Abstract
Histone post-translational modifications (PTM) alter the chromatin architecture, generating ‘open’ and ‘closed’ states, and these structural changes can modulate gene expression under specific cellular conditions. While methylation and acetylation are the best-characterized histone PTMs, citrullination by the protein arginine deiminases (PADs) represents another important player in this process. In addition to “fine tuning” chromatin structure at specific loci, histone citrullination can also promote rapid global chromatin decondensation during the formation of extracellular traps (ETs) in immune cells. Recent studies now show that PAD4-mediated citrullination of histone H1 at promoter elements can also promote localized chromatin decondensation in stem cells, thus regulating the pluripotent state. These observations suggest that PAD-mediated histone deimination profoundly affects chromatin structure, possibly above and beyond that of other PTMs. Additionally, these recent findings further enhance our understanding of PAD biology and the important contributions that these enzymes play in development, health, and disease.
Keywords: cancer, chromatin, deiminase, gene regulation, histone, pluripotency, rheumatoid arthritis, stem-cells
Introduction
Protein citrullination is catalyzed by the protein arginine deiminase (PAD) family of enzymes (Fig. 1A)[1]. There are five human PADs (PADs 1,2,3,4, and 6), and PADs 1-4 are known to hydrolyze the side chain guanidinium of an arginine residue to form citrulline (PAD6 does not appear to be active)[2]. This PTM leads to a loss of positive charge and alters the hydrogen-bonding pattern afforded by an arginine residue, which can alter protein-protein and protein-nucleic acid interactions [3, 4]. Despite significant homology amongst the five family members, they are functionally non-redundant, which likely relates in part to their restricted tissue distribution [5]. The PADs were first recognized to play a role in human disease when antibodies that specifically target citrullinated proteins were discovered in rheumatoid arthritis (RA) patients [6]. These anti-citrullinated protein antibodies (ACPA) remain the most specific diagnostic for RA and can be detected on average four to five years before clinical symptoms appear. These data suggest that dysregulated PAD activity is involved is disease initiation [6]. Outside of RA, dysregulated PAD activity or expression appears to be a hallmark of inflammatory diseases such as systemic lupus erythematosus, ulcerative colitis, atherosclerosis, and potentially cancer [7-11]. Given these associations, PADs represent therapeutic targets for a range of disease states [12, 13]. Consistent with this statement is the fact that the PAD inhibitor Cl-amidine, as well as derivatives, have shown efficacy in preclinical models of RA, systemic lupus erythematosus, ulcerative colitis, atherosclerosis, and breast cancer [7,9-11].
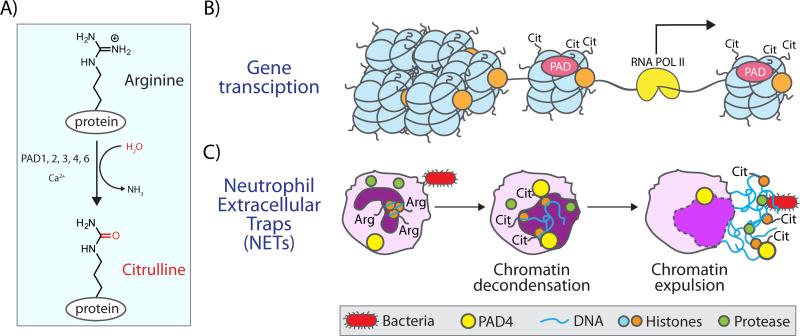
Figure 1.
Citrullination by PADs drives gene transcription and NET formation. A: Protein arginine deiminases (PADs) convert arginine to citrulline within proteins through a hydrolytic mechanism. B: PADs citrullinate core and linker histones, leading to changes to the chromatin architecture and altered gene transcription. C: Citrullination of histones in neutrophils by PAD4 leads to massive chromatin decondensation and the expulsion of DNA in response to bacterial challenge. This phenomenon produces neutrophil extracellular traps (NETs) through a process termed NETosis.
