Abstract
American ginseng (Panax quinquefolius) is originally grown in North America. Due to price difference and supply shortage, American ginseng recently has been cultivated in northern China. Further, in the market, some Asian ginsengs are labeled as American ginseng. In this study, forty-three American ginseng samples cultivated in the USA, Canada or China were collected and 14 ginseng saponins were determined using HPLC. HPLC coupled with hierarchical cluster analysis and principal component analysis was developed to identify the species. Subsequently, an HPLC-linear discriminant analysis was established to discriminate cultivation regions of American ginseng. This method was successfully applied to identify the sources of 6 commercial American ginseng samples. Two of them were identified as Asian ginseng, while 4 others were identified as American ginseng, which were cultivated in the USA (3) and China (1). Our newly developed method can be used to identify American ginseng with different cultivation regions.
Keywords: Panax quinquefolius, Cultivation region identification, HPLC, Hierarchical cluster analysis, Principal component analysis, Linear discriminant analysis
1. Introduction
Botanicals have been a foundation of traditional medicines for thousands of years and continue to be considered valuable materials for the health maintenance and disease treatment of human beings [1]. Among commonly used medicinal plants, ginseng has a long history of use and today is still one of the most widely used botanicals from East to West [2]. The ginseng root, available in white or red, is conventionally used. White ginseng is prepared by air-drying after harvest, and red ginseng is prepared by a steaming or heating process [3]. Ginseng products are commercially available as raw roots, capsules, liquid extracts, and teas.
The genus of ginseng is Panax, meaning “cure all” in Greek. Panax ginseng C.A. Meyer (commonly referred to as Asian ginseng) and Panax quinquefolius L. (also known as American ginseng, which is native in the United States and Canada) are the two most recognized species around the world [2, 4]. Another species is Panax notoginseng (Burk.) F.H. Chen, which is mainly cultivated in China [5, 6]. While notoginseng has acquired a favorable reputation for treatment of blood disorders, including blood stasis, bleeding, and blood deficiency based on traditional Chinese medicine theory [7], the more commonly used American ginseng and Asian ginseng have been considered as agents to reduce stress, enhance immune function and treat several chronic diseases, partially due to the antioxidant activities [8, 9]. However, since Asian ginseng has a “warm” or “heat” property based on the traditional Chinese medicine theory, Asian ginseng is only suitable for a limited patient population [10, 11]. In contrast, since American ginseng has much less of “heat” property, it can be used by most people in different age groups [10].
Since the appearance of these three ginseng species is similar, a specific ginseng species can be misidentified unintentionally or for economic gains. Due to price difference and/or supply shortage, American ginseng recently has been cultivated in northern China [12]. Using the wrong ginseng root is not only an unethical practice, but also may induce unpredicted therapeutic outcome. In addition, American ginseng root cultivated in China may have different therapeutic activities compared to American ginseng grown in North America. This is because different cultivation areas of the ginseng can have a variation in its ginsenoside composition and profile [12].
The major active components of ginseng are ginsenosides, a diverse group of steroidal saponins, which reportedly interact with a myriad of targets, producing an array of pharmacological responses [2, 8]. In a previous report, Harkey et al. performed a study to analyze the variability in commercial ginseng preparations and observed significant discrepancies in ginsenoside concentration between analytical results and the label claims [13]. Over the past ten years, our group has performed a number of investigations of ginseng compound isolation and analysis, and introduced new analytical approaches [2, 5, 6, 14, 15].
In the present study, an integrated approach combining an HPLC assay with multivariate statistical methodologies was developed in order to identify the adulterated American ginseng samples and determine the geographical locations of the cultivation. Our method was validated with further collected ginseng samples. This approach has been used to identify the origins of commercial ginseng samples. Our data showed that the newly established method can be used to accurately and effectively distinguish any adulteration from American ginseng with different cultivation areas.
2. Materials and methods
2.1. Chemicals
Standards for 14 ginsenosides, including 7 protopanaxadiol-type saponins (ginsenosides Rb1, Rb2, Rb3, Rc, Rd, Rg3, Rh2) and 7 protopanaxatriol-type saponins (notoginsenoside R1, ginsenosides Rg1, Re, Rf, Rg2, 20R-Rg2, Rh1), were obtained from Indofine Chemical Company (Somerville, NJ) or Delta Information Center for Natural Organic Compounds (Xuancheng, AH, China). All standards were of biochemical-reagent grade and at least 95% pure. HPLC-grade methanol and acetonitrile were purchased from Sigma-Aldrich (St. Louis, MO). Mili-Q water was supplied by a water purification system (US Filter, Palm Desert, CA). All solvents and samples were filtered through 0.2 μm membrane filters before analysis.
2.2. Herbal materials
To develop an efficaciously discriminant method, in the first collection, the four year old roots of American ginseng, cultivated in Wisconsin, USA (15 samples, labeled as US1-01 to US1-15); Ontario, Canada (6 samples, labeled as CA1-1 to CA1-06); and Jilin, China (10 samples, labeled as CH1-01 to CH1-10) were collected after harvesting. These samples were identified by Dr. Chong-Zhi Wang (Tang Center for Herbal Medicine Research at University of Chicago) as Panax quinquefolius L. roots. In addition, four standard Asian ginseng (Panax ginseng) samples (PG01 to PG04) and two standard notoginseng (Panax notoginseng) samples (PN01 and PN02) were obtained from the National Institutes for Food and Drug Control (Beijing, China) and also identified by Dr. Chong-Zhi Wang as their respective authenticity.
To validate the developed method, we secondly collected 12 American ginseng samples from the three cultivated areas, including from the USA (5 samples, labeled as US2-01 to US2-05), Canada (3 samples, labeled as from CA2-01 to CA2-03), and China (4 samples, labeled as from CH2-01 to CH2-04). At the same time, six commercial samples (MK01-06) labeled as American ginseng were purchased from supermarkets. Voucher specimens were deposited in the Tang Center for Herbal Medicine Research at the University of Chicago.
2.3. Sample preparation
Before extraction, fresh samples were lyophilized for 72 h to remove water. Dried roots were ground and passed through a 40-mesh screen. Root powder (~ 500 mg) was extracted with methanol in a Soxhlet extractor for 12 hours. The extract solution was condensed under vacuum, transferred into a 50 mL volumetric flask and diluted to volume with methanol. The concentration of the solution was equivalent to 10 mg/mL of crude herb. All the extract solutions were filtered through Millex 0.2 μm nylon membrane syringe filters (Millipore Co., Bedford, MA) and stored at −20°C until HPLC analysis.
