Summary
The unintegrated viral DNA synthesized during human immunodeficiency virus type 1 infection includes linear and circular forms. Circular forms of viral DNA are surrogate markers for nuclear import of viral DNA during virus replication as well as events surrounding the completion of reverse transcription. Analysis of 2-LTR circles is convenient and quantity of 2-LTR circle formed in directly proportional the amount of viral DNA imported into cell nucleus. In addition, correct synthesis of 2-LTR circles is an outcome of HIV-1 Gag-Pol function. Thus, quantitation and sequence analysis of 2-LTR circles have been very important to study structure function relationship of key viral proteins. In this chapter, we describe the methods of quantitation and analysis of 2-LTR circle junction isolated from HIV-1 infected cells.
Keywords: 2-LTR circle junction, Southern blot, Real-time PCR, Sequence analysis
1 Introduction
Following infection, HIV-1 reverse transcriptase (RT) converts viral RNA into double stranded DNA by exploiting its multifunctional enzyme activities (1). During productive infection, the viral cDNA then enters into the nucleus of infected cell accompanied by integrase (IN) and other viral proteins, in the form of pre-integration complexes (PICs) (1,2,3). In the host cell nucleus, viral cDNA integrates into the host chromosomal DNA in a reaction mediated by IN (4,5,6). Integrated DNA then serves as the template for viral RNA production. A fraction of cDNA which remains unintegrated become circularized, a reaction that is thought to be a dead end product since circular cDNA is not the substrate for the viral integrase. Circularization of viral DNA is believed to be mediated by cellular non-homologous end joining (NHEJ) pathway which includes several host proteins namely Ku, DNA-PK, XRCC4 or DNA ligase IV (7). These factors bind to the PICs and modulate the course of infection.
Circular DNA formation in the nucleus in the cell nucleus is accomplished by: 1) Joining of two ends of full-length cDNA having complete LTRs to form 2-LTR circle (7, 8, 9); 2) Formation of 1-LTR circle as a result of recombination between 2-LTRs (10). 3) Circularization of stalled reverse transcription complexes that lead to 1-LTR circle. 4) Integration of the linear cDNA into itself yielding an internally arranged form (9). The amount of 2-LTR circles generated is always less than that of 1-LTR circles and independent of cell line used. The ratio of 2-LTR circles to 1-LTR circles generally range from 0.16 to 0.43 (11). Using HIV-1 based vectors, it has been shown that 2-LTR circles accumulate in cells more slowly compared to full-length cDNA and reach a maximum abundance after 24 hours, consistent with the expected precursor-product relationship. The 2-LTR circles were also found to be notably stable in the host cell nucleus.
Though 2-LTR circle junctions are only a fraction of total viral cDNA in the infected cells, they contain enormous critical information regarding the fidelity of viral enzyme functions and or key steps during viral DNA synthesis. First, the amount of 2-LTR circle junctions accumulated in the nucleus is proportional to the amount of nuclear import of viral DNA since this form of viral cDNA is found only in the host nucleus. Secondly, 2-LTR circle DNA is a byproduct rather than an intermediate of the integration event. In addition, sequence analysis of 2-LTR circle junctions gives us insight into a number of crucial processes mediated by reverse transcriptase including completion of strand displacement synthesis, removal of tRNALys,3 primer, removal of plus strand PPT primer that were used during plus and minus strand DNA synthesis respectively.
2 Materials
2.1 Tissue culture, virus generation and Isolation of viral DNA from infected cell
-
i)
Dulbecco's Modified Eagle's Medium (DMEM) (Cellgro media, Kansas city, MO) supplemented with 10% fetal bovine serum (FBS, Gibco/BRL).
-
ii)
RPMI1640 medium (Gibco /BRL) supplemented with 10% FBS. CaPO4 mediated mammalian cell transfection reagent (Specialty media).
-
iii)
Filtration unit (Costar).
-
iv)
RNase free DNase I (10,000 U/ml, Roche diagnostics).
-
v)
Phosphate Buffered Saline (Without Ca/Mg, Cellgro medium).
