Abstract
A coumarin-modified pyrimidine nucleoside (1) has been synthesized using Cu(I)-catalyzed click reaction and incorporated into oligodeoxynucleotides (ODNs). Interstrand cross-links are produced upon irradiation of ODN containing 1 at 350 nm. Cross-linking occurs via [2+2] cycloaddition reaction with the opposing thymidine, 2′-deoxycytidine, or 2′-deoxyadenosine. A much higher reactivity was observed with dT than dC or dA. Irradiation of the dT-1 and dC-1 cross-linked products at 254 nm leads to a reversible ring-opening reaction, while such phenomenon was not observed with dA-1 adducts. The reversible reaction is ultrafast and complete within 50 s – 90 s. Consistent photoswitching behavior was observed over 6 cycles of irradiation at 350 nm and 254 nm. To the best of our knowledge, this is the first example of photoswitchable interstrand cross-linking formation induced by a modified pyrimidine nucleoside. Compound 1 is a novel tool for developing reversible DNA photoswitches which will lead to new applications in chemistry, biology, and nanotechnology.
Keywords: Photoswitchable DNA cross-linking, modified pyrimidine nucleoside, Coumarin, Click chemistry, Reversible [2+2]cycloaddition
DNA interstrand cross-links (ICLs) covalently link two DNA strands, which can block DNA replication, transcription, and any other processes requiring strand separation; thus, they have a strong impact on biological function of nucleic acids. Chemical agents capable of inducing ICLs have been developed for biological applications, such as for DNA damage and repair studies,[1–5] as anticancer agents,[6] for nucleic acid detection,[7,8] for targeting telomeric G-quadruplex structures,[9,10] and for construction of DNA nanomaterials.[11] Over the past few decades, several research groups have developed novel chemical methods for inducing ICL formation, such as photoirradiation, oxidation, reduction, fluoride induction, and H2O2 induction.[12] Among these methods, light induction is particularly important as it is a non-invasive method with high spatio-temporal resolution and control and offers the options of orthogonality. Recently, Freccero, Zhou and their coworkers reported photo-inducible quinone methide formation from biphenyl or binol quaternary ammonium salts[13–15] and formation of vinylidene–quinone methides from 2-alkynylphenols.[16] Quinone methides are highly reactive electrophiles which efficiently cross-link DNA double strands.[11, 13–15] Greenberg et al.[17–21] described several modified nucleosides that efficiently induced DNA interstrand cross-linking upon photoirradiation. Recently, we reported hypoxia-selective ICL formation from nitroimidazole-modified thymidine upon UV-irradiation.[22]
Photoactive molecules have also been studied for the construction of DNA-based reversible photoswitches and photo-manipulation of DNA.[23–28] For instance, psoralens, a class of naturally occurring photoreactive products, are capable of cross-linking duplex and triplex DNA.[25–28] They have been used as probes of nucleic acid structure and function, for therapeutic gene modulation, and for studying DNA damage and repair.[26–28] Coumarins showed many advantages such as high fluorescence quantum yield, large Stokes shift, excellent photostability, and low toxicity. Coumarin derivatives have been widely used in the fields of biology, medicine, cosmetics, and as fluorescent chemosensors for DNA, RNA, and protein detection.[29,30] However, there is no report about the photo activity of coumarin with DNA. Here, we report the first coumarin-modified nucleoside that induces photo-switchable DNA ICL formation upon UV irradiation. A coumarin-modified thymidine efficiently induces ICL formation upon photoirradiation at 350 nm while the cross-linking can be reversed by irradiation at 254 nm. The cross-linking site was determined by LC-MS/MS as well as by hydroxyl radical cleavage of gel-purified cross-linked DNA.