2.4. HPLC analysis
The HPLC system was a Waters 2965 instrument (Milford, MA), with a quaternary pump, automatic injector, a photodiode array detector (Model 996), and Waters Empower 2 software for peak identification and integration. The separation was carried out on a Prodigy ODS (2) column (5 μm, 250 × 3.2 mm I.D.) (Phenomenex, Torrance, CA) with a guard column (Phenomenex ProdigyTM, 3.0 × 4.0 mm I.D.). For HPLC analysis, a 20-μL sample was injected into the column and eluted at room temperature with a constant flow rate of 1.0 mL/min. Acetonitrile (solvent A) and water (solvent B) were used. Gradient elution started with 17.5% solvent A and 82.5% solvent B, changed to 22% A for 20 min, then changed to 26% A for 3 min and held for 19 min, changed to 36.5% A for 13 min, changed to 53.5% A for 9 min, changed to 71.5% A for 10 min, changed to 85% A for 2 min and held for 3 min. Finally, it was changed to 17.5% A for 5 min and held for 6 min to rebalance the separated column. The detection wavelength was set to 202 nm.
2.5. Calibration curves of ginsenoside standards
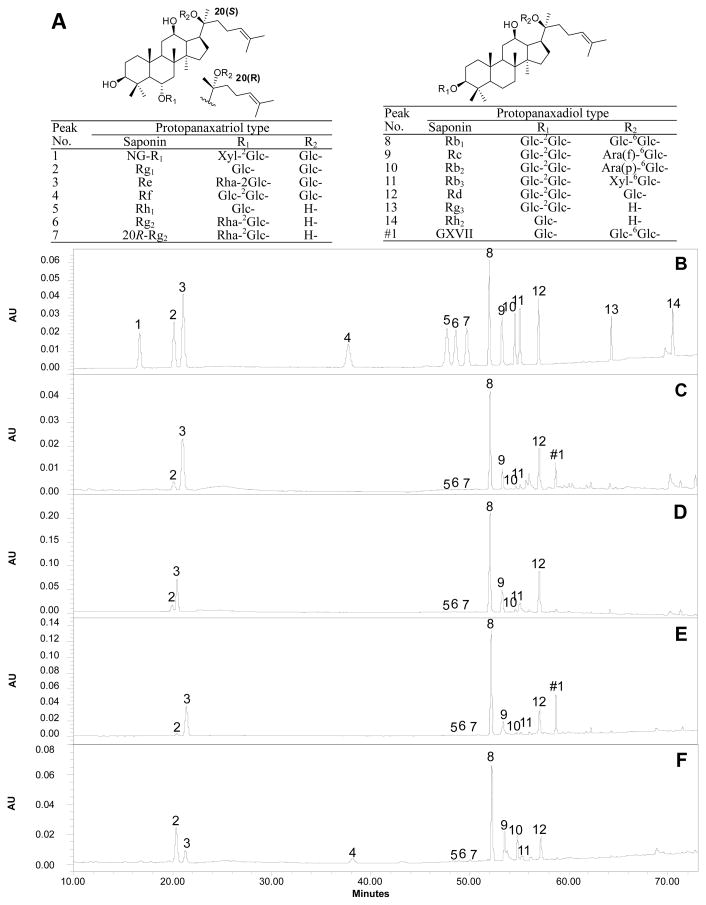
A stock solution of standards containing 14 ginsenosides (R1, Rg1, Re, Rf, Rg2, 20R-Rg2, Rh1, Rb1, Rc, Rb2, Rb3, Rd, Rg3 and Rh2) (Fig. 1A) in methanol was prepared and diluted to the concentrations of 10, 25, 50, 100 and 200 μg/mL. 20 μL of the mixed-standard solutions were injected in triplicate, and then the calibration curves were constructed by plotting the peak areas versus the amounts (mg) of each analyte. The contents (milligrams per gram herbs, mg/g) of 14 ginsenosides in these samples were calculated according to the standard curves of 14 ginsenosides.
Fig. 1.
HPLC analysis of ginseng samples. (A) Chemical structures of 14 tested ginseng saponins and GXVII (gypenoside XVII). Glc, β-D-glucopyranosyl; Xyl, β-D-xylopyranosyl; Rha, α-L-rhamnopyranosyl; Ara(f), α-L-arabinofuranosyl; Ara(p), α-L-arabinopyranosyl. Representative chromatograms of ginsenoside standards (B) and P. quinquefolius obtained from the USA (C), Canada (D) and China (E), and an adulterated sample (E).
2.6. Multivariate and data analysis
To identify the adulterated products and to distinguish the cultivation regions, three multivariate statistical methods, such as hierarchical cluster analysis (HCA), principal component analysis (PCA) and linear discriminant analysis (LDA), were performed with SPSS version 20.0 software (SPSS, Inc., Chicago, IL).
A dataset consisted of the tested saponin contents of 37 samples, including the initial collection of 31 American ginseng samples (US1-01 to US1-15, CA1-01 to CA1-06, and CH1-01 to CH1-14), four Asian ginseng samples (PG01-04) and two notoginseng samples (PN01-02), was used to generate the HCA dendrogram according to Ward’s method after standardized transformation into a range from 0 to 1. PCA was performed by applying a correlation matrix after by-step extracted pretreatment based on an Eigenvalue greater than 1. LDA of the above 31 American ginseng samples was performed by applying Wilk’s lambda method and a criterion of F value (Entry, 2; Removal, 1). To validate the developed method, a new dataset was generated with the second collection of 12 American ginseng samples.
This method has been used to characterize the authentication of six commercial samples (MK01-06). HCA was employed to identify adulteration of American ginseng. After excluded the adulteration, the grown places of the American ginseng samples were discriminated by LDA.
3. Results
3.1. Establishment of HPLC method for ginsenoside analysis
Compared to our previous study [15], 2 additional ginsenosides were determined, i.e. ginsenosides Rf and Rh2. In fact, notoginsenoside R1 and ginsenoside Rf were not detected in American ginseng roots; they are representative saponins in notoginseng (R1) and Asian ginseng (Rf). In this study, these two saponins were used to be marker compounds for the identification of a counterfeit.