-
vi)
Viral DNA isolation kit (DNeasy tissue kit, QIAGEN).
2.1. Southern blot hybridization of Viral DNA
-
i)
Hydrochloric acid 0.25 M
-
ii)
Denaturation solution: 1.5 M NaCl/0.5 M NaOH
-
iii)
Neutralization solution: 1.5 M NaCl/0.5 M Tris-Cl, pH 7.0
-
iv)
20x and 2x solutions of Saline- Sodium Citrate, 3M Sodium Chloride, 0.3 M Sodium Citrate, pH 7.0 (SSC)
-
v)
Whatman 3MM filter paper sheets
-
vi)
Nylon or nitrocellulose membrane
-
vii)
UV-transparent plastic wrap (Saran Wrap) for nylon membranes
-
viii)
UV trans-illuminator for nylon membranes.
2.3 Hybridization of transferred DNA
-
i)
DNA to be used as probe
-
ii)
Aqueous prehybridization/hybridization solution, room temperature and 68° C (Ambion)
-
iii)
2 x SSC/0.1% (w/v) SDS
-
iv)
0.2 x SSC/0.1% (w/v) SDS, room temperature and 42°C
-
v)
0.1 x SSC/0.1% (w/v) SDS, 68°C
-
vi)
2x and 6x SSC
-
vii)
Hybridization oven or 68°C water bath or incubator
-
viii)
Hybridization tube or sealable bag and heat sealer
-
ix)
Additional reagents and equipment for DNA labeling by nick translation or random oligonucleotide priming, measuring the specific activity of labeled DNA and separating unincorporated nucleotides from labeled DNA and autoradiography.
2.4 Real Time PCR
-
i)
Primers and Taqman Probes Specific for 2-LTR circle junction (12) Primer Sequences: Forward - MH535: 5'AACTAGGGAACCCACTGCTTAAG-3'; Reverse- MH536: 5'-TCCACAGATCAAGGATATCTTGTC-3'; Probe: MH603:5'-(FAM)ACACTACTTGAAGCACTCAAGGCAAGCTTT(TAMRA)-3'
-
ii)
PCR mix (Applied Biosystems): The TaqMan Universal PCR Master Mix is 2X in concentration. The mix is optimized for TaqMan reactions and contains AmpliTaq Gold DNA Polymerase, AmpErase Uracil N-Glycosylase, dNTPs with dUTP and optimized buffer components
-
iii)
384 well plate for PCR reaction (Applied Biosystems)
-
iv)
Plate sealer (Applied Biosystems).
2.5 Analysis of 2-LTR circle junction
-
i)
Primers specific for 2-LTR circle junctions: 2-LTR forword -- 5' GCC TGG AGC TCT GGC TAA 3' and 2-LTR Reverse – 5' GCC TTG TGT GGT AGA TCC A 3' (13) for first round PCR and MH535 and MH536 (Section 2.4. i.) for nested 2nd Round PCR.
-
ii)
TA cloning kit (Invitrogen)
-
iii)
Plasmid DNA isolation kit (QIAGEN)
-
iv)
DNA alignment software BioEdit (14) (BioEdit v7.0.7: www.mbio.ncsu.edu/BioEdit).
3. Methods
3.1 Transfection of proviral DNA
-
i)
Plate 293 cells in 100mm tissue culture dish. Plate should be 40% confluent within 24h.
-
ii)
Remove media from the plate 3–5 h before transfection and replace with fresh media.
-
iii)Transfect 293T cells with 10ug of proviral DNA by Ca-PO4 method using Mammalian cell transfection kit (Specialty Media) following manufacturers' protocol.
-
a)Approximately 106 cells were plated 24h before transfection.
-
b)Add fresh DMEM media containing 10% FBS to cells 3h before transfection.
-
c)In a polystyrene tube, place 0.5 ml 1x HBS (per transfection) and add 10μl phosphate solution. In a separate tube, mix 10 μg DNA in a total volume of 430 μl and add 60 μl CaCl2 solution to the DNA solution
-
d)Add DNA and CaCl2 mixture drop-wise to HBS solution and gently tap the tube and incubate for 20 minutes.