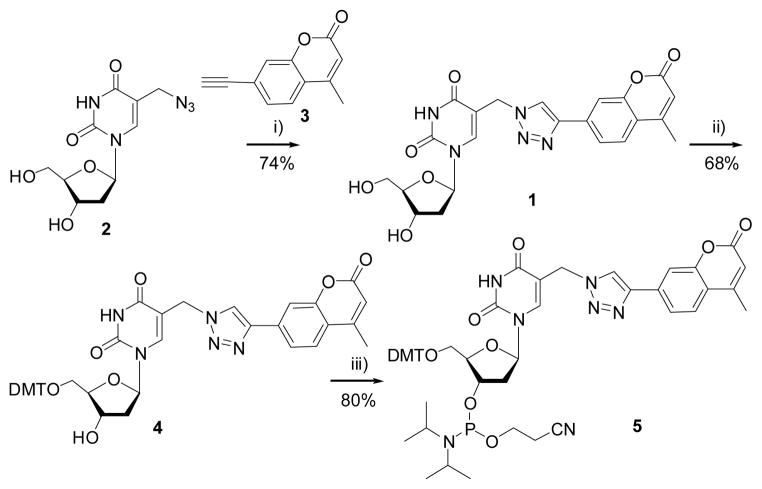
Coumarin moiety has been conjugated to thymidine by Cu-catalyzed azide-alkyne cycloaddition reaction using azide-modified thymidine 2 and an alkyne-modified coumarin 3.[18, 31] Compound 1 was converted to its phosphoramidite building block 5 under standard conditions (Scheme 1). Oligodeoxynucleotides (ODNs) containing 1 were synthesized via automated solid-phase synthesis using 5 and confirmed by MALDI-TOF-MS analysis (Supporting information (SI), Table 1).
Scheme 1.
Synthesis of compound 1 and its phosphoramidite building block 5. Reagents: i) Cu2SO4 and sodium ascorbate; ii) Dimethoxytrityl chloride (DMTCl), pyridine; iii) 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite, N,N-diisopropylethylamine and CH2Cl2.
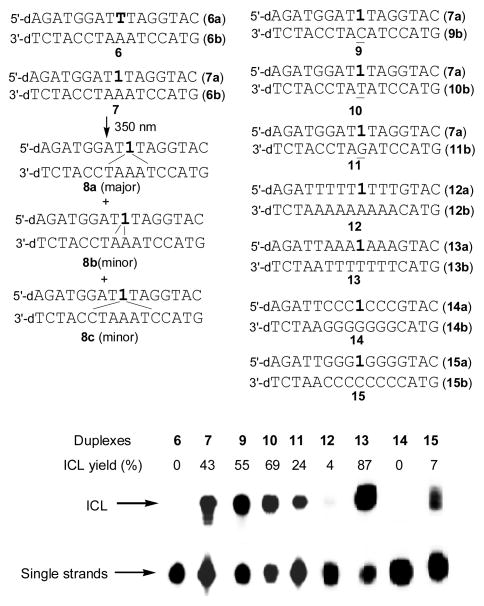
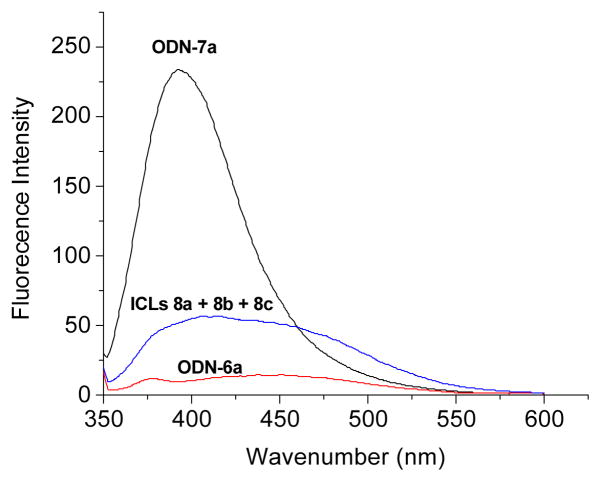
The DNA duplex was photo-irradiated at 350 nm for 50 min using a Rayonet Photochemical Chamber Reactor (Model RPR-100). A wavelength of 350 nm was chosen because near-UV light (> 300 nm) is compatible with living cells and is almost not absorbed by most biological molecules other than the coumarin-modified ODNs. Moreover, the coumarin moiety can be excited to singlet and triplet excitation states upon irradiation with 350-nm light. The UV-irradiation of duplex 7 resulted in a new band whose migration is severely retarded relative to unreacted oligonucleotide, indicative of formation of interstrand cross-linked material, while this was not observed with native DNA duplex 6 (Figure 1). The yield of ICL induced by 1 followed first-order kinetics with a rate constant (kICL) of 9.24 ± 0.25 × 10−4 s−1 (SI, Figure S1). The cross-linked product was isolated and characterized by mass spectrometry and fluorescence measurement. ODN 7a showed strong fluorescence with an emission maximum at 392 nm. However, greatly decreased fluorescence intensity was observed with the cross-linked product 8 (8a–8c) (Figure 2). We propose that the cross-link formation may lead to the destruction of the conjugation system of coumarin moiety.