Based on the chromatograms (Fig. 1B), these 14 saponins are divided into four groups: group 1, notoginsenoside R1, ginsenosides Rg1 and Re; group 2, ginsenosides Rf, Rg2, 20R-Rg2 and Rh1; group 3, ginsenosides Rb1, Rc, Rb2, Rb3, Rd; and group 4, ginsenosides Rg3 and Rh2. The saponins in groups 1, 3 and 4 were relatively easily separated. However, the saponins in group 2 were difficult to separate. As shown in Fig. 1B, ginsenoside Rf in group 2 was well-separated through an isocratic elution of 26% acetonitrile kept from 23 to 42 min, a baseline separation of other three ginsenosides (Rg2, 20R-Rg2 and Rh1) was achieved through a consequently increasing gradient elution from 26% to 36.5% of solvent A.
The linearity, regression and precision of 14 tested ginsenosides were performed according to our previous study [15]. The higher correlation coefficient values (R2 > 0.99) indicated good correlations between investigated saponin amounts and their peak areas within the test ranges (Supplemental Table 1).
Recovery experiments were conducted to determine the accuracy of the method for the quantification of the 14 ginsenosides. The recovered ginsenosides of an American ginseng root extract spiked at three levels of 10, 50, 100 μg/ml for 14 ginsenosides and were between 91.0~100.4% (n=3). The relative standard deviations (RSD) are less than 8.6% for any of these 14 ginsenosides.
3.2. Saponin composition in ginseng samples
The developed method was subsequently applied to simultaneously determine the contents of 14 ginsenosides in ginseng samples. Typical HPLC chromatograms of American ginseng obtained from three cultivation regions were shown in Fig. 1, including from the USA (Fig. 1C), Canada (Fig. 1D), and China (Fig. 1E). The major constituents of American ginseng are ginsenosides Rb1, Re and Rd, while notoginsenoside R1 and ginsenoside Rf were not detected. A new peak, which is labeled as #1 in Fig. 1, supposed as gypenoside XVII according to the previous study [5], was observed in samples from the USA and China. As shown in Table 1, the contents of Rb1, Re and Rd in American ginseng samples were 3.4–22.9 mg/g, 2.5–7.5 mg/g, and 1.5–12.0 mg/g, respectively.
Table 1.
Content of 14 ginseng saponins for investigated samples (mg/g)
| No. | R1 | Rg1 | Re | Rf | Rh1 | Rg2 | 20R-Rg2 | Rb1 | Rc | Rb2 | Rb3 | Rd | Rg3 | Rh2 | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| US1-01 | - | 0.88 | 7.08 | - | - | 0.21 | 0.09 | 22.88 | 1.60 | 1.01 | 2.74 | 6.58 | - | - | 43.06 |
| US1-02 | - | 1.97 | 5.31 | - | - | - | 0.05 | 12.85 | 3.36 | 0.64 | 0.72 | 5.11 | - | - | 30.02 |
| US1-03 | - | 0.67 | 2.70 | - | - | - | 0.01 | 3.42 | 1.68 | 0.21 | 0.28 | 2.48 | - | - | 11.45 |
| US1-04 | - | 0.92 | 4.73 | - | - | - | 0.04 | 12.02 | 3.62 | 0.51 | 0.58 | 4.31 | - | - | 26.72 |
| US1-05 | - | 0.88 | 7.36 | - | - | 0.22 | 0.12 | 22.68 | 3.39 | 1.18 | 1.57 | 6.30 | - | - | 43.70 |
| US1-06 | - | 1.54 | 6.23 | - | - | 0.14 | 0.10 | 13.24 | 4.31 | 0.66 | 3.33 | 8.20 | - | - | 37.74 |
| US1-07 | - | 0.99 | 4.50 | - | - | - | 0.03 | 10.56 | 3.01 | 0.48 | 0.64 | 6.52 | - | - | 26.73 |
| US1-08 | - | 2.86 | 5.05 | - | - | - | - | 23.59 | 2.73 | 0.52 | 0.37 | 9.01 | - | - | 44.13 |
| US1-09 | - | 1.31 | 7.54 | - | - | 0.20 | 0.11 | 17.39 | 4.87 | 0.77 | 0.99 | 7.45 | - | - | 40.62 |
| US1-10 | - | 0.78 | 4.27 | - | - | 0.26 | 0.04 | 15.08 | 2.84 | 0.80 | 2.00 | 8.34 | - | - | 34.42 |
| US1-11 | - | 2.78 | 4.99 | - | - | - | - | 23.63 | 2.69 | 0.51 | 3.86 | 8.85 | - | - | 47.31 |
| US1-12 | - | 0.72 | 4.28 | - | - | 0.25 | 0.03 | 15.25 | 2.98 | 0.68 | 1.87 | 8.33 | - | - | 34.38 |
| US1-13 | - | 1.02 | 4.62 | - | - | - | 0.05 | 11.29 | 3.24 | 0.45 | 0.50 | 6.75 | - | - | 27.92 |
| US1-14 | - | 1.30 | 7.54 | - | - | 0.20 | 0.11 | 17.05 | 4.52 | 0.73 | 0.98 | 6.78 | - | - | 39.20 |
| US1-15 | - | 1.78 | 5.24 | - | - | - | 0.08 | 21.39 | 0.93 | 0.65 | 1.22 | 3.71 | 0.26 | - | 35.26 |
| CH1-01 | - | 1.47 | 6.38 | - | - | - | 0.13 | 17.73 | 7.77 | 1.14 | 1.02 | 5.49 | - | - | 41.14 |
| CH1-02 | - | 0.64 | 4.84 | - | - | - | 0.03 | 11.69 | 4.89 | 0.61 | 0.49 | 4.66 | - | - | 27.85 |
| CH1-03 | - | 1.39 | 2.53 | - | - | - | - | 10.24 | 5.05 | 0.76 | 0.96 | 3.98 | - | - | 24.91 |