-
e)Add Calcium-phosphate precipitated DNA to the cells
-
f)12–16 hour post transfection change the media and supplement with fresh DMEM containing 10% FBS
-
a)
-
iv)
Replace media after 16–18h post tranfection.
3.2 Determination of Virus Production
-
i)
Collect supernatant from transfected cells and preserve at −70°C in 50 mM HEPES (pH 7.4) buffer until used.
-
ii)
Quantitate p24 production using HIV-1 p24 antigen ELISA kit (Perkin Elmer). HIV-1 p24 antigen assay is to be done by following method.
-
a)
Bring all reagents to room temperature (15–30°C) before use.
-
b)
Add 20 μl Triton X-100 to all wells except substrate blank.
-
c)
Add 200 μl of the 100 pg/ml stock and make 2-fold serial dilutions (eg. 50, 25, 12.5 pg/ml). Add positive control and cell-free culture supernatant to the designated wells. Mix well with pipette.
-
d)
Seal the plate and incubate for two hours at 37°C.
-
e)
Wash plate six times with 300μl diluted (1X) wash buffer. Blot well before addition of next reagent.
-
f)
Add 100 μl detector antibody to all wells except substrate blank.
-
g)
Seal plate and incubate for 60 minutes at 37 °C.
-
h)
Wash plate six times with 1x wash buffer as in step `e'.
-
i)
Within 15 minutes of use, dilute sufficient Streptavidin-HRP (SA-HRP) Concentrate to the 1:100 working concentration with Streptavidin-HRP diluent. Mix thoroughly.
-
j)
Add 100 μl diluted SA-HRP to all wells except substrate blank.
-
k)
Seal the plate and incubate 30 minutes at room temperature (15–30°C).
-
m)
Wash plate 6 times with 1x wash buffer as in step `e'.
-
n)
Prepare sufficient OPD Substrate (O-phenyl diamine) solution by dissolving 1 tablet in 11 ml substrate within 15 minutes of use. Vortex vigorously to assure complete dissolution and protect from light. The OPD substrate solution should be colorless to pale yellow. A yellow-orange color indicates that the reagent is contaminated and must be discarded.
-
o)
Add 100 μl OPD substrate solution to all wells including substrate blank.
-
p)
Seal plate and incubate 30 minutes at room temperature (15–30°C) in the dark.
-
q)
Stop the reaction by adding 100 μL of Stop Solution to all wells.
-
r)
Read the plate at 490 or 492 nm, blanking the plate reader on air (Consult plate reader Instruction Manual).
3.3 Virus infection and the isolation of HIV cDNA from infected cells
-
i)
Infect Jurkat cells (1×105 cells) with 60ng virus.
-
ii)
Incubate 2h at 37°C.
-
iii)
Wash with PBS and supplement with fresh media.
-
iv)Harvest cells at 2h, 24h and 48h post infection. Isolate DNA using DNeasy tissue kit (QIAGEN) following manufacturer's protocol as described below.
-
a)Resuspend cells in 200μl PBS and add 20μl Proteinase K to the cells.
-
b)Lyse the cells with 200 μl lysis buffer supplied with the kit.
-
c)Incubate at 56° for 10 minutes.
-
d)Add 200 μl 95% Ethanol to the tubes and mix by vortexing.
-
e)Apply mixture to spin column.
-
f)Centrifuge for 1 min at 8000 rpm (6000×g).
-
g)Place the cartridge to a fresh 2ml collection tube and add 0.5 ml Wash buffer AW1 supplied with the kit.
-
h)Centrifuge for 1 minute at 8000 rpm as above.
-
i)Put the spin column on a fresh collection tube and add 0.5 ml wash buffer AW2 supplied with the kit.
-
j)Centrifuge at 14, 000 rpm (20, 000×g) for 3 minutes.
-
k)Place the spin column inside a fresh 1.5ml Eppendorf tube.
-
l)Add 100–200 μl elution buffer and incubate at room temperature for 1 minute.