Figure 1.
ICL formation upon UV-irradiation at 350 nm for 50 min. Phosphorimage autoradiogram of 20% denaturing PAGE analysis of the ICL products: lane 1, unmodified duplex 6; lanes 2–5, duplexes 7 and 9–11 containing coumarin-modified ODNs hybridized with opposing sequences containing A, C, T, and G; lanes 6–9, duplexes 12–15 containing coumarin-modified ODNs hybridized with opposing sequences containing only As, Ts, Gs, and Cs in opposite sequences.
Figure 2.
Fluorescence emission spectra of coumarin-modified ODN 7a, ICLs (8a–c) induced by coumarin after photoirradiation, and the unmodified ODN 6a. The reaction mixture contained 5.0 μM DNA, 100 mM NaCl, and 10 mM phosphate buffer (pH 7.0).
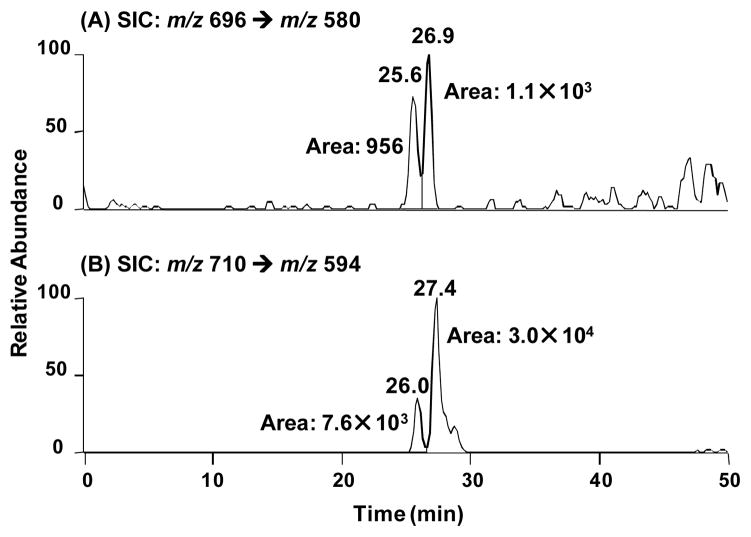
Initially, hydroxyl radical footprinting of gel-purified cross-linked DNA was used to determine which nucleotides were covalently bonded with one another.[17] Each ODN was either 5′- or 3′-32P-labeled. As expected, cross-linking occurred mainly with the position of 1 (SI Figure S2). However, the exact cross-linking sites in the complementary strand could not be determined by hydroxyl radical cleavage reaction possibly due to the presence of multiple reaction sites. To exploit this further, we used LC-MS and MS/MS to determine the identities of the nucleobase(s) in the opposite strand that is(are) cross-linked with the coumarin-modified thymidine moiety. To this end, we digested the isolated ICL products obtained from duplexes 7 and 9 with a cocktail of four enzymes to release the ICL as a dinucleoside remnant, following previously published procedures,[32] and subjected the resulting mixture to LC-MS and MS/MS analyses. Our LC-MS/MS results revealed, in the digestion mixture, the presence of dinucleosides, where the coumarin-derived thymidine is conjugated with a thymidine (dT), 2′-deoxycytidine (dC, detected as the deaminated product where the dC moiety was converted to a 2′-deoxyuridine, dU), and, to a lower extent, 2′-deoxyadenosine (dA) (Figure 3 and SI Figure S3–S9).