| CH1-04 | - | 3.34 | 3.73 | - | - | - | 0.08 | 8.97 | 3.11 | 0.51 | 0.48 | 1.88 | - | - | 22.09 |
| CH1-05 | - | 0.37 | 5.12 | - | - | - | 0.04 | 10.70 | 1.32 | 0.60 | 0.42 | 1.69 | 0.23 | - | 20.49 |
| CH1-06 | - | 0.42 | 3.46 | - | - | - | 0.04 | 11.47 | 1.58 | 0.35 | 0.48 | 1.68 | 0.25 | - | 19.73 |
| CH1-07 | - | 0.67 | 5.20 | - | - | - | 0.13 | 21.46 | 2.86 | 0.77 | 0.99 | 3.83 | 0.58 | - | 36.50 |
| CH1-08 | - | 3.45 | 3.67 | - | - | - | 0.07 | 9.11 | 4.84 | 0.58 | 1.15 | 2.10 | - | - | 24.97 |
| CH1-09 | - | 0.80 | 4.24 | - | - | - | 0.04 | 9.19 | 1.99 | 0.36 | 0.57 | 1.48 | 0.13 | 0.02 | 18.81 |
| CH1-10 | - | 0.55 | 5.07 | - | - | - | 0.08 | 10.69 | 1.43 | 0.36 | 0.55 | 1.45 | 0.17 | - | 20.35 |
| CA1-01 | - | 1.60 | 7.09 | - | - | - | 0.06 | 19.51 | 8.49 | 1.28 | 5.97 | 11.81 | - | - | 55.81 |
| CA1-02 | - | 0.47 | 3.87 | - | - | - | 0.01 | 10.60 | 1.36 | 0.17 | 4.80 | 2.08 | - | - | 23.37 |
| CA1-03 | - | 1.18 | 5.37 | - | - | - | 0.05 | 14.60 | 4.34 | 0.63 | 4.71 | 5.89 | - | - | 36.77 |
| CA1-04 | - | 2.49 | 7.32 | - | - | - | 0.08 | 19.73 | 8.69 | 1.32 | 5.93 | 12.04 | - | - | 57.58 |
| CA1-05 | - | 0.72 | 4.77 | - | - | - | 0.04 | 14.99 | 1.79 | 0.23 | 5.62 | 2.86 | - | - | 31.02 |
| CA1-06 | - | 0.83 | 4.61 | - | - | - | 0.05 | 10.42 | 2.06 | 0.27 | 1.96 | 1.58 | - | - | 21.77 |
| PG01 | - | 5.08 | 1.63 | 1.89 | 0.31 | - | 0.21 | 5.78 | 2.18 | 10.14 | 0.65 | 5.71 | - | - | 33.59 |
| PG02 | - | 5.07 | 1.87 | 1.61 | 0.27 | - | 0.28 | 5.60 | 2.61 | 12.53 | 0.86 | 6.64 | - | - | 37.33 |
| PG03 | - | 4.76 | 3.51 | 1.41 | 0.42 | 0.07 | 0.98 | 6.93 | 1.25 | 7.55 | 1.25 | 5.86 | - | - | 34.00 |
| PG04 | - | 5.11 | 1.03 | 1.22 | 0.19 | - | 0.05 | 5.41 | 0.72 | 3.54 | 0.34 | 2.14 | 0.17 | - | 19.94 |
| PN01 | 5.86 | 24.94 | 1.30 | - | 0.89 | 0.78 | 0.42 | 13.88 | 0.17 | 0.26 | 0.07 | 6.11 | - | - | 54.67 |
| PN02 | 5.81 | 24.70 | 1.31 | - | 0.95 | 0.66 | 0.42 | 13.77 | 0.17 | 0.25 | 0.03 | 6.06 | - | - | 54.13 |
| US2-01 | - | 0.82 | 3.08 | - | - | - | 0.02 | 12.88 | 1.98 | 0.36 | 0.37 | 4.68 | 0.19 | - | 24.39 |
| US2-02 | - | 0.95 | 5.07 | - | - | - | 0.04 | 11.77 | 3.62 | 0.52 | 0.59 | 4.23 | - | - | 26.78 |
| US2-03 | - | 0.40 | 3.45 | - | - | - | 0.03 | 11.75 | 1.65 | 0.28 | 0.37 | 1.65 | 0.27 | - | 19.85 |
| US2-04 | - | 0.88 | 3.38 | - | - | - | 0.01 | 5.97 | 2.46 | 0.35 | 0.41 | 4.21 | - | - | 17.67 |
| US2-05 | - | 1.65 | 6.38 | - | - | 0.18 | 0.10 | 13.77 | 4.50 | 0.94 | 2.20 | 8.43 | - | - | 38.16 |
| CH2-01 | - | 0.86 | 4.13 | - | - | - | 0.04 | 12.17 | 5.03 | 0.86 | 0.68 | 3.67 | 0.45 | - | 27.89 |
| CH2-02 | - | 0.67 | 4.18 | - | - | - | 0.03 | 12.71 | 3.79 | 0.51 | 0.42 | 2.02 | - | - | 24.32 |
| CH2-03 | - | 0.91 | 5.28 | - | - | - | 0.09 | 13.96 | 6.23 | 1.23 | 0.80 | 5.47 | 1.36 | - | 35.33 |
| CH2-04 | - | 1.31 | 2.62 | - | - | - | - | 10.13 | 5.38 | 1.14 | 0.83 | 3.83 | - | - | 25.24 |
| CA2-01 | - | 1.13 | 5.46 | - | - | - | 0.03 | 15.15 | 4.83 | 0.62 | 5.38 | 6.95 | - | - | 39.55 |
| CA2-02 | - | 0.66 | 4.33 | - | - | - | 0.05 | 11.10 | 1.67 | 0.35 | 2.69 | 1.93 | - | - | 22.78 |
| CA2-03 | - | 1.59 | 6.05 | - | - | - | 0.07 | 15.66 | 5.44 | 0.72 | 3.16 | 6.69 | - | - | 39.38 |
| MK01 | - | 4.73 | 1.00 | 1.17 | 0.17 | - | 0.05 | 5.39 | 0.90 | 3.03 | 0.63 | 2.12 | 0.18 | - | 19.37 |
| MK02 | - | 0.89 | 3.19 | - | - | - | 0.01 | 13.03 | 2.21 | 0.38 | 0.45 | 4.90 | 0.20 | - | 25.26 |
| MK03 | - | 2.46 | 3.31 | 1.65 | 0.43 | - | 1.35 | 6.43 | 1.47 | 15.25 | 9.50 | 6.33 | - | - | 48.17 |
| MK04 | - | 2.23 | 4.97 | - | - | - | 0.05 | 12.34 | 3.31 | 0.61 | 0.69 | 5.02 | - | - | 29.21 |
| MK05 | - | 0.61 | 4.25 | - | - | - | 0.09 | 9.20 | 1.92 | 0.62 | 0.53 | 1.60 | 0.22 | 0.02 | 19.07 |
| MK06 | - | 1.02 | 4.77 | - | - | - | 0.06 | 14.16 | 4.87 | 0.65 | 0.64 | 4.68 | - | - | 30.85 |
-, Not detected. US01-20, American ginseng, cultivated in Wisconsin, USA; CH01-14, American ginseng, cultivated in Jilin, China; CA01-06, American ginseng, cultivated in Ontario, Canada; PG1-4, Asian ginseng, standard samples, cultivated in China; PN1-2, notoginseng, standard samples, cultivated in China; MK01-06, samples purchased from marketplaces, labeled as American ginseng.