-
m)Centrifuge for 2 minutes at 14, 000 rpm (20, 000×g).
-
n)Store DNA at 4°C.
-
a)
-
v)
Analyze 2-LTR circle junction DNAs formed in the infected cells by restriction digestion of the cellular DNA isolated followed by Southern blot and the absolute DNA quantitation is performed by real-time PCR. A portion of the remaining DNA is used to do PCR amplification, cloning and sequencing.
3.4 Analysis of 2-LTR circle by Southern-blot analysis
3.4 Preparation of Viral DNA
-
i)
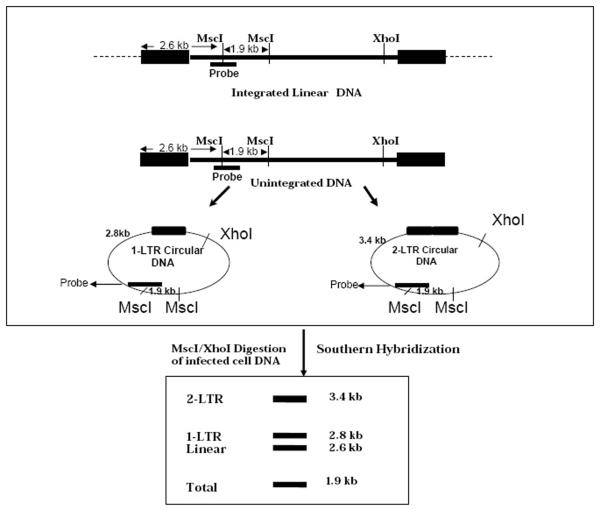
Digest the viral DNA (10 μg) isolated from infected cells with MscI and XhoI overnight. An XhoI site is present at position 8897 and two MscI sites at position 2619 and 4551 are present of R3B provirus. Double digestion of DNA with XhoI and MscI will produce fragments depending on whether viral DNA is present as integrated or un-integrated forms. Restriction digestion of viral DNA will also generate different fragments corresponding to the circular DNA, either 1-LTR circle or 2-LTR circle.
3.4.2 Agarose gel Electrophoresis of Viral DNA
Analyze Viral DNA digested with XhoI and MscI enzymes on 0.7% agarose gel to separate the different sized fragments generated. Carry out the electrophoresis in 1X TAE buffer for 5h at a constant 50V. Photograph the gel along with appropriate molecular size markers.
3.4.3 Transfer of DNA to Nylon membrane
Transfer viral DNA separated by agarose gel electrophoresis to a Hybond N+ membrane as described earlier (15).
3.4.4 Hybridization of Transferred viral DNA
-
i)Preparation of the Probe: Probe to be used for the hybridization is a 1.6kb fragment corresponding to nucleotide positions 1818 to 3498 of HXB2R3B. This fragment must be PCR amplified using the primers 5' AGA AGA AAT GAT GAC AGC 3' and 5 TGC CAG TTC TAG CTC TG 3' (16). PCR amplified DNA is labeled with radioactive 32P by random-primed DNA labeling kit following manufacturers' protocol (Roche Diagnostics).
-
a)Add to 25 ng template DNA (linear) sterile double distilled water to a final volume of 9 μl in a microfuge tube.
-
b)Denature the DNA by heating in a boiling water bath for 10 min at 95°C and chilling quickly on ice.
-
c)To the DNA, add 1 μl each of dATP, dGTP and dTTP from the stock, 2μl reaction mixture (hexanucleotide), 5 μl (50 μCi) of [α-32P] dCTP, 3000 Ci/mmol (aqueous solution) and the Klenow enzyme 1 μl (2U/μl).
-
2.Mix and centrifuge briefly. Incubate at 37°C for 30 minutes (Note: Longer incubation can increase the yield of labeled DNA).
-
3.Stop the reaction by adding 2 μl 0.2 M EDTA (pH 8.0) and/or by heating to 65 °C for 10 min.