Figure 3.
Selected-ion chromatograms (SICs) for monitoring the indicated transitions for dU-1 cross-link (A) and dT-1 cross-link(B) in the digestion mixture of ICL products generated from duplex 7 (7a•6b).
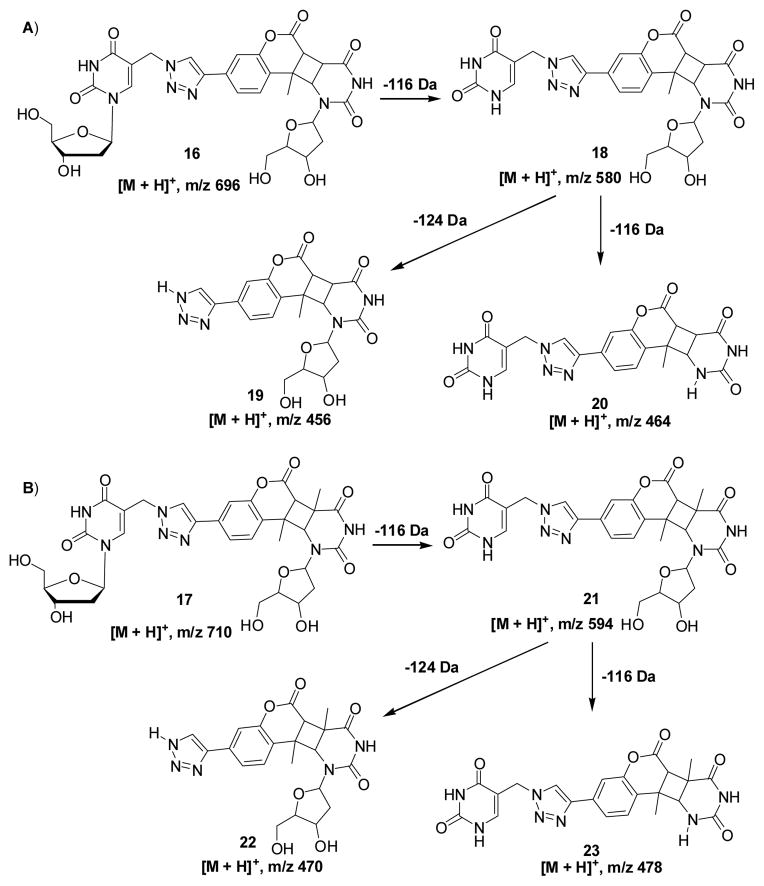
When 1 is opposite to four different nucleosides, the ICL yields decreased in the order of dT (10, 69%) > dC (9, 55%) > dA (7, 43%) > dG (11, 24%) (Figure 1). This result indicates that 1 has a higher reactivity towards pyrimidines than purines. In order to fully investigate the reactivity of 1 towards the four canonical nucleosides, we synthesized DNA duplexes 12–15 with only dA, dT, dG, or dC surrounding the photo-cross-linking site and examined the cross-linking efficiency and the rate constant for ICL formation. Among these duplexes, the highest ICL yield (87%) was observed with duplex 13 (dT), while no ICLs were found with duplex 14 (dG) (Figure 1). Although duplexes 12 (dA) and 15 (dC) resulted in about 4% and 7% ICLs, respectively, the reaction of dC or dA with 1 is much slower than that of dT with 1. The rate constants are in the order of kdT (3.5 ± 0.5 × 10−3) > kdC (1.2 ± 0.2 × 10−3) > kdA (0.8 ± 0.2 × 10−3) (SI Figure S10A–C). All these results supported that dT has a much higher reactivity than dC and dA towards the coumarin moiety. The LC-MS/MS analysis of the ICL products obtained from duplex 7 or 9 showed that the coumarin-derived thymidine mainly conjugated with dT in duplex 7 and with dC in duplex 9 (Figure 3 and SI, Figure S4–5). In addition, all cross-linked products are stable to heating in phosphate buffer or 1.0 M piperidine (SI, Figure S11). Collectively, based on the LC-MS/MS analysis and greatly decreased fluorescence intensity of the ICL adducts and the relative reactivities of 1 towards dT, dC, and dA, we propose that the [2+2] photocycloaddition reaction occurred, forming adduct 16 or 17 (Scheme 2). The MS/MS analysis for monitoring the [M+H]+ ion of dU-1 revealed two major peaks eluting at 25.6 and 26.9 min in the selected-ion chromatogram (SIC) for the m/z 696 (16) → 580 (18) transition, which monitors the neutral loss of a 2-deoxyribose (Scheme 2A). Product ions of m/z 456 (19) and 464 (20) were also found in the MS/MS due to neutral losses of 2-deoxyribose and thymine or the second 2-deoxyribose (SI Figure S6A, B, E). A similar fragmentation pathway was observed for dT-1 cross-linking product 17 (Scheme 2B and SI Figure S6C,D,F). It is precedented that the [2+2] cycloaddition reaction occured between dT and psoralen involving either the 3,4-double bond of the pyrone ring or the 4′,5′-double bond of the furan ring.