Citrullination and the role of PADs in immune system regulation and gene transcription
One well-established mechanism by which the PADs promote disease progression is by altering the antigenicity of target proteins such as fibrinogen and vimentin [14-17]. Antibodies to these citrullinated proteins then promote the formation of immune complexes, leading to an enhanced immune response and autoimmune disease [18, 19]. Another mechanism by which PADs likely promote disease progression is by altering the structure of target proteins, which, in turn, leads to changes in the organization and function of associated macromolecules. For example, recent studies have shown that PAD-mediated deimination of histones not only affects their tertiary structure but can also alter the global and local architecture of chromatin [4, 20, 21]. While direct links between PAD-mediated histone deimination and disease progression have yet to be definitively established, recent reports suggest that it is only a matter of time before these connection are made. For example, PAD2 and PAD4 have been shown to alter chromatin structure both globally during the formation of neutrophil extracellular traps (NETs), and locally at specific loci to regulate gene expression, and the associations between these two functions and disease progression are strong [9, 22-24].
Neutrophils, the major white blood cell, are among the first responders to an infection and play a key role in innate immunity. In response to a stimulus (e.g. lipopolysaccharide or LPS), a subset of neutrophils will undergo NET formation via the genome-wide citrullination of histones H1 and H3 [20, 25]. Modification of these proteins in cells undergoing NETosis leads to chromatin decondensation on a global scale, which initiates the unraveling of the chromatin and the consequent expulsion of DNA from the cell to form net-like structures that can ‘trap’ invading bacteria (Fig. 1). While NETosis is a defense mechanism against invading organisms, this response acts as a double-edged sword because it is thought to be a pro-inflammatory form of cell death. In chronic autoimmune diseases, this process is also aberrantly upregulated and likely plays an important role in the etiology of RA, systemic lupus erythematosus, ulcerative colitis, atherosclerosis, and even cancer [26-30]. In cancer and atherosclerosis, recent data indicate that aberrant NET formation promotes vascular inflammation, leading to thrombosis [9,30]. Recently, PAD2 was found to play a similar role in macrophages where its activity induces the formation of macrophage extracellular traps (METs), which leads inflammatory signaling in adipose tissue [31].
PAD-mediated histone deimination has also been demonstrated to “fine tune” chromatin structure to regulate gene transcription. Following up on findings from the Yamada group, which showed that histones are citrullinated by PAD4 [32], Coonrod and Allis, as well as the Kouzarides group, provided the first reports that histone citrullination could impact gene transcription [33, 34]. Specifically, they showed that citrullination of histones H3 and H4 was associated with decreased expression of genes under the control of the estrogen and thyroid receptors [33, 34]. Subsequently, Wang and colleagues reported that treatment of U2OS cells, an osteosarcoma cell line, with the pan-PAD inhibitor Cl-amidine leads to decreased PAD4 activity that was associated with the increased expression of p53 as well as several p53-dependent genes, including p21, PUMA, and OKL38. Ultimately, this treatment also results in the subsequent induction of apoptosis [35, 36]. Curiously, the forced overexpression of PAD4 in human Jurkat T cells also upregulates p53 activity, leading to cell cycle arrest and ultimately apoptosis [37]. The reason for this discrepancy may be due to cell line differences or forced overexpression of the enzyme. In addition to PAD4-mediated histone citrullination, PAD2 citrullinates histone H3 at Arg26 and this PTM induces the transcription of more than 200 genes under the control of the estrogen receptor [38]. In another study, citrullination of histone H3 induces the expulsion of heterochromatin protein 1α (HP1α), thereby creating an open chromatin state that modulates the expression of TNFα and IL8 [21]. In total, these studies demonstrate the important roles that histone citrullination plays in gene regulation in models of human disease.