-
5.Unincorporated deoxyribonucleoside triphosphates are removed by QIAGEN nucleotide purification kit. In brief, to the reaction mixture, add 5x volume of binding buffer and apply to the spin column. The spin column is centrifuged at 8000 rpm (6, 000×g) for 1 minute. Add 0.5 ml of wash buffer to the spin column and centrifuge for 1 minute. Another 0.5 ml of wash buffer is added to the column and centrifuged at 14, 000 rpm (20, 000×g) for 2 min. Finally probe was eluted at 20 μl volume of elution buffer (incorporation is 2 × 109 dpm/ μg of DNA).
-
a)
-
ii)
Pre-hybridization
Place the Nylon membrane (Hybond N+, Amersham Pharmacia) with transferred viral DNA into the hybridization bottle and add 10ml pre-hybridization buffer. Carry out pre-hybridization for 1h at 42°C.
-
iii)Hybridization
-
a)Denature radioactive probe at 95°C for 2 minutes. Add single stranded probe to hybridization bottle. Hybridize viral DNA with the radioactive probe at 42°C overnight (16 to 18h).
-
b)Remove the hybridization solution (into radioactive waste), and add an equal volume of 2x SSC/0.1% SDS. Incubate for 15 min at room temperature with shaking, changing the wash solution after 5 minutes.
-
c)Wash the membrane with 0.2x SSC/0.1% SDS and incubate with rotation for 15 minutes at room temperature, changing the wash solution after 5 minutes This is a low-stringency wash.
-
d)Pour off the final wash solution, rinse the membrane in 2x SSC at room temperature and pour off the wash buffer.
-
a)
-
iv)
Detection of Viral DNA
After washing, blot the membrane with a filter paper (3mm Whatman) to remove moisture. Air-dry the membrane for 10 minutes. Wrap the blot with saran-wrap prior to autoradiography. Expose the blot to X-ray film with an intensifying screen at –70°C for 1–2 days.
3.4.5 Interpretation of Southern Hybridization result
MscI and XhoI digestion of un-integrated viral DNA will generate fragments corresponding to linear, 1-LTR or 2-LTR circular DNA (see Figure 1). Fragments specific for 1-LTR or 2-LTR circular DNA could be easily detected by probe described above. 1-LTR circular DNA will produce 1.9 kb, 2.8 kb fragments whereas 2-LTR circle DNA will form 1.9kb and 3.4 kb fragments. By southern blot of viral DNA isolated from infected cells, a 2.6 kb fragment can also be detected which correspond to integrated or un-integrated linear DNA. Intensity of each band detected is directly proportional to the amount of viral DNA formed in the infected cell. Amounts of 1-LTR and 2-LTR circular DNA species increase over time after post-infection. Quantitative measurement of 1-LTR and 2-LTR circular DNA and the amount of linear DNA formed indicates that 55% of the total viral DNA imported into the nucleus are fully processed and integrated into the host nucleus whereas 35% and 5% DNA are circularized to form 1-LTR and 2-LTR circle junctions respectively. Remaining 10% DNA remains linear (16).
Figure 1.
Graphical outline of restriction digestion and Southern blot hybridization of HIV-1R3B infected cell DNA to analyze 2-LTR circle junction. The top panel shows the positions of the restriction sites on the linear and circular forms. The bottom panel shows the relative expected positions of the various bands on a Southern blot. Total refers to a band that is common to all restriction fragments.
3.5 Real-time PCR quantitation of 2-LTR circle Junctions
Quantify DNA copy numbers of reverse transcription intermediates synthesized within the HIV-1 infected cells by real-time PCR using TaqMan technology (Applied Biosystems). The primers and probes specific for 2-LTR circles were as described previously (12, 13). Perform Real time PCR on an ABI7700 apparatus (Applied Biosystems). Use approximately 500ng of infected cell DNA for each PCR reaction.
3.5.1 PCR amplification of 2-LTR circle junctions and cloning
-
a)
Construct a plasmid DNA p2LTR from HIV-1 infected cell DNA. PCR amplify 2-LTR junction sequence from wild-type HIV-infected Jurkat cell DNA and clone PCR product into the TA cloning vector pCR 2.1 (Invitrogen) to generate p2-LTR (see section 3.6 for details PCR protocol and cloning). Verify clones by sequencing.