[25] However, the structure of dA-1 adduct could not be determined at this point. In this vein, it is worth noting that, upon exposure to 254-nm UV light, thymine was found to undergo [2+2] cycloaddition with its neighboring 3′ adenine to give intrastrand cross-link products.[33, 34]
Scheme 2.
The proposed structures for the ICL products and the proposed major fragmentation pathways for the [M+H]+ ion of dU-1 (A) or dT-1 cross-link (B) observed in LC-MS/MS.
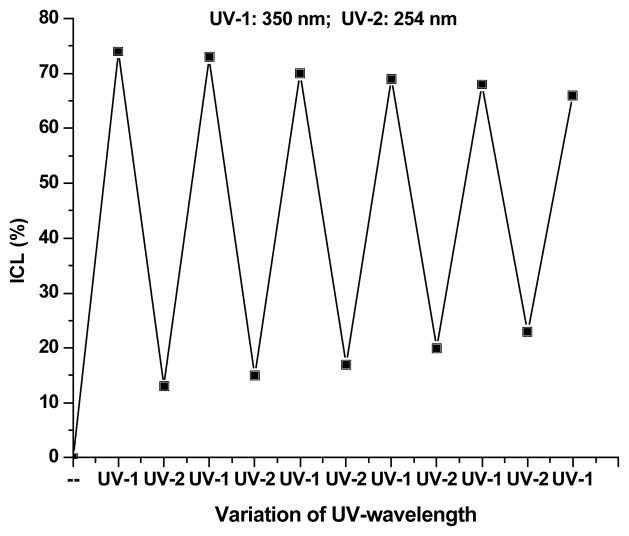
The ICL formation induced by 1 is photoswitchable. Irradiation of duplex 10 at 350 nm efficiently generated ICLs while the cross-linked product can be split into single-stranded DNAs by irradiation at 254 nm (Figure 4 and SI Figure S12). Cleavage of the ICL products to two single stranded ODNs was ultrafast and complete within 60 s by 254 nm irradiation (kcleave = 8.0 ± 0.5 × 10−2 s−1, t1/2 = 9.0 s, SI Figure 14A). This process was highly reversible and consistent switching behavior was validated over six cycles of irradiation at 350 nm for 50 min and 254 nm for 6 min (Figure 4 and SI Figure S12). A similar phenomenon was observed with duplex 7. This process was also confirmed by mass spectrometry. The MALDI-TOF-MS analysis showed that the cross-linked DNA products isolated from the 350 nm-irradiated duplex 7 were split to two single-standed ODNs upon 254 nm UV irradiation. The detected masses of two ODNs are 5185 and 4800 Da, which are identical to the coumarin-modified ODN 7a (Cal.: 5186.4 Da) and the complementary strand 6b (Cal.: 4800.2 Da) (SI, Figure S13). So far neither unsubstituted nor substituted pyrimidine nucleosides have ever been applied as components of coumarin photoswitches.[35,36] This feature would be one of the important milestones for the photoswitchable DNA interstrand cross-linking. The photochromic behavior of these compounds was observed via gel electrophoresis analysis followed by UV photo-irradiation. Six distinct absorption bands were investigated to verify the ratification of the exceptional photoswitchability.