PAD4 expression in stem cells drives the upregulation of pluripotency specific transcription factors
Adding to these roles, Christophorou et al. now report that the citrullination of histone H1 contributes to the epigenetic control of gene transcription and plays a key role in the maintenance of pluripotency [4]. This finding counters the prevailing view that modifications to histones H2A, H2B, H3 and H4, which make up the core nucleosomal particle, are primarily responsible for regulating gene transcription – modification of these proteins, via citrullination or lysine acetylation, is generally thought to weaken interactions between nucleosomal particles and DNA via charge neutralization. Although it is not known how histone H1 citrullination controls gene expression and contributes to the pluripotent state, this report provides important new insight into how PADs regulate chromatin architecture at a global level.
Pluripotent stem cells can differentiate along multiple cell lineages and adopt the characteristics of any cell in the body [39]. Such cells can either be derived from embryos (embryonic stem cells or ES cells) or, alternatively, these cells can be induced to form induced Pluripotent Stem cells (iPS). This technology introduces pluripotency factors, such as Oct4 (Pou5f1), Sox2, cMyc, and Klf4 [40], into differentiated cells thereby reprogramming the cells back to a pluripotent state. As a part of this process, the chromatin architecture is altered to facilitate the proper expression of specific genes, often transcription factors, which are required to maintain pluripotency. Given the known roles of the PADs in chromatin remodeling, Kouzarides and colleagues recently tested the hypothesis that PAD4-mediated histone citrullination may regulate pluripotency by modifying chromatin structure.
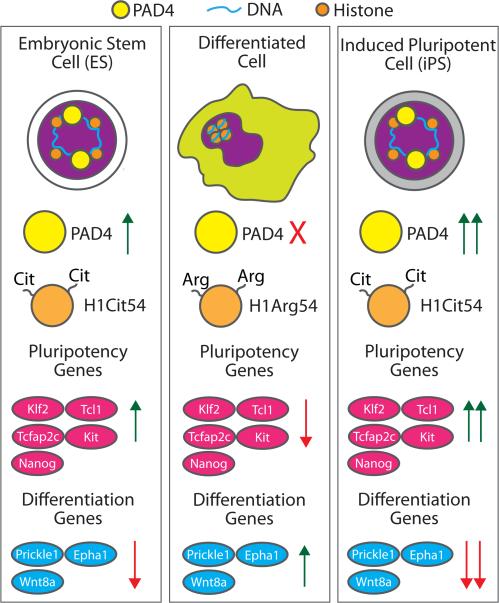
Initially, the authors examined the mRNA levels of PADs 1-4 in ES and iPS cells and showed that PAD4 mRNA levels are elevated in both mouse ES cells (ES Oct4-GIP) and reprogrammed NSO4G iPS cells, whereas they are decreased in NSO4G cells, a committed neural stem cell line. PAD4 expression also strongly correlated with the expression of genes that promote Nanog, Klf2, Tcl1, Tcfap2c, and Kit. By contrast, the levels of the PADs1-3 transcripts were variable between the different cell lines, leading the authors to focus on the role of PAD4 in stem cell biology. To examine whether the increased Nanog expression observed in PAD4 overexpressing cells was related to the histone deiminating activity of this enzyme, the authors evaluated the levels of citrullinated H3 and showed that increased levels of this PTM are correlative with higher levels of the pluripotency genes Klf2, Tcl1, Tcfap2C, and Kit. In total, these data suggest that histone H3 citrullination promotes a more open chromatin state that is permissive to the transcription of pluripotency genes. Importantly, PAD4 activity appears to be dependent on Nanog levels because, in its absence, histone H3 citrullination is also reduced. To further demonstrate that PAD4 activity is required to generate pluripotent stem cells, the authors treated mouse ES cells with the pan-PAD inhibitor Cl-amidine [41] and showed that decreased histone H3 citrullination was correlated with the decreased expression of Nanog, Tcl1, and Klf5. Further verifying the importance of PAD4 in maintaining pluripotency, Cl-amidine treatment also increased the expression of several differentiation genes, including Epha1, Prickle1, and Wnt8a. Given that Cl-amidine inhibits all of the PADs, the authors also used TDFA, a derivative that shows improved selectivity toward PAD4 [42], to show that inhibition of PAD4 activity reduces the number of pluripotent cells during early embryogenesis.