-
b)
Quantitate the pLTR DNA by taking the Absorbance260 of the purified DNA.
-
c)
Calculate DNA copy number of p2-LTR from the O.D. value. Use this DNA as positive control for the real-time PCR quantitation of 2-LTR circle junctions present in the HIV-1 infected cell DNA. Also use this DNA to generate the standard curve to calculate the copy number of unknown DNA.
3.5.2 Real time PCR to quantify 2-LTR circle junction
-
i)
Prepare real-time reaction cocktail by mixing 2x PCR mix (Applied Biosystems PE), 2-LTR specific probe and primers.
-
ii)
Dispense 30 μl of reaction mixture in a 200μl PCR tube.
-
iii)
Add 5 μl (500 ng) of virus-infected cell DNA to the mixture.
-
iv)
Add 10 μl of the mix into 384 wells plate in triplicate.
-
v)
Make a serial dilution of p2LTR DNA to obtain 108, 106, 104 and 102 copies/μl of this DNA.
-
vi)
Add 5ul of the diluted p2-LTR plasmid DNA of known copy numbers to 30 μl PCR mix.
-
vii)
Add 10 μl of the above mix to 384 well plate.
-
viii)
Cover the plate with plate sealer carefully avoiding any mixing.
-
ix)
Spin the plate for a short time at 4°C to bring the PCR reaction mix at the bottom.
-
x)
Place the plate into the real time PCR machine.
-
xi)Run Real time PCR reactions for the quantification of 2-LTR circles under the following conditions:
-
1 cycle:50°C, 2 minutes
-
1 cycle95°C, 10 minutes
-
40 cycle95°C, 15 seconds60°C, 1minute
-
3.5.3 Determination of 2-LTR circle junction copy number in the infected cells
-
i)
Generate a standard curve by plotting the Ct values (cycle threshold) obtained from real time PCR reaction of p2-LTR plasmid against the DNA copy number.
-
ii)
Calculate the copy numbers of 2-LTR circle DNA in the viral infected cellular DNA from the standard curve.
3.5.5 Interpretation of 2-LTR circle junction quantification results
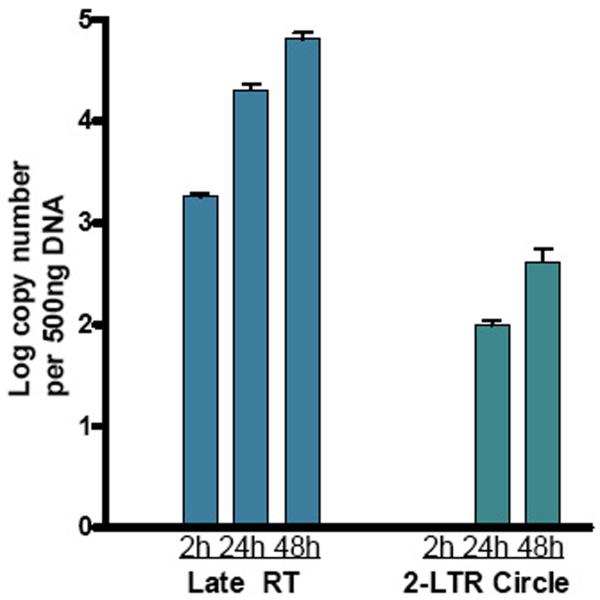
The standard curve generated is always linear when log copy of number (ranging from 2 to 8) of control DNA is plotted against the Ct values. 2-LTR copy number from experimental sample is obtained from the standard curve. It is possible by real-time PCR to quantify full-length viral DNA synthesized during infection. It is generally found that copy number of 2-LTR circle junction formed is 102 times less than full-length DNA (Figure 2).
Figure 2.