Figure 4.
Reversibility of the DNA interstrand photo-cross-linking by a coumarin-modified ODN duplex 10 over six cycles of 50 min UV irradiation at 350 nm and 6 min at 254 nm.
To determine whether dT-1, dC-1, and dA-1 cross-links are reversible, we isolated the ICLs formed from duplex 10 after one cycle of 350 nm irradiation (ICL-1), one cycle of 350 nm/254 nm irradiation (ICL-2), and those formed after 10 cycles of 350 nm/254 nm irradiation (ICL-20). We then digested the isolated ICL products with enzymes to release the ICL as a dinucleoside remnant and subjected the resulting mixture to LC-MS and MS/MS analyses, as described above. We then plotted the ratios of area of peaks found in the SICs for monitoring the loss of a 2-deoxyribose from the [M+H]+ ions of dC-1 (as deaminated form, i.e. 16, m/z 696 → 580 transition), dT-1 (17, m/z 710 → 594 transition), and dA-1 adducts (m/z 710 → 594 transition) versus that for the [M+H]+ ion of dA (SI Figure 27). The signals for the dT-1 and dU-1 cross-links were decreased from ICL-1 to ICL-2 and ICL-20, whereas a significant elevation of dA-1 signal was observed from ICL-1 and ICL-2 to ICL-20. These results suggested that the formation of dC-1 and dT-1, but not that of dA-1 is reversible. In this context, it is worth noting that, owing to the differences in ionization efficiencies for these dinucleosides (dA-1, due to the higher proton affinity of adenine than uracil or thymine, is expected to have much better ionization efficiency than dT-1 and dU-1 in the positive-ion mode), the peak area ratios displayed in Figure 5 do not reflect the relative levels of dT-1, dU-1 and dA-1 interstrand cross-links. To further substantiate this finding, we examined the reversibility of ICL formation from duplexes 12, 13, and 15. Our data showed that the cross-linked products formed from duplexes 13 (dT-1 cross-link) and 15 (dC-1 cross-link) via 350 nm irradiation were reversible by 254 nm irradiation (SI Figure 14B,C) while ICL products generated from duplex 12 (dA-1 cross-link) was not cleaved by irradiation at 254 nm (SI Figure 14D). The reversible photo-cross-linking reaction with duplexes 13 and 15 is ultrafast and complete within 90 s and 50 s, respectively (duplex 13: kdT-1-cleave = 5.0 ± 0.9 × 10−2 s−1, t1/2 = 13.0 s; duplex 15: kdC-1-cleave = 8.8 ± 0.4 × 10−2 s−1, t1/2 = 8.0 s) (SI Figure 14B,C). These data are consistent with our results from LC-MS and MS/MS analysis, and are also in line with the fact that the cyclobutane-type photoproduct formed between thymine and the psoralen moiety can be photoreversed upon irradiation with 254-nm,[37] whereas the UVC-induced dimeric TA photoproduct cannot be photoreversed.[34]
In conclusion, we have developed a novel strategy for photo-switchable DNA interstrand cross-linking using a coumarin-modified nucleoside (1) that can be easily prepared via “click” chemistry. Irradiation of ODN containing 1 at 350 nm produced a non-fluorescent DNA cross-linking product which can be reverted to the original fluorescent ODN via 254 nm irradiation. The mechanism involves the [2+2] photocycloaddition between coumarin moiety and dT, dC, or to a much lower extent, dA. The dT-1 and dC-1 adducts are photo-reversible while dA-1 adduct is not reversible. The reversible interstrand photo-cross-linking reaction is bioorthogonal, does not require additional chemical reagents, and can be controlled in space and time by choosing different irradiation wavelengths. We envision that the method is applicable for in situ DNA manipulation, such as photocontrolled inhibition of DNA transcription or DNA ligation and the regulation of DNA aptamer.[38] It provides a simple and versatile approach for developing reversible DNA fluorescent photoswitches, photoswitchable DNA nanomotor, and smart DNA nanostructures and nanodevices.