Citrullination of histone H1 drives chromatin decondensation
Having connected PAD4-mediated citrullination to pluripotency, Christophorou et al. then set out to identify PAD4 targets, which, upon citrullination, may regulate the pluripotent state. Using stable isotope labelling of amino acids in cell culture (SILAC), a number of nuclear proteins were found to be citrullinated in mouse ES cells, including Atrx, Dnmt3b, Trim28, and histone H1. Importantly, all of these proteins have been previously identified as playing a role in controlling pluripotency [43-46].
Given the well-established role of histone H1 in regulating chromatin structure, Christophorou et al. then focused on H1 for the rest of the study. Subsequent MS studies revealed that the histone H1 variants, H1.2, H1.3, H1.4, and H1.5 were citrullinated and that histone H1.2 was citrullinated at Arg54 (H1R54Cit), a residue that is conserved amongst the histone H1 variants and is present within the central DNA binding domain. Importantly, they also demonstrated that, upon mutation of this residue to alanine, histone H1 is released from chromatin, leading to the adoption of a more open state, which is consistent with other recent findings [22]. Additionally, the researchers found that inhibition of PAD4 expression or activity also decreases histone citrullination and generates a more compact chromatin state, which correlates with the down-regulation of pluripotency genes and the up-regulation of differentiation genes. By contrast, they found that treatment of permeabilized C2C12 cells with wild type PAD4 leads to nuclear swelling and chromatin decondensation, as demonstrated by an increased susceptibility to micrococcal nuclease digestion. In total, these studies show that PAD4 maintains stem cell pluripotency by promoting an open chromatin architecture via citrullination of histone H1.
Conclusions and outlook
These studies further our understanding of the role that PADs play in normal human physiology and demonstrate that, like other histone modifications (e.g. methylation and acetylation), protein citrullination plays a key role in regulating many chromatin templated activities, including gene regulation and chromatin decondensation. Whether this PTM plays a role in other aspects of chromatin biology (e.g. DNA replication and repair) remains an open question. Additionally, unlike methylation and acetylation, which are reversible histone modifications, enzymes that convert citrullinated residues back into arginine have not yet been identified. Given the clear role of histone citrullination in gene activation, it seems unlikely that the citrullination mark is irreversible in the context of chromatin, and several strategies can be envisioned to maintain the proper balance between arginine and citrulline. For example, it is possible that citrullinated histones are ejected from the nucleosome and replaced by non-citrullinated histone variants. Alternatively, it is also possible that unidentified ‘decitrullinase’ enzymes exist to convert citrulline back to arginine. There is chemical logic to the latter possibility because in the Urea Cycle, the free amino acids are readily interconverted. If decitrullinases exist, they would add another layer of complexity to chromatin biology.
A key remaining question is whether other PADs help regulate pluripotency and/or function at different stages of development. This question arises because Christophorou et al. showed that PADs 1, 2, and 3 were also expressed in pluripotent stem cells. However, high expression of these three enzymes was not observed in both ES and iPS cells, suggesting that the different PADs function at different stages of development. This hypothesis could be tested by knocking down each of the PAD isozymes by RNAi during ES cell differentiation to test how isozyme specific-depletion affects the balance between pluripotency and the differentiated state. These experiments will be especially important for PAD2, as this enzyme also enters the nucleus and citrullinates histones. Since PADs 1-4 are overexpressed in several forms of cancer, it is also possible that this enzyme family may facilitate the increased transcription of growth and migration promoting genes.
In summary, Kouzarides and colleagues uncover a fascinating new aspect of PAD biology that has profound implications for understanding how histone modifications modulate chromatin structure, and how these changes affect gene transcription, pluripotency, and perhaps other chromatin templated processes. The role of PAD4 in embryogenesis also has important implications for the development of PAD-targeted therapeutics because PAD inhibitors may affect cell fate decisions during embryogenesis. Whether this is the case is unknown, but will need to be accounted for as PAD inhibitors move into the clinic.