Quantitation of 2-LTR circle Junction DNA by Real-time PCR. Jurkat cells were infected with WT R3B virus and infected cells DNA was isolated at 2h, 24h and 48h post-infection. Virus infected cell DNA was analyzed for amount of 2-LTR circle junction DNA (green bar) and compared with the amount of late reverse transcription (RT) products (Blue bars). [Adapted from Mandal et al. (2006) Nucleic Acids Reseach. May 24;34(10):2853–63.]
3.6. Sequence Analysis of 2-LTR circle junctions
3.6.1
Viral DNA isolated from 48h infected cells is used as template to amplify 2-LTR circle junctions. A nested two-step PCR is required to amplify sufficient amounts of 2-LTR circle junction DNA for the purpose of cloning. Primers used for the first round PCR are 2-LTR forward and 2-LTR reverse and primers for the 2nd round are MH535 and MH536 (described in the Materials section). A 200bp DNA fragment can be amplified using primers specific for U3 and U5 sequences abutting the 2-LTR circle junction by this approach. Conditions to be employed for the PCR amplification is as follows:
Conditions for the 1st round PCR:
-
1cycle of
95°C, 5 minutes
-
35Cycle of
95°C, 15 seconds
50°C, 1 minute
72°C, 30 seconds
-
1Cycle of
72°C, 5 minutes
Use 2 μl of products from the 1st round of amplification as template for 2nd round PCR.
Conditions for the 2nd round PCR:
-
1 cycle of
95°C, 5 minutes
-
35 cycles of
95°C, 15 seconds
55°C, 1 minute
72°C, 30 seconds
-
1 cycle of
72°C, 5 minutes
3.6.2 Agarose gel analysis
-
a)
Five micro liters of the PCR product obtained after the 2nd step PCR are employed for analysis on 2% agarose gel for 1h at 100V.
-
b)
Stain the gel with ethidium bromide (EtBr) and monitor under UV trans-illuminator. Check for the DNA product of 189bp.
3.6.2 TA cloning of PCR Products
-
a)
Ligate PCR products to TA cloning vector (pCR 2.1, Invitrogen): Mix 1μl PCR product, 1μl vector, 1μl 10x buffer and 1μl ligase in a total volume of 10μl volume. Incubate the mixture at 16°C overnight.
-
b)
Transform DH5α cells with the ligation product.
-
c)
Clones obtained are characterized by EcoRI restriction digestion. Clones having insertion of ~188bp PCR product are confirmed by the release of the corresponding fragment.
3.6.3 Sequencing of 2-LTR circle junction
Determine the nucleotide sequence of about 50 clones to get enough information about the 2-LTR circle junctions. Sequence the clones using primers specific for TA cloning vector, adjacent to the cloning site.
3.6.4 Alignment of the 2-LTR circle junction sequences
Do a multiple alignment of all the sequence obtained by ClustalX program using BioEdit Software.
3.6.5 Statistical Analysis
Perform statistical analysis with the 2-LTR circle junction sequences to determine the significance in the difference between sequence distributions of different mutant viruses. Analyze the data by contingency table analysis. Subsets of pertinent groupings are followed up with the traditional two-by-two Fisher's exact tests.
3.6.6 Interpretation of 2-LTR circle junction sequence analysis result
2-LTR circle junctions are formed when full-length linear viral DNA fails to integrate into the host genome. In most cases, viral DNA fails to integrate mostly because of erroneous terminal sequences due to the error proneness of the processes involved. Sequence analysis of 2-LTR circle obtained from wild type HIV-1R3B infected cell DNA showed that almost 50% circle junctions were of `consensus' sequences (Figure 3) without revealing any specific defect – confirming an inefficient nature of the integration reaction. Some of the sequences were found to have terminal `GA' in the 3' U5 boundary. Majority of the remaining sequences have deletions at the U5 edge, the U3 edge or contain a deletion that spans the U5–U3 junction. Fewer sequences display the insertion of fragments or full PPT sequence between the U5 –U3 junction. Insertion of fragments of tRNA primer are also often observed. The internal insertions and deletions within the 2-LTR circle junction, which tend to involve single nucleotides are a result of error prone reverse transcription.
Figure 3.