Supplementary Material
Footnotes
We are grateful for financial support from the National Cancer Institute (1R15CA152914-01), UWM Research Growth Initiative (RGI101X234), and the Great Milwaukee Foundation (Shaw Scientist Award) to X. P. and the National Institute of Environmental Health Sciences (R01 ES019873) to Y. W. Thanks for support from the distinguished graduate student fellowship for Huabing Sun.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.((Please delete if not appropriate))
Contributor Information
Mohammad Mojibul Haque, Department of Chemistry and Biochemistry, University of Wisconsin Milwaukee, 3210 N. Cramer St., Milwaukee. WI 53211, USA.
Huabing Sun, Department of Chemistry and Biochemistry, University of Wisconsin Milwaukee, 3210 N. Cramer St., Milwaukee. WI 53211, USA.
Shuo Liu, Department of Chemistry, University of California Riverside, 332 Chemical Sciences Building, 501 Big Springs Road, Riverside, CA 92521-0403.
Prof. Yinsheng Wang, Department of Chemistry, University of California Riverside, 332 Chemical Sciences Building, 501 Big Springs Road, Riverside, CA 92521-0403
Prof. Xiaohua Peng, Email: pengx@uwm.edu, Department of Chemistry and Biochemistry, University of Wisconsin Milwaukee, 3210 N. Cramer St., Milwaukee. WI 53211, USA
References
- 1.Noll DM, Mason TM, Miller PS. Chem Rev. 2006;106:277. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dronkert MLG, Kanaar R. Mutat Res. 2001;486:217. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 3.Peng X, Ghosh A, Houten BV, Greenberg MM. Biochemistry. 2010;49:11. doi: 10.1021/bi901603h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng X, Pigli Y, Rice PA, Greenberg MM. J Am Chem Soc. 2008;130:12890. doi: 10.1021/ja805440v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weng M, Zheng Y, Jasti V, Champeil E, Tomasz M, Wang Y, Basu A, Tang M. Nucleic Acids Res. 2010;38:6976. doi: 10.1093/nar/gkq576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brulikova L, Hlavac J, Hradil P. Curr Med Chem. 2012;19:364. doi: 10.2174/092986712803414295. [DOI] [PubMed] [Google Scholar]
- 7.Peng X, Greenberg MM. Nucleic Acids Res. 2008;36:e31. doi: 10.1093/nar/gkn052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun H, Peng X. Bioconjugate Chem. 2013;24:1226. doi: 10.1021/bc4001678. [DOI] [PubMed] [Google Scholar]
- 9.Doria F, Nadai M, Folini M, Scalabrin M, Germani L, Sattin G, Mella M, Palumbo M, Zaffaroni N, Fabris D, Freccero M, Richter SN. Chem Eur J. 2013;19:78. doi: 10.1002/chem.201203097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doria F, Nadai M, Folini M, Di Antonio M, Germani L, Percivalle C, Sissi C, Zaffaroni N, Alcaro S, Artese A, Richter SN, Freccero M. Org Biomol Chem. 2012;10:2798. doi: 10.1039/c2ob06816h. [DOI] [PubMed] [Google Scholar]
- 11.Rusling DA, Nandhakumar IS, Brown T, Fox KR. Chem Commun. 2012;48:9592. doi: 10.1039/c2cc35407a. [DOI] [PubMed] [Google Scholar]
- 12.Cao S, Peng X. Curr Org Chem. 2014;18:1–15. [Google Scholar]
- 13.Antonio MD, Doria F, Mella M, Merli D, Profumo A, Freccero M. J Org Chem. 2007;72:8354. doi: 10.1021/jo7014328. [DOI] [PubMed] [Google Scholar]
- 14.Verga D, Nadai M, Doria F, Percivalle C, Antonio MD, Palumbo M, Richter SN, Freccero M. J Am Chem Soc. 2010;132:14625. doi: 10.1021/ja1063857. [DOI] [PubMed] [Google Scholar]