Figure 2.
PAD4 regulates gene expression during pluripotency and differentiation. Expression of PAD4 is increased in embryonic stem cells (ES) and induced pluripotent stem cells (iPS), but is virtually undetectable in differentiated mouse neural stem-cells. PAD4 expression correlates with increased expression of pluripotency specific proteins and a loss of cellular differentiation proteins. These changes are thought to arise by the PAD4 catalyzed citrullination of histone H1 at Arg54 (H1Arg54 → H1Cit54), which promotes localized chromatin decondensation, likely through the loss of histone H1 binding to DNA.
Acknowledgements
This work was supported by TSRI and in part by NIH grant GM079357 (PRT), as well as an Era of Hope Scholar Award (W871XWH-07-0372) (SAC).
Abbreviations
- ACPA
anti-citrullinated protein antibodies
- Cl-amidine
N-alpha-benzoyl-N5-(2-chloro-1-iminoethyl)-L-ornithine amide, which is a pan-PAD inhibitor
- ES
embryonic stem cell
- H1R54Cit
arginine at position 54 in histone H1 is converted to citrulline
- iPS
induced pluripotent stem cell
- LPS
lipopolysaccharide
- MET
macrophage extracellular trap
- NET
neutrophil extracellular trap
- PAD
protein arginine deiminase or peptidylarginine deiminase
- PTM
post-translational modification
- R54A
arginine 54 in histone H1 is mutated to an alanine
- RA
rheumatoid arthritis
- SILAC
stable isotope labelling of amino acids in cell culture
- TDFA
Threonine-Aspartate-N-alpha-benzoyl-N5-(2-fluoro-1-iminoethyl)-L-ornithine amide, which is a PAD4 specific inhibitor
References
- 1.Bicker KL, Thompson PR. The protein arginine deiminases: Structure, function, inhibition, and disease. Biopolymers. 2013;99:155–63. doi: 10.1002/bip.22127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raijmakers R, Zendman AJ, Egberts WV, Vossenaar ER, et al. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol. 2007;367:1118–29. doi: 10.1016/j.jmb.2007.01.054. [DOI] [PubMed] [Google Scholar]
- 3.Slack JL, Jones LE, Jr., Bhatia MM, Thompson PR. Autodeimination of protein arginine deiminase 4 alters protein-protein interactions but not activity. Biochemistry. 2011;50:3997–4010. doi: 10.1021/bi200309e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christophorou MA, Castelo-Branco G, Halley-Stott RP, Oliveira CS, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507:104–8. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohanan S, Cherrington BD, Horibata S, McElwee JL, et al. Potential role of peptidylarginine deiminase enzymes and protein citrullination in cancer pathogenesis. Biochem Res Int. 2012;2012:895343. doi: 10.1155/2012/895343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, et al. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101:273–81. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight JS, Zhao W, Luo W, Subramanian V, et al. Peptidylarginine deiminase inhibition is immunomodulatory and vasculoprotective in murine lupus. J Clin Invest. 2013;123:2981–93. doi: 10.1172/JCI67390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen CC, Isomoto H, Narumi Y, Sato K, et al. Haplotypes of PADI4 susceptible to rheumatoid arthritis are also associated with ulcerative colitis in the Japanese population. Clin Immunol. 2008;126:165–71. doi: 10.1016/j.clim.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Knight JS, Luo W, O'Dell AA, Yalavarthi S, et al. Peptidylarginine deiminase inhibition reduces vascular damage and modulates innate immune responses in murine models of atherosclerosis. Circ Res. 2014;114:947–56. doi: 10.1161/CIRCRESAHA.114.303312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chumanevich AA, Causey CP, Knuckley BA, Jones JE, et al. Suppression of colitis in mice by Cl-amidine: a novel peptidylarginine deiminase inhibitor. Am J Physiol Gastrointest Liver Physiol. 2011;300:G929–38. doi: 10.1152/ajpgi.00435.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McElwee JL, Mohanan S, Griffith OL, Breuer HC, et al. Identification of PADI2 as a potential breast cancer biomarker and therapeutic target. BMC Cancer. 2012;12:500. doi: 10.1186/1471-2407-12-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musse AA, Li Z, Ackerley CA, Bienzle D, et al. Peptidylarginine deiminase 2 (PAD2) overexpression in transgenic mice leads to myelin loss in the central nervous system. Dis Model Mech. 2008;1:229–40. doi: 10.1242/dmm.000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones JE, Causey CP, Knuckley B, Slack-Noyes JL, et al. Protein arginine deiminase 4 (PAD4): Current understanding and future therapeutic potential. Curr Opin Drug Discov Devel. 2009;12:616–27. [PMC free article] [PubMed] [Google Scholar]
