E.) from the NIH, and the Florence R.C. Murray Program (Children's Hospital of Philadelphia) (A.L.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–13. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 2.Takumi T, Nagamine Y, Miyake S, Matsubara C, Taguchi K, Takekida S, Sakakida Y, Nishikawa K, Kishimoto T, Niwa S, Okumura K, Okamura H. A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cells. 1999;4:67–75. doi: 10.1046/j.1365-2443.1999.00238.x. [DOI] [PubMed] [Google Scholar]
- 3.Zylka MJ, Shearman LP, Levine JD, Jin X, Weaver DR, Reppert SM. Molecular analysis of mammalian timeless. Neuron. 1998;21:1115–22. doi: 10.1016/s0896-6273(00)80628-5. [DOI] [PubMed] [Google Scholar]
- 4.Myers MP, Wager-Smith K, Wesley CS, Young MW, Sehgal A. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science. 1995;270:805–8. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 5.Gotter AL. A Timeless debate: resolving TIM's noncircadian roles with possible clock function. Neuroreport. 2006;17:1229–33. doi: 10.1097/01.wnr.0000233092.90160.92. [DOI] [PubMed] [Google Scholar]
- 6.Gotter AL, Manganaro T, Weaver DR, Kolakowski LF, Jr., Possidente B, Sriram S, MacLaughlin DT, Reppert SM. A time-less function for mouse timeless. Nat Neurosci. 2000;3:755–6. doi: 10.1038/77653. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Stuart RO, Qiao J, Pavlova A, Bush KT, Pohl M, Sakurai H, Nigam SK. A role for Timeless in epithelial morphogenesis during kidney development. Proc Natl Acad Sci U S A. 2000;97:10038–43. doi: 10.1073/pnas.97.18.10038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiao J, Li C, Zhu NL, Borok Z, Minoo P. Timeless in lung morphogenesis. Dev Dyn. 2003;228:82–94. doi: 10.1002/dvdy.10346. [DOI] [PubMed] [Google Scholar]
- 9.Gotter AL. Tipin, a novel timeless-interacting protein, is developmentally co-expressed with timeless and disrupts its self-association. J Mol Biol. 2003;331:167–76. doi: 10.1016/s0022-2836(03)00633-8. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, Newitt R, Aebersold R, Boone C, Brown GW, Hieter P. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15:1736–45. doi: 10.1091/mbc.E03-08-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noguchi E, Noguchi C, McDonald WH, Yates JR, 3rd, Russell P. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol Cell Biol. 2004;24:8342–55. doi: 10.1128/MCB.24.19.8342-8355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 14.Dalgaard JZ, Klar AJ. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell. 2000;102:745–51. doi: 10.1016/s0092-8674(00)00063-5. [DOI] [PubMed] [Google Scholar]
- 15.Lee BS, Grewal SI, Klar AJ. Biochemical interactions between proteins and mat1 cis-acting sequences required for imprinting in fission yeast. Mol Cell Biol. 2004;24:9813–22. doi: 10.1128/MCB.24.22.9813-9822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krings G, Bastia D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 2004;101:14085–90. doi: 10.1073/pnas.0406037101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 18.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–83. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 19.Nedelcheva MN, Roguev A, Dolapchiev LB, Shevchenko A, Taskov HB, Shevchenko A, Stewart AF, Stoynov SS. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J Mol Biol. 2005;347:509–21. doi: 10.1016/j.jmb.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 20.Noguchi E, Noguchi C, Du LL, Russell P. Swi1 prevents replication fork collapse and controls checkpoint kinase Cds1. Mol Cell Biol. 2003;23:7861–74. doi: 10.1128/MCB.23.21.7861-7874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–52. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lupardus PJ, Byun T, Yee MC, Hekmat-Nejad M, Cimprich KA. A requirement for replication in activation of the ATR-dependent DNA damage checkpoint. Genes Dev. 2002;16:2327–32. doi: 10.1101/gad.1013502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tercero JA, Longhese MP, Diffley JF. A central role for DNA replication forks in checkpoint activation and response. Mol Cell. 2003;11:1323–36. doi: 10.1016/s1097-2765(03)00169-2. [DOI] [PubMed] [Google Scholar]
- 24.Ward IM, Minn K, Chen J. UV-induced ataxia-telangiectasia-mutated and Rad3-related (ATR) activation requires replication stress. J Biol Chem. 2004;279:9677–80. doi: 10.1074/jbc.C300554200. [DOI] [PubMed] [Google Scholar]
- 25.Foss EJ. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics. 2001;157:567–77. doi: 10.1093/genetics/157.2.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Neill BM, Hanway D, Winzeler EA, Romesberg FE. Coordinated functions of WSS1, PSY2 and TOF1 in the DNA damage response. Nucleic Acids Res. 2004;32:6519–30. doi: 10.1093/nar/gkh994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sommariva E, Pellny TK, Karahan N, Kumar S, Huberman JA, Dalgaard JZ. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol Cell Biol. 2005;25:2770–2784. doi: 10.1128/MCB.25.7.2770-2784.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suter B, Tong A, Chang M, Yu L, Brown GW, Boone C, Rine J. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2004;167:579–91. doi: 10.1534/genetics.103.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warren CD, Eckley DM, Lee MS, Hanna JS, Hughes A, Peyser B, Jie C, Irizarry R, Spencer FA. S-phase checkpoint genes safegard high-fidelity sister chromatid cohesion. Mol Cell Biol. 2004;15:1724–1735. doi: 10.1091/mbc.E03-09-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marston AL, Tham WH, Shah H, Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–70. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- 31.Xu H, Boone C, Klein HL. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol Cell Biol. 2004;24:7082–90. doi: 10.1128/MCB.24.16.7082-7090.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan RC, Chan A, Jeon M, Wu TF, Pasqualone D, Rougvie AE, Meyer BJ. Chromosome cohesion is regulated by a clock gene paralogue TIM-1. Nature. 2003;423:1002–9. doi: 10.1038/nature01697. [DOI] [PubMed] [Google Scholar]