Sequence analysis of HIV-1 2-LTR circle junction DNA. 2-LTR circle junction sequences obtained from HIV-1 R3B infected cell DNA were aligned by multiple alignment. The 3' U5, 5' U3 and circle junction sequence is indicated. Deletions within the 3'U5 or 5'U3 region are indicated by dashes. Insertion within the junction sequence is mentioned by number of bases. [Adapted from Mandal et al. (2006) Nucleic Acids Reseach. May 24; 34(10):2853–63.]
4. Notes
The number of producer cells to be plated for transfection is important depending on the time of supernatant collection. Seed ~1×106 293T cells on a 100 mm tissue culture dish so that, on the day transfection, the confluency of the cells reaches about 30–40%.
Viral supernatant can be collected as early as 6h post- transfection or as late as 48h post-transfection. Supernatants collected at some of the early time points (24h) may have higher infectivity per ng of virus produced, but the total amount of virus produced will be less than at peak p24 production. On the other hand, if virus supernatant is collected at very late time points, the total virus production may be increased, but the proportion of infectious virus particles decreases.
For p24 antigen quantitation, note that 200μl cell-free culture supernatant generally contains p24 above the range of measurement. Serial dilutions of up to 1000-fold would be helpful.
For Southern blot analysis of 2-LTR circle junctions, restriction sites are specific for the R3B molecular clone. These restriction sites may vary for different molecular clones, isolates or subtypes of HIV-1.
For quantitation of 2-LTR circle junction by real time PCR, infect 1×105 Jurkat cells with 50–60 ng of p24 equivalent of virus supernatant which gives m.o.i of ~1.
References
- 1.Goff SP. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 2001;3:517–52. doi: 10.1002/1521-2254(200111)3:6<517::AID-JGM234>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 2.Bukrinsky MI, Haffar OK. HIV-1 nuclear import: in search of a leader. Front Biosci. 1997;2:d578–d587. doi: 10.2741/a213. [DOI] [PubMed] [Google Scholar]
- 3.Cullen BR. Journey to the center of the cell. Cell. 2001;105:697–700. doi: 10.1016/s0092-8674(01)00392-0. [DOI] [PubMed] [Google Scholar]
- 4.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Harbour laboratory press, Cold Spring Harbour; NY: 1997. [PubMed] [Google Scholar]
- 5.Hansen MST, Carteau S, Hoffman C, Li L, Bushman F. Retroviral cDNA integration: mechanism, applications and inhibition. In: Setlow JK, editor. Genetic Engineering. Principles and Methods. Vol 20. Plenum Press; New York, NY: 1998. pp. 41–62. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky MI, Sharova N, McDonald TL, Pushkarskaya T, Tarpley WG, Stevenson M. Association of integrase, matrix, andreverse transcriptase antigens of human immunodeficiency virus type 1 withviral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA. 1993;90:6125–6129. doi: 10.1073/pnas.90.13.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, Martin SL, Bushman FD. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20(12):3272–81. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown PO. Integration. In: Coffin JM, Hughes SH, Vermus HE, editors. Retroviruses. Cold Spring harbor laboratory press; New York: 1997. [PubMed] [Google Scholar]
- 9.Shoemaker C, Goff S, Gilboa E, Paskind M, Mitra SW, Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: implications for retrovirus integration. Proc Natl Acad Sci U S A. 1980;77(7):3932–6. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farnet CM, Haseltine WA. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol. 1991;65(12):6942–52. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kilzer JM, Stracker T, Beitzel B, Meek K, Weitzman M, Bushman FD. Roles of host cell factors in circularization of retroviral DNA. Virology. 2003;314(1):460–7. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 12.Butler SL, Hansen MS, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7(5):631–4. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 13.Julias JG, Ferris AL, Boyer PL, Hughes SH. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J Virol. 2001;75(14):6537–46. doi: 10.1128/JVI.75.14.6537-6546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall TA. BioEdit : a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- 15.Brown T. Southern Blotting. In: Ausubel FM, editor. Current protocols in molecular biology. 1999. [DOI] [PubMed] [Google Scholar]
- 16.Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101(2):173–85. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]