- 14.Fan LY, He DY, Wang Q, Zong M, et al. Citrullinated vimentin stimulates proliferation, pro-inflammatory cytokine secretion, and PADI4 and RANKL expression of fibroblast-like synoviocytes in rheumatoid arthritis. Scand J Rheumatol. 2012;41:354–8. doi: 10.3109/03009742.2012.670263. [DOI] [PubMed] [Google Scholar]
- 15.Bay-Jensen AC, Karsdal MA, Vassiliadis E, Wichuk S, et al. Circulating citrullinated vimentin fragments reflect disease burden in ankylosing spondylitis and have prognostic capacity for radiographic progression. Arthritis Rheum. 2013;65:972–80. doi: 10.1002/art.37843. [DOI] [PubMed] [Google Scholar]
- 16.Okumura N, Haneishi A, Terasawa F. Citrullinated fibrinogen shows defects in FPA and FPB release and fibrin polymerization catalyzed by thrombin. Clin Chim Acta. 2009;401:119–23. doi: 10.1016/j.cca.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Shelef MA, Bennin DA, Mosher DF, Huttenlocher A. Citrullination of fibronectin modulates synovial fibroblast behavior. Arthritis Res Ther. 2012;14:R240. doi: 10.1186/ar4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harre U, Georgess D, Bang H, Bozec A, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest. 2012;122:1791–802. doi: 10.1172/JCI60975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sokolove J, Zhao X, Chandra PE, Robinson WH. Immune complexes containing citrullinated fibrinogen costimulate macrophages via Toll-like receptor 4 and Fcgamma receptor. Arthritis Rheum. 2011;63:53–62. doi: 10.1002/art.30081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leshner M, Wang S, Lewis C, Zheng H, et al. PAD4 mediated histone hypercitrullination induces heterochromatin decondensation and chromatin unfolding to form neutrophil extracellular trap-like structures. Front Immunol. 2012;3:307. doi: 10.3389/fimmu.2012.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma P, Azebi S, England P, Christensen T, et al. Citrullination of histone H3 interferes with HP1-mediated transcriptional repression. PLoS Genet. 2012;8:e1002934. doi: 10.1371/journal.pgen.1002934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dwivedi N, Neeli I, Schall N, Wan H, et al. Deimination of linker histones links neutrophil extracellular trap release with autoantibodies in systemic autoimmunity. FASEB J. 2014 doi: 10.1096/fj.13-247254. in press, doi: 10.1096/fj.13-247254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li P, Li M, Lindberg MR, Kennett MJ, et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. The Journal of experimental medicine. 2010;207:1853–62. doi: 10.1084/jem.20100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Li M, Stadler S, Correll S, et al. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J Cell Biol. 2009;184:205–13. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dwivedi N, Radic M. Citrullination of autoantigens implicates NETosis in the induction of autoimmunity. Ann Rheum Dis. 2014;73:483–91. doi: 10.1136/annrheumdis-2013-203844. [DOI] [PubMed] [Google Scholar]
- 26.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, Gizinski A, et al. NETs are a source of citrullinated autoantigens and stimulate inflammatory responses in rheumatoid arthritis. Sci Transl Med. 2013;5:178ra40. doi: 10.1126/scitranslmed.3005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakkim A, Furnrohr BG, Amann K, Laube B, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci USA. 2010;107:9813–8. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savchenko AS, Inoue A, Ohashi R, Jiang S, et al. Long pentraxin 3 (PTX3) expression and release by neutrophils in vitro and in ulcerative colitis. Pathol Int. 2011;61:290–7. doi: 10.1111/j.1440-1827.2011.02651.x. [DOI] [PubMed] [Google Scholar]