- 33.de Laat WL, Jaspers NG, Hoeijmakers JH. Molecular mechanism of nucleotide excision repair. Genes Dev. 1999;13:768–85. doi: 10.1101/gad.13.7.768. [DOI] [PubMed] [Google Scholar]
- 34.Wold MS. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 35.Chini CC, Chen J. Human claspin is required for replication checkpoint control. J Biol Chem. 2003;278:30057–62. doi: 10.1074/jbc.M301136200. [DOI] [PubMed] [Google Scholar]
- 36.Ünsal-Kaçmaz K, Mullen TE, Kaufmann WK, Sancar A. Coupling of human circadian and cell cycles by the timeless protein. Mol Cell Biol. 2005;25:3109–16. doi: 10.1128/MCB.25.8.3109-3116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita M, Yamada C, Tsurumi T, Hanaoka F, Matsuzawa K, Inagaki M. Cell cycle- and chromatin binding state-dependent phosphorylation of human MCM heterohexameric complexes. A role for cdc2 kinase. J Biol Chem. 1998;273:17095–101. doi: 10.1074/jbc.273.27.17095. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Lu X, Peterson CA, Legerski RJ. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol. 1995;15:5396–5402. doi: 10.1128/mcb.15.10.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 41.Dimitrova DS, Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J Cell Sci. 2002;115:4037–51. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- 42.Melo J, Toczyski D. A unified view of the DNA-damage checkpoint. Curr Opin Cell Biol. 2002;14:237–45. doi: 10.1016/s0955-0674(02)00312-5. [DOI] [PubMed] [Google Scholar]
- 43.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 44.Zhao H, Piwnica-Worms H. ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol. 2001;21:4129–39. doi: 10.1128/MCB.21.13.4129-4139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang ME, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell. 2005;17:709–20. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci U S A. 2003;100:11529–34. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shackelford RE, Kaufmann WK, Paules RS. Oxidative stress and cell cycle checkpoint function. Free Radic Biol Med. 2000;28:1387–404. doi: 10.1016/s0891-5849(00)00224-0. [DOI] [PubMed] [Google Scholar]
- 48.Helt CE, Cliby WA, Keng PC, Bambara RA, O'Reilly MA. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J Biol Chem. 2005;280:1186–92. doi: 10.1074/jbc.M410873200. [DOI] [PubMed] [Google Scholar]
- 49.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–19. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes and Development. 2002;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 51.de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10:479–82. doi: 10.1016/s0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 52.Hopkins KM, Auerbach W, Wang XY, Hande MP, Hang H, Wolgemuth DJ, Joyner AL, Lieberman HB. Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol Cell Biol. 2004;24:7235–48. doi: 10.1128/MCB.24.16.7235-7248.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takai H, Tominaga K, Motoyama N, Minamishima YA, Nagahama H, Tsukiyama T, Ikeda K, Nakayama K, Nakanishi M, Nakayama K. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(−/−) mice. Genes Dev. 2000;14:1439–47. [PMC free article] [PubMed] [Google Scholar]
- 54.Barnes JW, Tischkau SA, Barnes JA, Mitchell JW, Burgoon PW, Hickok JR, Gillette MU. Requirement of mammalian Timeless for circadian rhythmicity. Science. 2003;302:439–42. doi: 10.1126/science.1086593. [DOI] [PubMed] [Google Scholar]
- 55.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 56.Hsu DS, Zhao X, Zhao S, Kazantsev A, Wang RP, Todo T, Wei YF, Sancar A. Putative human blue-light photoreceptors hCRY1 and hCRY2 are flavoproteins. Biochemistry. 1996;35:13871–7. doi: 10.1021/bi962209o. [DOI] [PubMed] [Google Scholar]
- 57.Özgür S, Sancar A. Purification and properties of human blue-light photoreceptor cryptochrome 2. Biochemistry. 2003;42:2926–32. doi: 10.1021/bi026963n. [DOI] [PubMed] [Google Scholar]
- 58.Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, Kyriacou CP, Hastings MH. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–15. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 59.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–4. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 60.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–30. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 61.Barr SM, Leung CG, Chang EE, Cimprich KA. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr Biol. 2003;13:1047–51. doi: 10.1016/s0960-9822(03)00376-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.