- 15.Wang P, Liu R, Wu X, Ma H, Cao X, Zhou P, Zhang J, Weng X, Zhang X, Qi J, Zhou X, Weng L. J Am Chem Soc. 2003;125:1116. doi: 10.1021/ja029040o. [DOI] [PubMed] [Google Scholar]
- 16.Doria F, Percivalle C, Freccero M. J Org Chem. 2012;77:3615. doi: 10.1021/jo300115f. [DOI] [PubMed] [Google Scholar]
- 17.Hong IS, Greenberg MM. J Am Chem Soc. 2005;127:3692. doi: 10.1021/ja042434q. [DOI] [PubMed] [Google Scholar]
- 18.Hong IS, Ding H, Greenberg MM. J Am Chem Soc. 2006;128:485. doi: 10.1021/ja0563657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong IS, Greenberg MM. J Am Chem Soc. 2005;127:10510. doi: 10.1021/ja053493m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong IS, Ding H, Greenberg MM. J Am Chem Soc. 2006;128:2230. doi: 10.1021/ja058290c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding H, Greenberg MM. J Org Chem. 2010;75:535. doi: 10.1021/jo902071y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuang Y, Sun H, Blain JC, Peng X. Chem Eur J. 2012;18:12609. doi: 10.1002/chem.201201960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahova H, Jäschke A. Angew Chem Int Ed. 2013;52:3186. doi: 10.1002/anie.201209943. [DOI] [PubMed] [Google Scholar]
- 24.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Annu Rev Biochem. 1985;54:1151. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 25.Takasugi M, Guendouz A, Chassignol M, Decout JL, Lhomme J, Thuong NT, Hélène C. Proc Natl Acad Sci USA. 1991;88:5602. doi: 10.1073/pnas.88.13.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shahid KA, Majumdar A, Alam R, Liu ST, Kuan JY, Sui XF, Cuenoud B, Glazer PM, Miller PS, Seidman MM. Biochemistry. 2006;45:1970. doi: 10.1021/bi0520986. [DOI] [PubMed] [Google Scholar]
- 27.Majumdar A, Muniandy PA, Liu J, Liu JL, Liu ST, Cuenoud B, Seidman MM. J Biol Chem. 2008;283:11244. doi: 10.1074/jbc.M800607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thazhathveetil AK, Liu ST, Indig FE, Seidman MM. Bioconjug Chem. 2007;18:431. doi: 10.1021/bc060309t. [DOI] [PubMed] [Google Scholar]
- 29.Wagner BD. Molecules. 2009;14:210. doi: 10.3390/molecules14010210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Cai L, Chen Z. Coumarin-Derived Fluorescent Chemosensors. In: Wang Wen., editor. Advances in Chemical Sensors. 2012. pp. 121–150. [Google Scholar]
- 31.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew Chem Int Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KM, Price NE, Wang J, Fekry MI, Dutta S, Seiner DR, Wang Y, Gates KS. J Am Chem Soc. 2013;135:1015. doi: 10.1021/ja308119q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bose SN, Davies RJ, Sethi SK, McCloskey JA. Science. 1983;220:723. doi: 10.1126/science.6836308. [DOI] [PubMed] [Google Scholar]
- 34.Zhao X, Nadji S, Kao J, Taylor JS. Nucl Acids Res. 1996;24:1554. doi: 10.1093/nar/24.8.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshimura Y, Fujimoto K. Org Lett. 2008;10:397–400. doi: 10.1021/ol7026784. [DOI] [PubMed] [Google Scholar]
- 36.Fujimoto K, Yamada A, Yoshimura Y, Tsukaguchi T, Sakamoto T. J Am Chem Soc. 2013;135:16161. doi: 10.1021/ja406965f. [DOI] [PubMed] [Google Scholar]
- 37.Cimino GD, Shi YB, Hearst JE. Biochemistry. 1986;25:3013. doi: 10.1021/bi00358a042. [DOI] [PubMed] [Google Scholar]
- 38.Brieke C, Rohrbach F, Gottschalk A, Mayer G, Heckel A. Angew Chem Int Ed. 2012;51:8446. doi: 10.1002/anie.201202134. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.