To determine the correct species of commercial samples, their HPLC chromatograms were compared with that of American ginseng. The chromatograms of 4 commercial samples were very similar to the collected American ginseng samples, while notoginsenoside R1 and ginsenoside Rf were not detected. However, ginsenoside Rf was detected in 2 samples (labeled as American ginseng, Fig. 1F). The sample presented in Fig. 1F does not show a typical chromatograph of American ginseng according to USP [16], and is similar to the saponin distribution of the authorized Asian ginseng samples (shown in Table 1). Moreover, the peak of ginsenoside Rg1 is higher than that of ginsenoside Re in American ginseng (Figs. 1C–E), but it was the opposite in the counterfeit samples (Fig. 1F). Thus, based on the similarity of chromatogram fingerprint and existence of Rf, the two counterfeit samples could be Asian ginseng.
3.3. Adulteration identification with hierarchical cluster analysis (HCA) and principal component analysis (PCA)
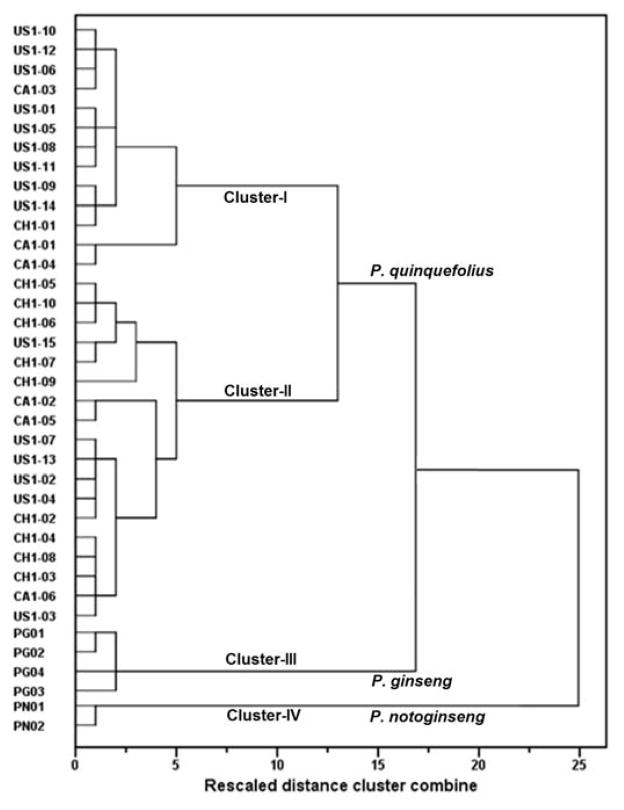
To accurately identify possible adulterated samples, HCA and PCA were used to quantitatively evaluate the diversity among American ginseng, Asian ginseng and notoginseng. Firstly, HCA was performed using Ward’s method after a normalizing treatment of the data to visualize the differences and/or similarities among samples through linkage distances. As shown in Fig. 2, the 31 American ginseng samples collected from three cultivation regions were merged into two clusters (Cluster-I and Cluster-II); 4 Asian ginseng and 2 notoginseng samples were merged into Cluster-III and Cluster-IV, respectively. The result indicated that the developed HCA method could separate American ginseng from Asian ginseng and notoginseng.
Fig. 2.
Dendrograms of hierarchical cluster analysis (HCA) using Ward’s method after standardization (range from 0 to 1). HCA was generated from the content of 14 ginsenosides in the tested samples. Samples included: American ginseng cultivated in the USA (US), Canada (CA) and China (CH); standard herbs Asian ginseng (PG) and notoginseng (PN).
Secondly, with PCA, an unsupervised method for pattern recognition, was employed to distinguish the diversities among three Panax species. After unit variance scaling and mean centering, all data were displayed as scores and loadings in a coordinate system of principal components resulting from data dimensionality reduction. As result, the first three principal components (PC 1-3) account for over 69% of the variation (Fig. 3A). A three-dimensional PC score plot clearly discriminated the three Panax species (Fig. 3B). Furthermore, a two-dimensional PCA score plot (PC 1–2, Fig. 3C) was able to discriminate the three Panax species, thus simplifying data management. The above results suggested that the authenticity of American ginseng could be determined through HPLC analysis coupled with HCA and PCA.
Fig. 3.
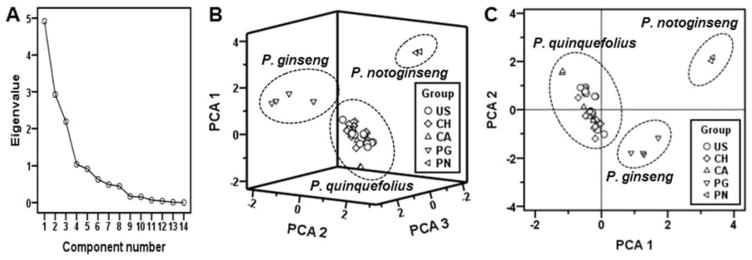
HPLC-principal component analysis (PCA) of ginseng samples. (A) Score plot of 14 components. (B) Three-dimensional diagrams of three PCAs (PCA1, PCA2, and PCA3) based on the different classification. (C) Two-dimensional diagrams of PCA1 and PCA2. Samples included: American ginseng cultivated in the USA (US), Canada (CA) and China (CH); standard herbs Asian ginseng (PG) and notoginseng (PN).