- 29.Borissoff JI, Joosen IA, Versteylen MO, Brill A, et al. Elevated levels of circulating DNA and chromatin are independently associated with severe coronary atherosclerosis and a prothrombotic state. Arterioscler Thromb Vasc Biol. 2013;33:2032–40. doi: 10.1161/ATVBAHA.113.301627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demers M, Krause DS, Schatzberg D, Martinod K, et al. Cancers predispose neutrophils to release extracellular DNA traps that contribute to cancer-associated thrombosis. Proc Natl Acad Sci USA. 2012;109:13076–81. doi: 10.1073/pnas.1200419109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohanan S, Horibata S, McElwee JL, Dannenberg AJ, et al. Identification of macrophage extracellular trap-like structures in mammary gland adipose tissue: a preliminary study. Front Immunol. 2013;4:67. doi: 10.3389/fimmu.2013.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagiwara T, Nakashima K, Hirano H, Senshu T, et al. Deimination of arginine residues in nucleophosmin/B23 and histones in HL-60 granulocytes. Biochem Biophys Res Commun. 2002;290:979–83. doi: 10.1006/bbrc.2001.6303. [DOI] [PubMed] [Google Scholar]
- 33.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, et al. Histone deimination antagonizes arginine methylation. Cell. 2004;118:545–53. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Wysocka J, Sayegh J, Lee YH, et al. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–83. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 35.Yao H, Li P, Venters BJ, Zheng S, et al. Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem. 2008;283:20060–8. doi: 10.1074/jbc.M802940200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li P, Yao H, Zhang Z, Li M, et al. Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol. 2008;28:4745–58. doi: 10.1128/MCB.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu GY, Liao YF, Chang WH, Liu CC, et al. Overexpression of peptidylarginine deiminase IV features in apoptosis of haematopoietic cells. Apoptosis. 2006;11:183–96. doi: 10.1007/s10495-006-3715-4. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Bolt M, Guertin MJ, Chen W, et al. Peptidylarginine deiminase 2-catalyzed histone H3 arginine 26 citrullination facilitates estrogen receptor alpha target gene activation. Proc Natl Acad Sci USA. 2012;109:13331–6. doi: 10.1073/pnas.1203280109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaspar-Maia A, Alajem A, Meshorer E, Ramalho-Santos M. Open chromatin in pluripotency and reprogramming. Nat Revi Mol Cell Biol. 2011;12:36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–7. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 41.Luo Y, Arita K, Bhatia M, Knuckley B, et al. Inhibitors and inactivators of protein arginine deiminase 4: functional and structural characterization. Biochemistry. 2006;45:11727–36. doi: 10.1021/bi061180d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones JE, Slack JL, Fang P, Zhang X, et al. Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem Biol. 2012;7:160–5. doi: 10.1021/cb200258q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong LH, McGhie JD, Sim M, Anderson MA, et al. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 2010;20:351–60. doi: 10.1101/gr.101477.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leitch HG, McEwen KR, Turp A, Encheva V, et al. Naive pluripotency is associated with global DNA hypomethylation. Nat Struct Mol Biol. 2013;20:311–6. doi: 10.1038/nsmb.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu G, Kim J, Xu Q, Leng Y, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–48. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Cooke M, Panjwani S, Cao K, et al. Histone h1 depletion impairs embryonic stem cell differentiation. PLoS Genet. 2012;8:e1002691. doi: 10.1371/journal.pgen.1002691. [DOI] [PMC free article] [PubMed] [Google Scholar]