3.4. Cultivation region discrimination with linear discriminant analysis (LDA)
To distinguish the cultivation locations of American ginseng, a LDA method was developed. LDA has been used in economic analysis and social research [17, 18]. In this study, Fisher’s linear discriminant functions were generated based on the saponin contents in 31 American ginseng samples, which were collected from three countries. Three discriminant equations for the differently cultivated places are shown as below:
Where, DUS is expressed as the function of American ginseng samples cultivated in USA, DCH is from China, and DCA is from Canada. To achieve the best separation, Ci represents the content of the first five most important ginsenoside i (i = 20R-Rg2, Rc, Rb3, Rd, Rg3,). These equations were constructed using the Euclidean distance in the LDA algorithm in order to classify unknown samples and the Wilk’s lambda method, a stepwise algorithm, to extract the most important variables [19, 20].
To visualize the difference among the cultivation regions, a two-dimensional plot of the individual scores for the discriminant functions was demonstrated in Fig. 4. The tested samples from the three-cultivation regions were mainly concentrated on three separate centroids. By calculating the maximum value of the above three equations, all the tested samples were re-catalogued (Supplemental Table 2). It suggested that the developed functions could be utilized to discriminate the cultivation locations of American ginseng.
Fig. 4.
Canonical discriminant functions of linear discriminant analysis (LDA) based on 14-ginsenoside contents in American ginseng samples which were firstly collected from three cultivation regions. Significance between the canonical discriminant functions is P < 0.01 by Chi-square test. Samples included: American ginseng cultivated in the USA (US), Canada (CA) and China (CH).
3.5. Further verification of used techniques
To validate our identification method, we secondly collected 12 American ginseng samples from three cultivation regions and determined their saponin content. Using two Asian ginseng and two notoginseng samples as outgroups, HCA and PCA actively separate all American ginseng from Asian ginseng and notoginseng (Fig. 5A). Next, using our established LDA method, samples from different cultivation regions were separated clearly, in which the original sources were correctly classified (Fig. 5B). These results suggested that HPLC coupled with HCA or PCA can be used to identify the three species of the Panax genus. Then, HPLC-LDA method can be used to distinguish the cultivation regions of American ginseng.
Fig. 5.
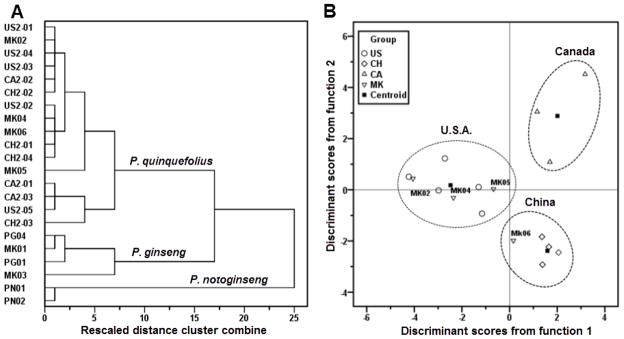
Method validation and identification of commercial samples. (A) HPLC-HCA analysis. (B) HPLC-LDA analysis. Samples include secondly collected American ginseng cultivated in three regions (USA, US2-01 to US2-05; Canada, CA2-01 to CA2-03; China, CH2-01 to CH2-04) and commercial samples labeled as American ginseng (MK01-06).
3.6. Identification of commercial samples
Our methods were used to identify the sources of commercial American ginseng. Using HPLC-HCA, among six samples purchased from different supermarkets, two of them were merged into the cluster of Asian ginseng (MK01 and MK03), while the other four samples were merged into American ginseng cluster (Fig. 5A and Supplemental Table 2). Similar results were also observed with HPLC-PCA analysis. This result is consistent with our initial judgment by comparison of the HPLC fingerprint. It suggested that our developed HCA and PCA methods can be used to identify adulteration of American ginseng from commercial sources.
After the exclusion of adulterants, the cultivation regions of the rest of the 4 American ginseng samples were quantitatively described by HPLC-LDA. As shown in Fig. 5B (and Supplemental Table 2), MK02, MK04 and MK05 were aggregated into the USA group, while MK06 was clustered into the China group. Results indicated that among the American ginseng samples, MK02, MK04 and MK05 were grown in the USA, while MK06 was cultivated in China. Our HPLC-LDA method successfully distinguished cultivation regions of commercial available American ginseng samples.
4. Discussion
Ginseng, one of the most internationally popular herbal medicines and dietary supplements, belongs to the genus Panax (family Araliaceae) that consists of approximately 14 species of slow-growing perennial plants with fleshy roots [21, 22]. Of which two ginseng species, American ginseng and Asian ginseng, are extensively used as a tonic to improve the overall health of human beings [23, 24]. However, these species have different natures. Based on traditional Chinese medicine theory, Asian ginseng is “hot”, and only few of people can use this herb. Since American ginseng is “cool”, most people can use this herb to promote vitality, assist the body functions, improve the immune system and protect against stress [25, 26]. Thus, the price of American ginseng was significantly higher than that of Asian ginseng and other ginsengs.
American ginseng originally grows in the USA and Canada. In recent years, this herb has been cultivated in northern China [12]. With the increasing market demand and profit temptation, the phenomena that American ginseng is substituted and/or adulterated by other cheaper species appears to be due to a considerable price difference in North America and China. Furthermore, in the market, American ginseng cultivated in China is often labeled as cultivated in North America. Therefore, accurate identification the species of American ginseng and discrimination of cultivation region are essential for quality control. Several methods have been used to identify the source, including chemistry-based techniques, and genetic analysis [27, 28]. However, it is difficult to identify counterfeits of American ginseng, especially to distinguish their cultivation regions.
In this study, a HPLC method was developed to simultaneously determine 14 ginseng saponins. Differences of chemical composition were observed between American ginseng and Asian ginseng. A pivotal evidence is the presence of ginsenoside Rf in Asian ginseng and its absence in American ginseng [29, 30], a compound that contributes to antinociception and regulates lipoprotein metabolism [31, 32]. In addition, the ratios of Rg1/Rb1 and Rb2/Rb1 have been widely used to differentiate these two ginsengs. Ratios of Rg1/Rb1 less than 0.3 and Rb2/Rb1 less than 0.4 are indicative of American ginseng [16]. In contrast, significantly higher values of both ratios are characteristic of Asian ginseng (Supplemental Table 2) [33, 34]. Since notoginseng (another species in the same genus) is commonly used in China, this species was selected as an outgroup in multivariate analysis.
In order to accurately identify American ginseng and adulterants, HPLC coupled with HCA and PCA analysis was developed. Asian ginseng and notoginseng can be discriminated by either of the two methods. The identification result using chromatography fingerprint supported our HCA and PCA analysis.
Regarding American ginseng samples, after further comparison, some small differences were found. Gypenoside XVII, a neuroprotective compound, which is labeled as #1 in Fig. 1 [5, 35], is found distinctly in American ginseng samples cultivated in Wisconsin and China, but not obviously in samples from Canada. However, because of the high similarity of chromatograms among samples from three agricultural areas, the traditional HPLC fingerprint cannot be used to identify cultivation regions, it is necessary to introduce novel method to achieve this objective.
HPLC-LDA method was established in this study to discriminate the cultivation regions of American ginseng. LDA is a supervised classification technique based on the linear discriminant functions, which maximizes the ratio of interclass variance and minimizes the ratio of intraclass variance [19]. As shown in Fig. 4, tested samples collected from three regions were concentrated on three separate centroids, suggesting the cultivation locations of American ginseng could be identified by this method.
To validate our developed methods, we secondly collected American samples from three cultivation regions, and determined the content of ginsenosides. Using our developed HCA and PCA methods, all the American ginseng samples were separated from Asian ginseng and notoginseng. By using LDA, their areas of cultivation were accurately discriminated (Fig. 5). Our developed methods were employed in the discrimination of samples purchased from the marketplaces. Two of the six commercial samples (labeled as American ginseng) were identified as Asian ginseng, while four others were identified as American ginseng, which were cultivated in the USA (3 samples) and China (1 sample).
In conclusion, to improve the current quality insurance technology of American ginseng, HPLC coupled with multivariate analyses were developed. HPLC-HCA or -PCA can be used to identify adulterants of American ginseng. Moreover, an HPLC-LDA method was established to discriminate cultivation regions such as USA, Canada and China. The methods have been successfully applied in the identification of commercial ginseng samples. Integrated HPLC and multivariate analysis supplies both quantitative determination and quantitative identification, our method could be a critical complement for current quality control studies of American ginseng.
Supplementary Material
Highlights.
It is difficult to distinguish origins of American ginseng, especially cultivation locations
We developed HPLC coupled with HCA and PCA methods to identify the Panax species
We established an HPLC-LDA method to identify American ginseng cultivation regions
This method was successfully applied to identify the sources of commercial samples
Acknowledgments
This work was supported in part by the NIH/NCCAM grants P01 AT004418 and K01 AT005362; the Natural Science Foundation of Jiangsu Province (BK2008194), Jiangsu Overseas Research & Training Program for University Prominent Young & Middle-aged Teachers and Presidents, Science and Technology Project of Department of Traditional Chinese Medicines in Jiangsu Province, China (LZ11163).
Footnotes
Conflicts of Interest
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ernst E. The risk-benefit profile of commonly used herbal therapies: Ginkgo, St. John’s Wort, Ginseng, Echinacea, Saw Palmetto, and Kava. Ann Intern Med. 2002;136:42–53. doi: 10.7326/0003-4819-136-1-200201010-00010. [DOI] [PubMed] [Google Scholar]
- 2.Qi LW, Wang CZ, Yuan CS. Isolation and analysis of ginseng: advances and challenges. Nat Prod Rep. 2011;28:467–495. doi: 10.1039/c0np00057d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang CZ, Aung HH, Ni M, Wu JA, Tong R, Wicks S, et al. Red American ginseng: ginsenoside constituents and antiproliferative activities of heat-processed Panax quinquefolius roots. Planta Med. 2007;73:669–674. doi: 10.1055/s-2007-981524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shergis JL, Zhang AL, Zhou W, Xue CC. Quality and risk of bias in Panax ginseng randomized controlled trials: a review. Am J Chin Med. 2013;41:231–252. doi: 10.1142/S0192415X13500171. [DOI] [PubMed] [Google Scholar]
- 5.Wan JY, Liu P, Wang HY, Qi LW, Wang CZ, Li P, et al. Biotransformation and metabolic profile of American ginseng saponins with human intestinal microflora by liquid chromatography quadrupole time-of-flight mass spectrometry. J Chromatogr A. 2013;1286:83–92. doi: 10.1016/j.chroma.2013.02.053. [DOI] [PubMed] [Google Scholar]
- 6.Qi LW, Wang HY, Zhang H, Wang CZ, Li P, Yuan CS. Diagnostic ion filtering to characterize ginseng saponins by rapid liquid chromatography with time-of-flight mass spectrometry. J Chromatogr A. 2012;1230:93–99. doi: 10.1016/j.chroma.2012.01.079. [DOI] [PubMed] [Google Scholar]
- 7.Ng TB. Pharmacological activity of sanchi ginseng (Panax notoginseng) J Pharm Pharmacol. 2006;58:1007–1019. doi: 10.1211/jpp.58.8.0001. [DOI] [PubMed] [Google Scholar]
- 8.Attele AS, Wu JA, Yuan CS. Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol. 1999;58:1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 9.Gu C, Qiao J, Zhu M, Du J, Shang W, Yin W, et al. Preliminary evaluation of the interactions of Panax ginseng and Salvia miltiorrhiza Bunge with 5-fluorouracil on pharmacokinetics in rats and pharmacodynamics in human cells. Am J Chin Med. 2013;41:443–458. doi: 10.1142/S0192415X13500328. [DOI] [PubMed] [Google Scholar]
- 10.Sengupta S, Toh SA, Sellers LA, Skepper JN, Koolwijk P, Leung HW, et al. Modulating angiogenesis: the yin and the yang in ginseng. Circulation. 2004;110:1219–1225. doi: 10.1161/01.CIR.0000140676.88412.CF. [DOI] [PubMed] [Google Scholar]
- 11.Yap KY, Chan SY, Weng Chan Y, Sing Lim C. Overview on the analytical tools for quality control of natural product-based supplements: a case study of ginseng. Assay Drug Dev Technol. 2005;3:683–699. doi: 10.1089/adt.2005.3.683. [DOI] [PubMed] [Google Scholar]
- 12.Sun J, Chen P. Differentiation of Panax quinquefolius grown in the USA and China using LC/MS-based chromatographic fingerprinting and chemometric approaches. Anal Bioanal Chem. 2011;399:1877–1889. doi: 10.1007/s00216-010-4586-7. [DOI] [PubMed] [Google Scholar]
- 13.Harkey MR, Henderson GL, Gershwin ME, Stern JS, Hackman RM. Variability in commercial ginseng products: an analysis of 25 preparations. Am J Clin Nutr. 2001;73:1101–1106. doi: 10.1093/ajcn/73.6.1101. [DOI] [PubMed] [Google Scholar]
- 14.Wang CZ, Wu JA, McEntee E, Yuan CS. Saponins composition in American ginseng leaf and berry assayed by high-performance liquid chromatography. J Agric Food Chem. 2006;54:2261–2266. doi: 10.1021/jf052993w. [DOI] [PubMed] [Google Scholar]
- 15.Wang CZ, Ni M, Sun S, Li XL, He H, Mehendale SR, et al. Detection of adulteration of notoginseng root extract with other Panax species by quantitative HPLC coupled with PCA. J Agric Food Chem. 2009;57:2363–2367. doi: 10.1021/jf803320d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.USP. USP35-NF30 (United States Pharmacopoeia 35 - National Formulary 30) United States Pharmacopeial; Rockville: 2012. pp. 1175–1176. [Google Scholar]
- 17.Hsiao TH, Tsai SM, Chou ST, Pan JY, Tseng YC, Chang HP, et al. Sex determination using discriminant function analysis in children and adolescents: a lateral cephalometric study. Int J Legal Med. 2010;124:155–160. doi: 10.1007/s00414-009-0412-1. [DOI] [PubMed] [Google Scholar]
- 18.Yarnold PR, Hart LA, Soltysik RC. Optimizing the classification performance of logistic-regression and fisher discriminant analyses. Educ Psychol Meas. 1994;54:73–85. [Google Scholar]
- 19.Sarbu C, Nascu-Briciu RD, Kot-Wasik A, Gorinstein S, Wasik A, Namiesnik J. Classification and fingerprinting of kiwi and pomelo fruits by multivariate analysis of chromatographic and spectroscopic data. Food Chem. 2012;130:994–1002. [Google Scholar]
- 20.Mot AC, Soponar F, Sarbu C. Multivariate analysis of reflectance spectra from propolis: geographical variation in Romanian samples. Talanta. 2010;81:1010–1015. doi: 10.1016/j.talanta.2010.01.052. [DOI] [PubMed] [Google Scholar]
- 21.Li CY, Lau DTW, Dong TTX, Zhang J, Choi RCY, Wu HQ, et al. Dual-index evaluation of character changes in Panax ginseng C. A. Mey stored in different conditions. J Agric Food Chem. 2013;61:6568–6573. doi: 10.1021/jf400456w. [DOI] [PubMed] [Google Scholar]
- 22.Case MA, Flinn KM, Jancaitis J, Alley A, Paxton A. Declining abundance of American ginseng (Panax quinquefolius L.) documented by herbarium specimens. Biol Conserv. 2007;134:22–30. [Google Scholar]
- 23.Wang CZ, Calway T, Yuan CS. Herbal medicines as adjuvants for cancer therapeutics. Am J Chin Med. 2012;40:657–669. doi: 10.1142/S0192415X12500498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hwang JW, Oh JH, Yoo HS, Lee YW, Cho CK, Kwon KR, et al. Mountain ginseng extract exhibits anti-lung cancer activity by inhibiting the nuclear translocation of NF-kappaB. Am J Chin Med. 2012;40:187–202. doi: 10.1142/S0192415X12500152. [DOI] [PubMed] [Google Scholar]
- 25.Wang CZ, Yuan CS. Potential role of ginseng in the treatment of colorectal cancer. Am J Chin Med. 2008;36:1019–1028. doi: 10.1142/S0192415X08006545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlag EM, McIntosh MS. The relationship between genetic and chemotypic diversity in American ginseng (Panax quinquefolius L.) Phytochemistry. 2013;93:96–104. doi: 10.1016/j.phytochem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 27.Choi HI, Kim NH, Lee J, Choi BS, Kim KD, Park JY, et al. Evolutionary relationship of Panax ginseng and P. quinquefolius inferred from sequencing and comparative analysis of expressed sequence tags. Genet Resour Crop Evol. 2013;60:1377–1387. [Google Scholar]
- 28.Wen J, Zimmer EA. Phylogeny and biogeography of Panax L. (the ginseng genus, araliaceae): inferences from ITS sequences of nuclear ribosomal DNA. Mol Phylogenet Evol. 1996;6:167–177. doi: 10.1006/mpev.1996.0069. [DOI] [PubMed] [Google Scholar]
- 29.Kim DH. Chemical diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng. J Ginseng Res. 2012;36:1–15. doi: 10.5142/jgr.2012.36.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi KT. Botanical characteristics, pharmacological effects and medicinal components of Korean Panax ginseng C A Meyer. Acta Pharmacol Sin. 2008;29:1109–1118. doi: 10.1111/j.1745-7254.2008.00869.x. [DOI] [PubMed] [Google Scholar]
- 31.Nemmani KV, Ramarao P. Ginsenoside Rf potentiates U-50,488H-induced analgesia and inhibits tolerance to its analgesia in mice. Life Sci. 2003;72:759–768. doi: 10.1016/s0024-3205(02)02333-0. [DOI] [PubMed] [Google Scholar]
- 32.Lee H, Gonzalez FJ, Yoon M. Ginsenoside Rf, a component of ginseng, regulates lipoprotein metabolism through peroxisome proliferator-activated receptor alpha. Biochem Biophys Res Commun. 2006;339:196–203. doi: 10.1016/j.bbrc.2005.10.197. [DOI] [PubMed] [Google Scholar]
- 33.Yuan CS, Wang CZ, Wicks SM, Qi LW. Chemical and pharmacological studies of saponins with a focus on American ginseng. J Ginseng Res. 2010;34:160–167. doi: 10.5142/jgr.2010.34.3.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura S, Sugimoto S, Matsuda H, Yoshikawa M. Medicinal flowers. XVII. New dammarane-type triterpene glycosides from flower buds of American ginseng, Panax quinquefolium L. Chem Pharm Bull. 2007;55:1342–1348. doi: 10.1248/cpb.55.1342. [DOI] [PubMed] [Google Scholar]
- 35.Meng X, Wang M, Sun G, Ye J, Zhou Y, Dong X, et al. Attenuation of Abeta-induced parallel autophagic and apoptotic cell death by gypenoside XVII through the estrogen receptor-dependent activation of Nrf2/ARE pathways. Toxicol Appl Pharmacol. 2014 doi: 10.1016/j.taap.2014.03.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.